Abstract
Previous studies suggested an association between 1425G/A polymorphism in PRKCH and stroke risk, but the results were inconsistent. To obtain a more precise estimation, we carried out a meta-analysis to analyze the effect of 1425G/A SNP in PRKCH on stroke risk. We searched PubMed, ISI Web of Science, Chinese Biomedical Database, China National Knowledge Infrastructure and WANFANG Data for all eligible case–control studies through April 2014. The odds ratios (ORs), together with the 95 % confidence intervals (CIs), were calculated to evaluate the strength of association between 1425G/A SNP and stroke risk. Overall, seven eligible studies involving a total of 4,574 cases and 5,471 controls were included in our meta-analysis. The results showed that the variant genotypes of 1425G/A polymorphism in PRKCH were significantly associated with a higher risk of stroke in all genetic models (GA vs. GG: OR 1.35, 95 % CI 1.24–1.47, P < 0.001; AA vs. GG: OR 1.50, 95 % CI 1.24–1.82, P < 0.001; GA/AA vs. GG: OR 1.37, 95 % CI 1.26–1.49, P < 0.001; AA vs. GA/GG: OR 1.35, 95 % CI 1.12–1.62, P = 0.002; A vs. G: OR 1.29, 95 % CI 1.21–1.39, P < 0.001). In the subgroup analysis, significantly increased risks were also observed for ischemic stroke, larger sample size (>1,000) and population-based studies. The result of our meta-analysis indicated that the 1425G/A SNP in PRKCH may contribute to susceptibility of stroke, especially for ischemic stroke.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stroke is a common neurological disease and a leading cause of death worldwide (Go et al. 2013; Kubo et al. 2007). The stroke patient will have a very low quality of life suffering from disability, cognitive dysfunction and many other complications (Kiyohara et al. 2003), leading to a serious burden to the whole society (Strong et al. 2007; Lopez et al. 2006).
Ischemic stroke and hemorrhagic stroke are the two main subtypes of stroke (Markus 2011). Like many other complex diseases, genetic factors have been considered as an important risk contributor to the development of stroke (Johnston et al. 2009). Twins and family studies have proven a substantial genetic contribution to the risk of stroke (Jerrard-Dunne et al. 2003; Flossmann et al. 2004; Bak et al. 2002; Liao et al. 1997).
Over the past 20 years, the candidate gene association studies and classical linkage approaches were widely adopted to identify genes involved in stroke occurrence and recurrence. However, few associations have been consistently replicated. Recently, genome-wide association study (GWAS) has emerged as a powerful new tool to identify many susceptibility variants with moderate genetic risk on various complex diseases (Hirschhorn and Daly 2005). In 2007, Kubo et al. reported that a nonsynonymous SNP (1425G/A, rs2230500) in PRKCH could increase the risk of cerebral infarction (Kubo et al. 2007). Thereafter, the original report has been followed by many publications in an attempt to replicate this finding; some confirmed the association (Serizawa et al. 2008; Wu et al. 2009; Song et al. 2008), whereas others failed to replicate it (Cheng et al. 2009, 2012). The association between this SNP with stroke risk has been a research focus and has drawn increasing attention. Because a single study might have been underpowered to detect the overall effects, a quantitative synthesis of the accumulated data from different studies was deemed more important to provide evidence on the association of this SNP with stroke risk. Thus, we carried out a meta-analysis on all eligible case–control studies to estimate the overall stroke risk of PRKCH 1425G/A polymorphism as well as to quantify the between-study heterogeneity and potential bias.
Materials and Methods
Literature Search and Study Selection
We searched PubMed, ISI Web of Science, Chinese Biomedical Database, China National Knowledge Infrastructure and WANFANG Data for all eligible case–control studies through April 2014. The associated medical subject headings and terms were as follows: stroke, ischemic stroke or hemorrhagic stroke in combination with PRKCH, 1452G/A SNP, Protein kinase Cη or rs2230500. No language restrictions were imposed. Abstracts, reviews or editorials were not included.
Studies included in our meta-analysis had to meet the criteria as follows: (1) had neuroimaging (CT or MRI) result to confirm the diagnosis of stroke, (2) case–control studies and (3) evaluated the association between 1425G/A in PRKCH and stroke susceptibility. Studies were excluded if: (1) the age of patients was under 18, and (2) original genotype data were not reported. As for duplicate publications, we chose the one with a larger sample size.
Data Extraction
Two investigators (S.L. and Z.Z.) independently extracted all data according to the selection criteria and reached a consensus on all of the items. The following characteristics were sought: the first author’s last name, the year of publication, country, sample size and the genotype distribution of samples in case and control groups, the minor allelic frequencies (MAF) in control groups, source of control, genotyping method and matching criteria.
Statistical Analysis
We first examined whether the genotype distribution in control group was consistent with Hardy–Weinberg equilibrium (HWE) by chi-square test for each study. The odds ratios (ORs), together with the 95 % confidence intervals (CIs), were then calculated to evaluate the strength of association between 1425G/A SNP in PRKCH and stroke risk. The statistical significance of summary OR value was evaluated by Z test, and P value <0.05 was considered to be statistically significant. To estimate the association between 1425G/A SNP and stroke risk, we first evaluated the risks of the AA and AG genotypes separately in comparison with the wild-type GG homozygote. Then, we estimated the dominant (AA/AG vs. GG) and recessive (AA vs. AG/GG) effects of the variant A allele on the occurrence of stroke, respectively. Meanwhile, we also estimated the risk of variant A allele on stroke risk compared with G allele. In addition, stratified analyses were performed by subtype, country, sample size and source of control.
Heterogeneity within and between subgroups were evaluated with the Q test and I 2 statistics (Higgins and Thompson 2002). Heterogeneity exists if P value of Q test <0.05. If no significant heterogeneity existed, the fixed-effects model (the Mantel–Haenszel method) was adopted to calculate the summary OR value. Otherwise, the random-effects model (the DerSimonian and Laird method) was used. The stratified analyses were performed to test the robustness of the association.
Cumulative meta-analysis was adopted to update a genetic effect from all studies and to measure the changes of the influence on diseases as new evidence accumulated (Lau et al. 1992; Zhang et al. 2012). Therefore, we could evaluate the variation trend of the effect about 1425G/A SNP on stroke through cumulative meta-analysis.
Sensitivity analyses were also performed to estimate the stability of the results. A single study included in our meta-analysis was deleted each time, and then, all the rest studies were analyzed to reflect the influence of the individual study to the pooled ORs. Besides, in order to assess the potential publication bias, we used funnel plots and Egger’s linear regression test. All analyses were done with Stata software version 12 (StataCorp LP, College Station, TX, USA).
Results
Literature Search and Study Characteristics
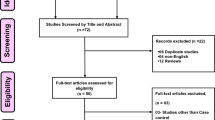
Figure 1 showed the flow diagram of the literature search. Total searches yielded 25 entries. Ten studies were excluded because of obvious irrelevance. Fifteen potentially relevant studies were reviewed. Another eight studies were excluded after reading the title and abstract for the following reasons: not exploring the association between 1425G/A SNP and stroke (n = 5), reviews (n = 2), analyzed the recurrence of stroke (n = 1). As a result, seven studies were included in our meta-analysis. The characteristics of each study are summarized in Table 1. These seven included studies were published between 2007 and 2012. Totally, 4,574 cases and 5,471 controls were included in our meta-analysis. Of the seven studies, four studies used frequency-matched controls to the cases by the age and sex. A classic polymerase chain reaction assay was performed in all of the seven studies. In addition, five of them studied ischemic stroke, one focusing on cerebral hemorrhage and another one including all subtypes of stroke (Wu et al. 2009). The distribution of genotypes in selected studies is shown in Table 2. The genotype distributions among the controls of all studies were in accordance with HWE.
Quantitative Synthesis
Overall, the variant genotypes of 1425G/A polymorphism in PRKCH were associated with a significantly higher risk of stroke in different genetic models. As shown in Table 3, the variant genotypes GA and AA were associated with a significantly higher risk of stroke in a dose–response manner, compared with the wild-type homozygote GG (OR 1.35, 95 % CI 1.24–1.47 for GA and 1.50, 1.24–1.82 for AA; P trend < 0.001). In addition, significant associations were also observed in dominant model (Fig. 2), recessive model and allele contrast model (OR 1.37, 95 % CI 1.26–1.49, OR 1.35, 95 % CI 1.12–1.62 and OR 1.29, 95 % CI 1.21–1.39, respectively).
In the stratified analysis by subtype, significantly increased risks were observed for ischemic stroke (GA vs. GG: OR 1.34, 95 % CI 1.22–1.47; AA vs. GG: OR 1.54, 95 % CI 1.24–1.90; dominant model: OR 1.36, 95 % CI 1.24–1.49; recessive model: OR 1.38, 95 % CI 1.12–1.70; allele model: OR 1.29, 95 % CI 1.20–1.39), especially for lacunar infarction (GA vs. GG: OR 1.46, 95 % CI 1.29–1.64; AA vs. GG: OR 2.16, 95 % CI 1.13–4.12; dominant model: OR 1.49, 95 % CI 1.32–1.67; recessive model: OR 1.81, 95 % CI 1.00–3.27; allele model: OR 1.50, 95 % CI 1.23–1.84), but only with borderline statistical significance for hemorrhagic stroke (GA vs. GG: OR 1.35, 95 % CI 1.02–1.77; dominant model: OR 1.35, 95 % CI 1.04–1.75; allele model: OR 1.27, 95 % CI 1.02–1.57).
In addition, in the stratified analysis by sample size, statistically significantly elevated risk was also observed, and this elevated risk was more pronounced among studies with sample size >1,000 (P heterogeneity = 0.58 for dominant model, Table 3). Moreover, significant association was observed among studies using the population-based controls, but not the hospital-based controls (Table 3). The cumulative meta-analysis for the dominant model showed a trend of the association as the evidence accumulated (Fig. 3).
Sensitivity Analyses
Since the MAF in Cheng’s study was much lower than that in other studies, we removed this study for sensitivity analyses first. As a result, the pooled OR (95 % CI) was 1.37 (1.26–1.49). Then, we dropped one study each time and analyzed the rest studies to observe the change of pooled ORs. The range of pooled ORs was from 1.35 to 1.39, indicating that the result of this meta-analysis was stable.
Publication Bias
The funnel plot did not show any obvious asymmetry (Fig. 4). Egger’s test also indicated no evidence of publication bias (P = 0.852).
Discussion
The present meta-analysis explored the association between 1425G/A polymorphism in PRKCH and risk of stroke. We found that this variant was associated with significant increase in overall stroke risk.
PKCη, decoded by PRKCH, belongs to the atypical isoform of protein kinase C (PKC) family since it is regulated by diacylglycerol and phospholipids (Spitaler and Cantrell 2004; Kubo et al. 2007). PKCη plays a very important role in signal transduction pathways essential for the activation and homeostasis of immune responses (Spitaler and Cantrell 2004). PKCη is highly abundant in T cells and has an important role in T cell stimulation (Fu et al. 2011). In addition, PKCη participates in the process of atherosclerosis and the nonsynonymous SNP (1425G/A) in PRKCH causes enhancement of PKC activity, which may increase stroke risk (Kubo et al. 2007).
In the subgroup analysis by subtype, significantly higher stroke risk was observed in ischemic stroke but with borderline statistical significance in hemorrhagic stroke. Moreover, in the subtype analysis of ischemic stroke, this effect was more pronounced in lacunar infraction. Lacunar infraction, a subtype of ischemic stroke, was first reported to have a significant association with 1425G/A SNP in PRKCH by Kubo et al. In our stratified analysis, the variant genotypes were associated with a significantly higher risk of lacunar infraction in all genetic models. The borderline significant association in hemorrhagic stroke was likely due to the small number of cases, which led to a lower statistical power to estimate this association.
In our present study, we also found that the association between the 1425G/A polymorphism and stroke risk among studies using the population-based controls was stronger than that among studies with hospital-based controls. Compared with population-based studies, hospital-based ones have intrinsic selection biases for the reason that hospital-based controls may not represent the general population very well, particularly when the genotypes under investigation were associated with the disease-related conditions that the hospital-based controls may have. Thus, selecting appropriate and representative population-based controls has an important role in reducing bias for such genotype association analysis.
Our meta-analysis added to the evidence that 1425G/A polymorphism played a part in the process of stroke. In addition, the cumulative meta-analysis was also adopted to evaluate the trend of the association as evidence accumulated. However, some limitations should be addressed. First, most of the studies had a relatively small sample size. Thus, larger well-designed studies should be conducted to further confirm all these results. Second, the studies included in our meta-analysis were all done in Asian populations, and no study with Caucasian populations was reported, which might be due to the low MAF (Traylor et al. 2012). According to HapMap database, the MAF of 1425G/A SNP is reported to be 0.239 in Japanese, 0.178 in Han Chinese Population in Beijing and 0.008 in CHEP samples (Utah residents with ancestry from northern and western Europe) (Kubo et al. 2007). Thus, more evidence was needed from other populations to further investigate the association between the PRKCH 1425G/A polymorphism and stroke risk.
In conclusion, this meta-analysis provided evidence that the variant 1425G/A SNP in PRKCH was associated with a significant higher risk of stroke. As studies among the Europeans and Africans are currently limited, further studies including a wider spectrum of subjects should be conducted to explore the role of this functional variant in other populations, which should lead to better, comprehensive understanding of the association between the PRKCH 1425G/A polymorphism and stroke risk.
References
Bak, S., Gaist, D., Sindrup, S. H., Skytthe, A., & Christensen, K. (2002). Genetic liability in stroke: A long-term follow-up study of Danish twins. Stroke, 33(3), 769–774.
Chen, Y. C., Chen, P., Wu, Y. R., Shie, S. S., Chen, S. T., Lee-Chen, G. J., et al. (2012). Protein kinase Ceta polymorphism and the susceptibilities to intracerebral hemorrhage in the Taiwan population. Neuroscience Letters, 528(2), 170–173.
Cheng, H., Wang, F., Ding, X., Ding, H., & Song, X. (2009). Association of PRKCH gene with lacunar infarction in a local Chinese Han population. Neuroscience Letters, 464(2), 146–149.
Flossmann, E., Schulz, U. G., & Rothwell, P. M. (2004). Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke, 35(1), 212–227.
Fu, G., Hu, J., Niederberger-Magnenat, N., Rybakin, V., Casas, J., Yachi, P. P., et al. (2011). Protein kinase C eta is required for T cell activation and homeostatic proliferation. Science Signaling, 4(202), ra84.
Go, A. S., Mozaffarian, D., Roger, V. L., Benjamin, E. J., Berry, J. D., Borden, W. B., et al. (2013). Heart disease and stroke statistics—2013 update: A report from the American Heart Association. Circulation, 127(1), e6–e245.
Higgins, J. P., & Thompson, S. G. (2002). Quantifying heterogeneity in a meta-analysis. Statistics in Medicine, 21(11), 1539–1558.
Hirschhorn, J. N., & Daly, M. J. (2005). Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics, 6(2), 95–108.
Jerrard-Dunne, P., Cloud, G., Hassan, A., & Markus, H. S. (2003). Evaluating the genetic component of ischemic stroke subtypes: A family history study. Stroke, 34(6), 1364–1369.
Johnston, S. C., Mendis, S., & Mathers, C. D. (2009). Global variation in stroke burden and mortality: Estimates from monitoring, surveillance, and modelling. The Lancet Neurology, 8(4), 345–354.
Kiyohara, Y., Kubo, M., Kato, I., Tanizaki, Y., Tanaka, K., Okubo, K., et al. (2003). Ten-year prognosis of stroke and risk factors for death in a Japanese community: The Hisayama study. Stroke, 34(10), 2343–2347.
Kubo, M., Hata, J., Ninomiya, T., Matsuda, K., Yonemoto, K., Nakano, T., et al. (2007). A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nature Genetics, 39(2), 212–217.
Lau, J., Antman, E. M., Jimenez-Silva, J., Kupelnick, B., Mosteller, F., & Chalmers, T. C. (1992). Cumulative meta-analysis of therapeutic trials for myocardial infarction. New England Journal of Medicine, 327(4), 248–254.
Liao, D., Myers, R., Hunt, S., Shahar, E., Paton, C., Burke, G., et al. (1997). Familial history of stroke and stroke risk. The Family Heart Study. Stroke, 28(10), 1908–1912.
Lopez, A. D., Mathers, C. D., Ezzati, M., Jamison, D. T., & Murray, C. J. (2006). Global and regional burden of disease and risk factors, 2001: Systematic analysis of population health data. The Lancet, 367(9524), 1747–1757.
Markus, H. S. (2011). Stroke genetics. Human Molecular Genetics, 20(R2), R124–R131.
Serizawa, M., Nabika, T., Ochiai, Y., Takahashi, K., Yamaguchi, S., Makaya, M., et al. (2008). Association between PRKCH gene polymorphisms and subcortical silent brain infarction. Atherosclerosis, 199(2), 340–345.
Song, X., Wang, F., Zhou, Y., Li, X., Shen, X., & Ding, X. (2008). Study on the association between SNP 1425G/A in protein kinase Cη gene and genetic susceptibility of cerebral infarction. Chinese Journal of Neurology, 41, 339–342.
Spitaler, M., & Cantrell, D. A. (2004). Protein kinase C and beyond. Nature Immunology, 5(8), 785–790.
Strong, K., Mathers, C., & Bonita, R. (2007). Preventing stroke: Saving lives around the world. The Lancet Neurology, 6(2), 182–187.
Traylor, M., Farrall, M., Holliday, E. G., Sudlow, C., Hopewell, J. C., Cheng, Y. C., et al. (2012). Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): A meta-analysis of genome-wide association studies. The Lancet Neurology, 11(11), 951–962.
Wu, L., Shen, Y., Liu, X., Ma, X., Xi, B., Mi, J., et al. (2009). The 1425G/A SNP in PRKCH is associated with ischemic stroke and cerebral hemorrhage in a Chinese population. Stroke, 40(9), 2973–2976.
Zhang, Z., Xu, G., Liu, D., Fan, X., Zhu, W., & Liu, X. (2012). Angiotensin-converting enzyme insertion/deletion polymorphism contributes to ischemic stroke risk: A meta-analysis of 50 case–control studies. PLoS One, 7(10), e46495.
Acknowledgments
This study was supported by National Natural Science Foundation of China (31200938, 81220108008), Natural Science Foundation of Jiangsu Province (BK2011021) and Natural Science Foundation of Jinling Hospital (2012009).
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Lingli Sun and Zhizhong Zhang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Sun, L., Zhang, Z., Ma, M. et al. A Quantitative Assessment of the Association Between 1425G/A Polymorphism in PRKCH and Risk of Stroke. Neuromol Med 16, 814–820 (2014). https://doi.org/10.1007/s12017-014-8330-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-014-8330-x