Abstract
In many neuroscience and clinical studies, accurate and automatic segmentation of subcortical structures is an important and difficult task. Multi-atlas-based segmentation method has been focus of considerable research due to its promising performance. In general, this technique first employs deformable image registration to construct the correspondences between pre-labeled atlas images and the target image. Then, using the acquired deformation field, labels in the atlas are propagated to the target image space. Obviously, anatomical differences between the target image and atlas images possibly affect the image registration accuracy, thus influencing the final segmentation performance. Another limitation is that the label propagation in most conventional multi-atlas based methods is implemented under a voxel-wise strategy, which cannot adequately utilize the local label information to determine the final label of the target sample. In this paper, we propose a patch-wise label propagation method based on multiple atlases for MR brain segmentation. First, each image patch is characterized by patch intensities and abundant texture features, to increase the accuracy of the patch-based similarity measurement. To determine the weights of the training patches for representing the test sample, a patch-based sparse coding procedure is employed. In the label propagation stage, to alleviate possible misalignment from the registration stage, we perform a patch-wise label propagation strategy in a nonlocal manner to predict the final label for each target sample. To evaluate the proposed segmentation method, we comprehensively implement our proposed method by conducting hippocampus segmentation on the ADNI data set. Experimental results demonstrate the effectiveness of the proposed method and show that the proposed method outperforms two conventional multi-atlas-based methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Magnetic resonance (MR) plays an important role in pathology, neuro-analysis, and clinical application [1,2,3,4]. Accurate segmentation of anatomical structure is usually required in many medical image analyses. Take the connectome applications for instance, before constructing the brain connectivity network, multiple regions which describe the network architecture of the brain should be identified in numerous brain MR images. However, the huge amounts of clinical MR data make the manual segmentation of MR images’ time-consuming and tedious. Therefore, the development of automatic segmentation methods has raised considerable interests in the field of medical image analysis.
Multiple atlases with pre-labeled images have proven to be effective for determining the region of interest (ROI) in the target image [5,6,7,8]. By fusing the propagated labels of multiple atlases in the target image space, multi-atlas based methods can obtain accurate and robust segmentation results. Specifically, in a typical multi-atlas-based segmentation method, each atlas image is first registered to the target image and a warping function can be obtained. Then, the estimated warping function is further used for propagating the associated atlas label to the target image space. Finally, the segmentation result of the target image can be achieved by fusing all the propagated atlas labels. Towards the multi-atlas based methods, the learning-based methods using multiple atlases have attracted great attention in recent years [9,10,11]. In the learning-based scheme, the segmentation problem is regarded as a classification task, in which each voxel in the target image is separated as the ROI or background by training a classifier. Some well-known learning methods such as support vector machine (SVM), Adaboost, and artificial neural networks have been widely adopted in the learning-based classification [12,13,14,15,16]. Particularly, sparse representation classifier for image segmentation, whose basic idea is that the target sample can be represented as a linear combination of the pre-labeled atlas image samples, has provided promising and powerful results [17]. The basic assumption behind the above technique is that, if the local appearances or other features are strongly similar, the target image point should bear the same label as the atlas image point. However, some details could be lost, and local variability cannot be obtained during the registration procedure. In addition, the definition of patch-based similarity is often handcrafted, which may not be sufficient for different regions of the brain structure. Another limitation is the label fusion technique which usually employs a voxel-wise label propagation strategy to determine the final label of the target image. This approach does not take the surrounding label information into account, which is sensitive to the registration error.
In this work, we propose a patch-wise label propagation method along with a sparse coding scheme, where the weight of each sample is driven by the sparse coding procedure. Here, patch-wise refers to the process in which each voxel in an atlas is assigned with one label patch, instead of a single label, for label propagation. Specifically, the target image is first linear registered to each atlas. Then, for each voxel to be segmented in the target image, voxels with similar features are considered to generate a training set from the voxels of registered atlases. Features we used in this paper are extracted from image patch with abundant texture information and intensity information. After that, the weight of each training sample is determined using the sparse encoding procedure. To preserve local anatomical structure information in the segmentation, we utilize label patches as the structured class label. That is, the label patch centered at the voxel on the label map of the corresponding registered atlas is extracted as the structured class label of the voxel. The training set then consists of feature representations for sampled voxels and their corresponding structured class labels (i.e., label patches centered at the corresponding voxels). Finally, the final label of the target voxel can be determined by applying the weight of each training sample on the structured patch labels.
Below, we first introduce the multi-atlas-based segmentation method as the basis of the proposed method in Sect. 2. Then, Sect. 3 presents the details of sparse coding scheme along with a patch-wise label propagation. Finally, the experimental results are shown in Sect. 4, and conclusion is drawn in Sect. 5.
2 Multi-atlas based segmentation method
The goal of our patch-wise label propagation using multiple atlases is to accurately label each voxel in the target image as either a positive (i.e., region of interest) or a negative (i.e., background) voxel. We first introduce the typical multi-atlas-based segmentation method which serves as the basis of our method.
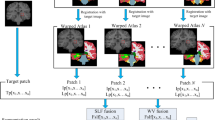
Given a target image I to be segmented and N atlases \(\tilde {A}=\{ {\tilde {A}_i}=({\tilde {I}_i},{\tilde {L}_i})\left| {i=1,2, \ldots ,N} \right.\}\), where \({\tilde {I}_i}(i=1,2, \ldots ,N)\) represents an atlas image and \({\tilde {L}_i}(i=1,2, \ldots N)\) is its corresponding segmentation label with a value of + 1 indicating ROI and − 1 indicating background. In the multi-atlas-based segmentation method, each atlas image is first spatially registered to the target image, thereby the warping function can be obtained. Then, the associated atlas labels are further propagated to the target image space based on the acquired warping function. Finally, all the propagated atlas labels are fused to generate a segmentation result for the target image using a specific label propagation strategy. Let \(A=\{ {A_i}=\left( {{I_i},{L_i}} \right)\left| {i=1,2, \ldots N} \right.\}\) denotes the set of N atlas images and their corresponding labels which have been registered to the target image. To alleviate possible misalignment from registration, most patch-based label propagation methods are performed in a nonlocal manner. Figure 1 shows an overview of a typical patch-based segmentation method in the multi-atlas scenario.
Particularly, to label a patch of the target image, the candidate training set from voxels of registered atlases within a spatial neighborhood of the target voxel (blue solid block in Fig. 1) is generated. Then, using the patch similarity as weights, all possible candidate patches from different atlases are fused to estimate the final label of the target voxel using a specific fusion strategy. Specifically, for each target image voxel x, all the atlas patches within a certain search neighborhood \(V\) are used to compute the patch-wise similarities with the target image patch. Each patch is arranged into a column vector. The estimation of the label for the target voxel \(x\) can be obtained by the following:
where \(L({x_{s,j}})\) is the label of voxel \({x_{s,j}}\) at location \(j\) in atlas \(s\), \(V\) is the a relatively small search neighborhood, and \(w\left( {x,{x_{s,j}}} \right)\) is the weight assigned to \(L({x_{s,j}})\). As the description above, the weights for different atlases are typically determined by a similarity measure of patch appearance between the atlas image patches of \({x_{s,j}}\) and the target image patch of \(x\), which can be obtained as follows:
where \(P\left( x \right)\) and \(P\left( {{x_{s,j}}} \right)\) are the cubic patches centered at \(x\) and \({x_{s,j}}\), respectively. \(|| \cdot |{|_2}\) represents the L2 norm between each intensity of the elements of the patches \(P\left( x \right)\) and \(P\left( {{x_{s,j}}} \right)\). \({\sigma _x}\) is the decay parameter controlling the strength of penalty in the exponential way. If the structural similarity \(ss\) is less than the threshold \({\text{th}}\), the weight of this patch will not be computed. Finally, the label of the target voxel can be determined as follows:
3 Proposed method
The proposed method mainly consists of feature extraction, local-search based sparse coding, and label propagation. In the following subsections, we will discuss the detail of each step of the proposed method.
3.1 Feature extraction
Most conventional multi-atlas-based methods often use handcrafted features (e.g., image intensity) as the identification of patch-based similarity. However, since the structures of MR brain images usually share similar intensity pattern, only using the handcrafted features may be not sufficient for segmentation. Thereby, it is necessary to develop an effective feature extraction method to strengthen the description for each voxel. In this paper, along with the patch-based intensity information, we also extract abundant texture information. Specifically, given a sampled voxel \({\varvec{z}}\), the intensity information is extracted using the patch-wise strategy. The texture information that we extract in this paper includes:
-
Outputs of the first-order difference filters (FODs):
$$\left\{ {H\left( {{\varvec{z}}+{\varvec{u}}} \right) - H\left( {{\varvec{z}} - {\varvec{u}}} \right),~{\varvec{u}}=\left( {r\cos \theta \sin \phi ,r\sin \theta \sin \phi ,r\cos \phi } \right)} \right\}.$$(4)
-
Outputs of the second-order difference filters (SODs):
$$\left\{ {H\left( {{\varvec{z}}+{\varvec{u}}} \right)+H\left( {{\varvec{z}} - {\varvec{u}}} \right) - 2H\left( {\varvec{z}} \right),~{\varvec{u}}=\left( {r\cos \theta \sin \phi ,r\sin \theta \sin \phi ,r\cos \phi } \right)} \right\}.$$(5)
-
Outputs of 3D Hyper plan filters:
$$\left\{ {{{{\varvec{\Psi}}}_1}*\left( {H\left( {{C_{3,3,1}}\left( {{\varvec{z}}+{\varvec{u}}} \right)} \right) - H\left( {{C_{3,3,1}}\left( {{\varvec{z}} - {\varvec{u}}} \right)} \right)} \right),~{\varvec{u}}=\left( {0,0,1} \right),{\text{~}}{{{\varvec{\Psi}}}_1}=\left[ {\begin{array}{*{20}{c}} 1&1&1 \\ 1&1&1 \\ 1&1&1 \end{array}} \right]} \right\}.$$(6)
-
Outputs of 3D Sobel filters:
$$\left\{ {{{{\varvec{\Psi}}}_2}*\left( {H\left( {{C_{3,3,1}}\left( {{\varvec{z}}+{\varvec{u}}} \right)} \right) - H\left( {{C_{3,3,1}}\left( {{\varvec{z}} - {\varvec{u}}} \right)} \right)} \right),{\varvec{u}}=\left( {0,0,1} \right),~{{{\varvec{\Psi}}}_2}=\left[ {\begin{array}{*{20}{c}} 1&2&1 \\ 2&3&2 \\ 1&2&1 \end{array}} \right]} \right\}.$$(7)
-
Outputs of Laplacian filters:
$$\mathop \sum \limits_{{{{\varvec{z}}_1} \in {O_p}\left( {\varvec{z}} \right)}} \left( {H\left( {{{\varvec{z}}_1}} \right) - H\left( {\varvec{z}} \right)} \right),{O_p}\left( {\varvec{z}} \right) \subseteq {C_{3,3,3}}\left( {\varvec{z}} \right).$$(8)
-
Outputs of range-difference filters:
$${\text{ma}}{{\text{x}}_{{{\varvec{z}}_1} \in {O_p}({\varvec{z}})}}\left( {H\left( {{{\varvec{z}}_1}} \right)} \right) - \mathop {\hbox{min} }\limits_{{{{\varvec{z}}_1} \in {O_p}({\varvec{z}})}} \left( {H\left( {{{\varvec{z}}_1}} \right)} \right),{O_p}\left( {\varvec{z}} \right) \subseteq {C_{3,3,3}}\left( {\varvec{z}} \right).$$(9)where \({C_{a,b,c}}\left( {\varvec{z}} \right)\) represents a cube centered at \({\varvec{z}}\) with size of \(a \times b \times c\), \({\varvec{u}}\) is the offset vector, \(r\) is the length of \({\varvec{u}}\), \(\theta\) and \(\phi\) are two rotation angles of \({\varvec{u}}\), \({O_p}\left( {\varvec{z}} \right)\) denotes the voxels in the \(p\)-neighborhood of \({\varvec{z}}\), and \(*\) denotes convolution operation. In the texture features, FODs and SODs detect intensity change along a line segment. Here, we set \(r \in \left\{ {1,2,3} \right\}\), \(\theta \in \left\{ {0,\pi /4,\pi /2,3\pi /4} \right\}\), and \(\phi \in \left\{ {0,\pi /4,\pi /2} \right\}\). 3D Hyperplane filters and 3D Sobel filters are the extensions of FODs and SODs in the plane. Filters along two other directions are also implemented. Laplacian filters are isotropic and detect the second-order intensity changes. Range-difference filters compute the difference between maximal and minimal values in a given neighborhood for each voxel. In this paper, we determine the size of a neighborhood \(~p \in \left\{ {7,19,27} \right\}\).
By comparing the features similarity between the target sample and the multi-atlas samples, we can employ a pre-selection method to generate the candidate training set, thus obtaining the coding dictionary which can be used in the sparse coding procedure of the next step.
3.2 Local-search-based sparse coding
To encode an image sample for each voxel of the target image, a set of training samples should be identified from the registered atlas images. Since linear image registration cannot achieve perfect alignment of all voxels across images, the direct usage of the corresponding voxel of the voxel considered in the atlases as the training sample is not appropriate. To achieve better correspondence between the target voxel and voxels in atlases, we employ a local-search strategy to find the best match in each atlas for the target voxel and thus obtain the coding dictionary.
Specifically, given a target voxel \(x\), we first define a neighborhood \(V(x)\) centered at the target voxel with the size \(\omega \times \omega \times \omega\) of all registered atlas samples. This can produce \(N \times \omega \times \omega \times \omega\) candidate training samples \(\{ ({\vec {f}_{i,j}},{l_{i,j}})\left| {i=1,2, \ldots ,N} \right.,j \in V(x)\}\) from \(N\) registered atlases, where \({\vec {f}_{i,j}}\) represents a feature vector extracted from voxel \(j\) of the \(i\)th atlas based on the feature extraction method which has been discussed in the above subsection, and \({l_{i,j}} \in \{ +1, - 1\}\) is the label information of the corresponding atlas image sample.
One of the critical problems in sparse representation is that how to effectively construct the coding dictionary. A small neighborhood size will result in a small dictionary, but may not be reliable enough for encoding the target sample. On the other hand, a large neighborhood size will lead to a redundant dictionary, which may increase the computational time and also introduce some irrelevant patches. To address this issue, we perform a pre-selection strategy based on a threshold strategy before sparse coding procedure. The feature similarity between patches is used as the criterion for patch selection, which can be determined by the following well-known structural similarity (\({\text{ss}}\)) measure:
where \(\mu\) is the mean and \(\sigma\) is the standard deviation of patches centered at the target voxel \(x\) and voxel j of the ith atlas.
The coding dictionary is further used to represent the target sample via sparse representation. Figure 2 shows the sparse coding procedure. Specifically, given a target image, it is assumed that each patch in the target image can be sparsely represented by a linear combination of the constructed coding dictionary. To estimate the label of the patch centered at \(x\) of the target image, a set of sparse weights, \(w\left( {x,{x_j}} \right)\), is calculated by minimizing the following nonnegative problem:
where \({\vec {f}_x}\) is a feature vector containing the intensity features and texture features extracted at x from the target image. The first term in Eq. (11) enables the feature vector \({\vec {f}_x}\) which should be represented by the coding dictionary. The second term enhances the sparsity on the weights via \({l_1}\) regularization, and the last term ensures that similar patches should have similar weights. \({\lambda _1}\) and \({\lambda _2}\) are the weights to balance three terms in Eq. (11). After the sparse coding procedure, we can obtain the sparse weights, and the weights are further used to perform the patch-wise label propagation to get the final segmentation results.
3.3 Patch-wise label propagation
To preserve local anatomical structure information in the segmentation, we perform label propagation procedure in a patch-wise manner. For the voxel-wise label propagation, we can directly extract voxel labels from the label map according to the selected atlas samples, and use them as class labels to conduct label fusion. However, for local-search-based sparse coding procedure, each voxel is assigned with one image patch. To preserve local anatomical structure in the label map, we extend the class label \({y_j} \in {\mathbb{R}}~(j=1, \ldots ,M)\) of each voxel to the structured class label patch \({\mathcal{Y}_j} \in {{\mathbb{R}}^{t \times t \times t}}\) corresponding to the label patch centered at the voxel, where \(t \times t \times t\) is the size of the label patch and \(M\) is the number of samples. Let \({{\varvec{\Omega}}}\) denote the voxel set of the label patch of the dictionary samples. Then, the estimation of the label patch centered at the target voxel \(x\) can be obtained as follows:
Here, \({\mathcal{Y}_j}(a)\) is the corresponding label patch centered at the voxel \(x\) on the label maps of the jth sample, \({D_x}\) is the coding dictionary of the voxel \(x\), and \(w\left( {x,{x_j}} \right)\) is the sparse weights calculating by the sparse coding procedure. When testing the neighbor voxels of the voxel \(x\), their corresponding label patches are also estimated, which can load the overlapped estimations for the voxel \(x\) from the estimated label patches of its neighbor voxels. Therefore, we adopt an averaging method to fuse the overlapped estimations and use it as the final label estimation \(L\left( x \right)\). Compared with voxel-wise label propagation, patch-wise label propagation uses the whole label patches to estimate the final label, which can take advantage of local anatomical structure information in the segmentation.
4 Experiments
Many neuroscience studies have shown that the hippocampus structure plays a crucial role in human memory and orientation. Besides, hippocampus dysfunction involves a variety of diseases, such as Alzheimer’s diseases, schizophrenia, dementia, and epilepsy [18]. However, due to its small size, high variability, low contrast, and the discontinuous boundaries in MR brain images, the hippocampus structure is especially difficult to be segmented. To evaluate the performance of the proposed method, we conduct several experiments for hippocampus segmentation. All 3D MR scans are obtained from the publicly available ADNI database (https://ida.loni.usc.edu). The manual hippocampal segmentations are regarded as the ground truth from the preliminary release of EADC–ADNI Harmonized Protocol training labels, which can be downloaded from http://www.hippocampal-protocol.net/SOPs/label.php. In the experiments, 29 Normal Control (NC) subjects, 34 Mild Cognitive Impairment (MCI) subjects, and 37 Alzheimer’s disease (AD) subjects are randomly selected from the ADNI data set. The detailed demographic information of the selected subjects is summarized in Table 1. We first perform the preprocessing procedure for all selected subjects, including: (1) skull-stripping using a learning-based meta-algorithm to remove nonbrain tissues [19], (2) N4-based bias field correction [20], and (3) histogram matching to normalize the intensity range [21]. A leave-one-out cross-validation (LOOCV) strategy is used to evaluate the performance of our method. To align each atlas image to the target image, we perform affine registration using FLIRT in the FSL toolbox, with 12 degrees of freedom and default parameters.
To quantitatively evaluate the performance of the proposed segmentation method, we compare the segmentation results \({O_s}\) with the ground truth \({O_g}\). Specifically, we use the dice coefficient (DC) and Hausdorff distance (HD) for evaluation. DC is a comprehensive similarity metric which measures the overlap degree between ROI \({O_s}\) and ROI \({O_g}\), and HD measures the surface distance between two ROIs:
where \(\left| \cdot \right|\) is the volume of particular ROI, and \(d\left( {a,b} \right)\) represents the Euclidean distance between \(a\) and \(b\). Theoretically, segmentation results with higher DC and lower HD represent better segmentation performance.
To determine the optimal parameters, we conduct a set of experiments. We set the patch size with 5 × 5 × 5 based on the empirically validated in our experiments. In the sparse coding procedure, \({\lambda _1}\) and \({\lambda _2}\) are set to be 0.1 and 0.01, respectively. The above parameters are fixed throughout all the experiments. For the candidate training set, the search neighborhood \({V_x}\) is selected from 5 × 5 × 5 to 13 × 13 × 13, with a step of 2 × 2 × 2. The number of samples in the coding dictionary is set from 200 to 1000, with a step of 200. For the patch-wise label propagation, we select the side length of label patches from 1 to 9 with a step of 2.
4.1 Influence of elements in the proposed method
Some important elements and parameter setting of the proposed method are investigated in this section. Such elements or parameters include patch-based intensity features and texture features, search neighborhood size, number of pre-selected samples in the sparse coding dictionary, and patch-wise label propagation.
We first evaluate the performance of the features used in our method including intensity features and multiple texture descriptors. To evaluate each feature type, we test our method with only using intensity patches (Intensity), texture descriptors (Texture), and the combination of intensity patch and texture descriptors (Intensity + Texture). Moreover, we vary the size of search neighborhood from 5 × 5 × 5 to 13 × 13 × 13, and use DC measure by performing voxel-wise label propagation to evaluate the performance. Figure 3 shows the mean DC for the left and right hippocampus segmentation for three feature types plotted against different neighborhood size.
From Fig. 3, we can see that, compared to the Intensity and Texture features, the combination of two features consistently outperforms across different search sizes. Only using texture descriptors as the feature representation leads to the worst performance. Though only using intensity features can lead to a better result, however, it is still not as good as that using both intensity features and texture features. It indicates that the texture information extracted from the atlas images does help the sparse coding to enhance the segmentation performance. We can also see that, when using search size of 11 × 11 × 11, all three types of features yield a better performance than that of the other search size. In the following experiments, we use search size of 11 × 11 × 11 to investigate other elements or parameter of the proposed method.
Table 2 lists the mean of DC and HD for the left and right hippocampus segmentation for the Intensity, Texture, and Intensity + Texture features. We can see that the combination of intensity and texture features achieves the best performance in terms of DC and HD.
In the sparse coding procedure, we also investigate the influence of the number of samples in the sparse coding dictionary. Pre-selection can improve the robustness of sparse coding by excluding the unrelated samples as well as speed up the coding procedure. Figure 4 shows the mean DC curve by our method using a different number of selected samples. It can be seen in the figure that increasing the number of training samples can improve segmentation accuracy. To balance the segmentation accuracy and computational time, we set 800 as the number of selected samples.
To evaluate patch-wise label propagation performed in our method, we further analyze the influence of label patch size on segmentation performance. Specifically, we varied the label patch size from 1 × 1 × 1 to 9 × 9 × 9 with a step of 2 for each side. When 1 × 1 × 1 label patch is used, patch-wise label propagation becomes voxel-wise label propagation. Figure 5 shows the mean DC using different label patch sizes.
From Fig. 5, it is apparent that when 7 × 7 × 7 label patch is used, our method yields the best performance (0.874 for left hippocampus and 0.883 for right hippocampus). It demonstrates the superiority of our method using patch-wise label propagation, compared with the use of voxel-wise label propagation. With the increasing of label patch size from 1 × 1 × 1 to 7 × 7 × 7, the segmentation accuracy is gradually improved. However, for patch size of 9 × 9 × 9, the DC decreases slightly. Thus, label patch size of 7 × 7 × 7 is preferred for our method.
4.2 Comparison with conventional methods
In this section, we compared our method with two conventional multi-atlas-based segmentation methods, i.e., patch-based method by nonlocal weighting (nonlocal-PBM) [22] and sparse patch-based labeling method (sparse-PBM) [18]. We set the search neighbor size to 11 × 11 × 11 and the label patch size for label propagation to 7 × 7 × 7. For sparse coding, we selected 800 samples to construct the coding dictionary. Figure 6 shows box plots of segmentation performance measures in terms of DC and HD for nonlocal-PBM, sparse-PBM, and our method, respectively. For visual inspection, Fig. 7 shows the segmentation results for a subject randomly chosen from the data set. It is evident that, compared to the nonlocal-PBM and sparse-PBM methods, the segmentation result of our method is more similar to the ground truth. For the sparse-PBM, over-smoothing appears in some regions, which result in losing some detailed information, as indicated by white arrows in Fig. 7. From the experimental results of Figs. 6 and 7, we can see that both the qualitative and quantitative results indicate that the proposed method performs consistently better than other segmentation methods.
Comparison of segmented hippocampus regions by different methods. One subject was randomly chosen from each data set (red: results of manual segmentation, yellow: results by nonlocal-PBM, blue: results by sparse-PBM, purple: results by our method). The first row shows segmentation results produced by different methods; the second row shows their corresponding surface rendering results. Our method shows the best segmentation performance (especially for the area depicted by white arrows)
5 Conclusion
In this paper, we propose a patch-wise label propagation for MR brain segmentation. Specifically, each image patch is first characterized by patch intensities and abundant texture features, to increase the presentation of the patch-based similarity. The experiment results in subsection 4.1 demonstrated that the texture features provide contribution in improving the segmentation accuracy. Then, each weight of the training samples for representing the target sample is determined based on a sparse coding procedure. In the label propagation stage, to preserve local anatomical structure information for the segmentation and alleviate possible misalignment from the registration stage, we utilize label patches as the structured class label to perform label propagation. The segmentation performance is evaluated based on the ADNI data set for hippocampus segmentation. Compared to the traditional multi-atlas-based segmentation methods, the experimental results show that the proposed method can achieve better segmentation performance. However, we just conduct our method on hippocampus segmentation in this paper. In the future, we will test our method on other significant anatomical structures of human brain, such as corpus callosum, amygdala, and insula.
References
Bertolino, A., Frye, M., Callicott, J.H., Mattay, V.S., Rakow, R., Shelton-Repella, J., et al.: Neuronal pathology in the hippocampal area of patients with bipolar disorder: a study with proton magnetic resonance spectroscopic imaging. Biol. Psychiat. 53(10), 1177–1194 (2003)
Zu, C., Wang, Z., Zhang, D., Shen, D., Wu, G.: Robust multi- atlas label propagation by deep sparse representation. Pattern Recogn. 63, 511–517 (2017)
Zhang, J., Liu, M., An, L., Gao, Y., Shen, D.: Alzheimer’s Disease diagnosis using landmark-based features from longitudinal structural MR images. IEEE J. Biomed. Health Inform. 21(6), 1607–1616 (2017)
Wang, Y., Zhang, P., An, L., Ma, G., Kang, J., Wu, X., et al.: Predicting standard-dose pet image from low-dose pet and multimodal MR images using mapping-based sparse representation. Phys. Med. Biol. 61(2), 791–812 (2016)
Aljabar, P., Heckemann, R.A., Hammers, A., Hajnal, J.V., Rueckert, D.: Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46(3), 726–738 (2009)
Zu, C., Jie, B., Liu, M., Chen, S., Shen, D., Zhang, D.: Label-aligned multi-task feature learning for multimodal classification of Alzheimer’s disease and mild cognitive impairment. Brain Imaging Behav. 10(4), 1148–1159 (2015)
Wang, L., Gao, Y., Feng, S., Li, G., Gilmore, J.H., Lin, W., et al.: Links: learning-based multi-source integration framework for segmentation of infant brain images. Neuroimage 108, 160–172 (2015)
Wang, H., Suh, J.W., Das, S.R., Pluta, J.B.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–23 (2013)
Hao, Y., Wang, T., Zhang, X., Duan, Y., Yu, C., Jiang, T., et al.: Local label learning (lll) for subcortical structure segmentation: application to hippocampus segmentation. Hum. Brain Mapp. 35(6), 2674–2697 (2014)
Tu, Z., Bai, X.: Auto-context and its application to high-level vision tasks and 3d brain image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 32(10), 1744–1757 (2010)
Darko, Z., Glocker, B., Criminisi, A.: Atlas encoding by randomized forests for efficient label propagation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 66–73. Springer, Berlin (2013)
Zhang, J., Liang, J., Zhao, H.: Local energy pattern for texture classification using self-adaptive quantization thresholds. IEEE Trans Image Process 22(1), 31–42 (2013)
Fan, Y., Shen, D.: Integrated feature extraction and selection for neuroimage classification. In: Proceedings of SPIE—The International Society for Optical Engineering, San Diego, CA, vol. 7259, pp. 155–160 (2009)
Freund, Y., Schapire, R.E.: A decision-theoretic generalization of on-line learning and an application to boosting. J Comput. Syst. Sci. 55(7), 119–139 (1999)
Zhang, J., Liu, M., Shen, D.: Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Trans. Image Process. 26(10), 4753–4764 (2017)
Gao, Y., Zhang, H., Zhao, X., Yan, S.: Event classification in microblogs via social tracking. ACM Trans. Intell. Syst. Technol. (TIST) 8(3), Article No. 35 (2017)
Wang, Y., Ma, G., An, L., Shi, F., Zhang, P., Lalush, D.S., et al.: Semisupervised tripled dictionary learning for standard-dose PET image prediction using low-dose PET and multimodal MRI. IEEE Trans. Biomed. Eng. 64(3), 569–579 (2017)
Tong, T., Wolz, R., Coupé, P., Hajnal, J.V., Rueckert, D.: Segmentation of mr images via discriminative dictionary learning and sparse coding: application to hippocampus labeling. Neuroimage 76(1), 11–23 (2013)
Feng, S., Li, W., Dai, Y., Gilmore, J.H., Lin, W., Shen, D.: Label: pediatric brain extraction using learning-based meta-algorithm. Neuroimage 62(3), 1975–1986 (2012)
Tustison, N.J., Avants, B.B., Cook, P.A., Zheng, Y., Egan, A., Yushkevich, P.A., et al.: N4itk: improved n3 bias correction. IEEE Trans. Med. Imaging. 29(6), 1310–1320 (2010)
Madabhushi, A., Udupa, J.K.: New methods of mr image intensity standardization via generalized scale. Med. Phys. 33(9), 3426–3434 (2006)
Coupé, P., Manjón, J.V., Fonov, V., Pruessner, J., Robles, M., Collins, D.L.: (2011). Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage 54(2), 940–954
Acknowledgements
This work is supported by the by National Natural Science Foundation of China (NSFC61701324), Science and Technology Department of Sichuan Province 2016JZ0014, and Open Fund Project of Fujian Provincial Key Laboratory of Information Processing and Intelligent Control (Minjiang University) (No. MJUKF201715).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, Y., Zu, C., Ma, Z. et al. Patch-wise label propagation for MR brain segmentation based on multi-atlas images. Multimedia Systems 25, 73–81 (2019). https://doi.org/10.1007/s00530-017-0577-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00530-017-0577-2