Abstract
PDE4D polymorphism (SNP83/rs966221) was reported to be associated with the susceptibility to ischemic stroke (IS), however, the results were inconclusive. An electronic search of Embase, PubMed, CNKI and Wan Fang Date was performed to identify relevant studies published throughout April 2017. A total of 26 studies were enrolled in the analysis. No significant association between the rs9662221 polymorphism and IS was observed in the overall analysis. Nevertheless, in the subgroup analysis, our results showed a significant association between the SNP83 polymorphism and IS in CC+ CT vs. TT (OR = 1.19, 95% CI: 1.02–1.38), CT vs.TT (OR = 1.14, 95% CI: 1.01–1.29) and C vs. T (OR = 1.25, 95% CI: 1.06–1.48) in Asian population. But we did not found any association in CC vs. CT + TT (OR = 1.2, 95% CI: 0.9–1.61) and CC vs. TT (OR = 1.26, 95% CI: 0.91–1.75) in the Asian populations. Meantime, no significant correlations were observed under the five genetic model in Caucasian population (p > 0.05). In conclusion, our meta-analysis demonstrated that the SNP83 polymorphism in the PDE4D gene might contribute to IS susceptibility especially in Asian populations. Whereas the relationship of the polymorphism to the disease in Caucasian population was still in controversial. In future, additional well designed studies with larger sample sizes are still required to further elucidate this association.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Ischemic stroke (IS) is a most common cause of death and long-term disability which accounting for approximately 85% of all types strokes (Kotlęga et al. 2017). Being one of the most devastating consequences of atherosclerotic disease, ischemic stroke can be caused by any or the combination of multifactorial disease, including age, hypertension, diabetes mellitus, smoking, obesity and so on (Munshi and Kaul 2008). Of all the risk factors, genetic susceptibility to ischemic stroke was considerate to play a vital role in the development of IS since first shown in twin pedigree studies in 1992 (Bak et al. 2002), however, until now, the genetic pathogenesis of stroke remains unclear.
Phosphodiesterase 4D (PDE4D) is a member of the superfamily of cyclic nucleotide phosphodiesterases and is involved in degradation of cyclic adenosine monophosphate (cAMP) which is a key signaling molecule involved in the inflammatory responses of vascular cells (Das et al. 2016). Researches showed that inhibitors of PDE4D can increase cAMP levels and adhesion in vascular endothelial cells and decrease migration of vascular smooth muscle cells (Jorgensen et al. 2015). Thus it had been postulated to contribute to vascular diseases in the pathogenesis of atherosclerosis. Since Gretarsdottir et al. identified SNP83 (rs966221) in PDE4D as a susceptible gene for ischemic stroke in Caucasian population, a large number of studies reported the association of single nucleotide polymorphisms across this gene with the disease (Gretarsdottir et al. 2003). Xue et al. had also identified that SNP83 as a genetic risk factor for atherothrombotic strokes in a Chinese population (Xue et al. 2009). Nevertheless, results have been contradictory. Quarta et al. reported that there were no evidences of association between SNP83 and IS in a genetically homogenous population from Sardinia (Quarta et al. 2009). Meantime, Matsushita et al. also reported the same results in Japanese cohort (Matsushita et al. 2009). Therefore, to confirm the association between SNP83 and the risk of IS, we performed a meta-analysis of case-control studies by pooling all eligible studies to evaluate the overall risk and influence of ethnic factors to this disease.
Materials and methods
To identify all relevant publications focus on the risk for Is and SNP83 polymorphism, we conducted a comprehensive literature search of electronic databases, including the Pubmed, Embase, China National Knowledge Infrastructure (CNKI) and Wan Fang Data. Eligible case-control studies were extracted with the last search update on April 1, 2017. The following terms were used: “ischemic infarction OR cerebrovascular disease OR stroke OR cerebrovascular disorders OR ischemic stroke” in combination with “PDE4D OR phosphodiesterase 4D OR rs966221 OR SNP83” in combination with “polymorphism OR variant OR mutation”. The reference lists of retrieved studies and recent reviews were also manually searched for further relevant studies. In addition, if the genotype data for the SNP83 polymorphism was not illuminated in the original studies, we send an email to obtain full data for the meta-analysis.
Inclusion and exclusion criteria
Studies were considered eligible if they met the following criteria: (1) evaluation of the association between PDE4D SNP83 and risk of ischemia stroke; (2) the study provided sufficient information of allele or genotype frequencies; (3) the study was a case-control study; Exclusion criteria: (1) duplication of previous publications; (2) comment, review or editorial; (3) study without detailed genotype data. A study reporting the results for different subpopulations was treated as separate studies.
Data extraction
The data of the eligible studies were extracted in duplicate by two investigators (Peng Wang and Fei Yang) independently with a standard data-collection form. The following data was collected from each study if available: (1) first author’s name; (2) country of origin; (3) years of publication; (4) ethnicity of the individuals (categorized as Caucasians or Asians); (5) Gender and mean age in cases and controls; (6) Hardy-Winberg equilibrium; (7) numbers of cases and controls; (8) genotyping method; (9) counts of cases and controls for each genotype. If there were multiple publications from the same group, only the one with largest study was included. The discrepancies were resolved through discussion. If the dissent still existed, the third investigators would be invited to resolve the dispute.
Quality assessment
The quality of the included studies was evaluated by using the quality scoring criteria modified from Newcastle–Ottawa scale (NOS) for genetic association studies (Zhang et al. 2017). Total score of quality assessment ranged from zero to nine stars, and six or more stars are rated as high quality. Disagreement was settled through discussion among the investigators.
Statistics analysis
Hardy-Weinberg equilibrium was evaluated for each study by chi-square test in the control groups and P value <0.05 was considered significant disequilibrium. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated to evaluate the strength of the association between SNP83 polymorphisms and susceptibility to IS. In the overall and subgroup meta-analysis, Pooled ORs were obtained from combination of single study for the dominant model (CC + CT vs. TT), recessive model (CC vs. CT + TT), homozygote model (CC vs. TT), heterozygote model (CT vs. TT) and allelic model (C vs. T). Heterogeneity was evaluated by Q statistic (significance level of p < 0.1) and I2 statistic (greater than 50% as evidence of significant inconsistency). Once, Q-test >0.10 or I2 < 50%, the fixed effect model (Mantel–Haenszelmethod) was used to calculate the pooled ORs, otherwise, the random-effect model (DerSimonian–Laird method) was employed in the study. Subgroup analysis was conducted according to the different ethnicities in the study population. Publication bias was estimated by Begg’s funnel plot. Sensitivity analyses were performed to evaluate the effect of each study on the combined ORs. All statistical analyses were performed with the software Stata 12.0 (STATA Corporation, College Station, TX, USA).
Results
Characteristics of studies
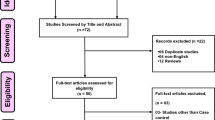
A flowchart of the literature search is presented in Fig. 1. After a preliminary online search, 175 potentially relevant articles were identified for further detailed evaluation. Among which 23 duplicated citations were removed, and 152 citations were included for further review by title and abstract screening. Finally, 36 articles were selected for full-text review and 26 cautions were enrolled in the analysis. Of all the studies, 7 were Caucasians and 19 were Chinese population. These studies were published between 2003 and 2015. Multiple genotyping methods were performed in the studies, including PCR-RFLP, TaqMan, DNA sequencing and so on.
A total of 9832 patients were from IS case group and 13,354 were enrolled in control group. The distribution of genotypes in the controls of all studies was consistent with Hardy-Weinberg equilibrium (HWE) except 6 studies. The NOS results showed that the score ranged from 3 to 8 which indicated that the methodological quality of these selected articles was generally good. The characteristics of involved articles were summarized in Tables 1 and 2.
Meta-analysis results
The heterogeneity was identified by Q-test and I-squared statistic among genetic models. As is showed in the Fig. 2, serious heterogeneity were found under dominant model (I2 = 71.5%), recessive model (I2 = 78.6%), homozygote model (I2 = 77.6%), heterozygote model (I2 = 53.4%) and allelic model (I2 = 84.2%), thus the random model was employed in the analysis. Our results reveled that there were no significant associations between SNP83 and IS under genetic model of CC + CT vs. TT (OR = 1.09, 95% CI:0.96–1.24), CC vs. CT + TT (OR = 1.1,95% CI: 0.91–1.34), CC vs. TT (OR = 1.11, 95% CI: 0.88–1.4), CT vs. TT (OR = 1.08, 95% CI:0.97–1.20) and C vs. T (OR = 1.12, 95% CI: 0.99–1.27).
Subgroups based on ethnicity were utilized to further analyze the relationship of polymorphism with IS. In Chinese populations, IS was proved to be correlated with SNP83 polymorphism under CC+ CT vs. TT (OR = 1.19, 95% CI:1.02–1.38), CT vs. TT (OR = 1.14, 95% CI:1.01–1.29) and C vs. T (OR = 1.25, 95% CI:1.06–1.48) Nevertheless, no significant association were found in CC vs. CT + TT (OR = 1.2, 95% CI:0.9–1.61) and CC vs. TT (OR = 1.26, 95% CI:0.91–1.75). In addition, we did not observed any correlation of SNP83 polymorphism with IS in all the five genetic models(CC+ CT vs. TT: OR = 0.87, 95% CI: 0.69–1.11; CC vs. CT + TT: OR = 0.95, 95% CI:0.84–1.07; CC vs.TT:0.87, 95% CI:0.66–1.13; CT vs.TT: OR = 0.9, 95% CI:0.72–1.12; C vs. T: OR = 0.92, 95% CI:0.8–1.05) in the Caucasian populations.
Sensitivity analysis
A sensitivity analysis by sequentially removing each eligible study at a time was used to assess the influence of each individual study on the pooled OR. Our results indicated that there was not any single study influence the quality of the pooled ORs (Fig. 3).
Publication bias
Publication bias was evaluated using the Begg’s funnel plot. No significant evidence of publication bias was observed in the five genetic models (CC + CT vs. TT: p = 0.853; CC vs. CT + TT: p = 0.731; CC vs. TT: p = 0.544; CT vs. TT: p = 0.812; C vs. T: p = 0.692) (Fig. 4).
Discussion
PDE4D is a large gene spanning >1.5 Mb on chromosomal region 5q12 and has 8 splice variants, 22 exons and several hundreds of SNPs (Song et al. 2015; Song et al. 2017). It has been shown to effects on brain function, pulmonary hypertension and vascular smooth cells migration (Mika and Conti 2016). It is more of a clinical syndrome rather than a disease due to numerous clinical, genetic, and lifestyle risk factors. Work over the past few decades has shown that PDE4D are present in the brain regions such as the amygdala, prefrontal cortex, hippocampus, and nucleus accumbens (Yang et al. 2012). The distribution pattern suggests that they may serve distinct roles in the central nervous system and provide a theoretical basis for the separation of therapeutic and adverse effects of PDE4 inhibitors.
The association of specific PDE4D single-nucleotide polymorphisms (SNPs) with stroke is initially identified in an Icelandic population and further evaluated in both animal models and human beings (Liu et al. 2013). Since the first study reported the associations of PDE4D SNPs and ischemic stroke risk, an increasing amount of research has been subsequently published to study the relationship of PDE4D SNP with IS, especially SNP 83(Gretarsdottir et al. 2003). Yoon et al. retrieved 15 publications with 19,318 subjects, failed to find a linkage between SNP83 and IS in the overall population (Yoon et al. 2011). However, Yan et al. comprised of 8878 cases and 12,306 controls in a review had shown that association between SNP83 and IS in the overall population and in the Asian and Chinese populations, but not among Caucasians (Yan et al. 2014). Till now there was still not a consistency conclusion. In our study, we examined the association between SNP83 and ischemic stroke using case-control and population-based cohort. We had found that there were significant association of SNP83 with IS risk under the dominant model, heterozygote model and allelic model. Nevertheless, we didn’t observed any correlation of the polymorphism with IS in the Caucasian. Our results were in accordance with the former meta-analysis which was done by Yan et al. (Yan et al. 2014). However, other studies had also reported with opposite conclusions, such as Matsushita et al., who failed to replicate the results in Japanese populations (Matsushita et al. 2009). Whereas, Staton et al. and Gretarsdottir et al. found that SNP83 was associated with an increased risk of ischemic stroke in Caucasians (Gretarsdottir et al. 2003; Staton et al. 2006).
Although we had got the positive results in Asian population, however, the conclusions should be treated with caution. The HWE results showed that there were 6 studies with their p < 0.05 which included Chinese population studies and Caucasian populations. However, in the stratified analysis by HWE, we had observed that the results were mostly unchanged. In addition, the heterogeneity was detected under five genetic models in this meta-analysis. This could be caused by various reasons, such as the small sample size, different diagnostic criteria and disunity detection methods. Meantime, most studies (n = 19) enrolled in our analysis were done in Asian populations and there were only 7 articles explored the PDE4D polymorphism with IS in Caucasian. This can partly explain the reasons why we cannot found any correlations of SNP83 with IS in Caucasian populations. In addition, though significant difference was observed in at least 3 genetic models, the results were still worth deliberated. Most of the studies were cross with the invalid line in the analysis of dominant model, heterozygote model and allelic model, thus the significance drawn were without enough persuasion. Besides, there was a lack of sufficient data regarding the patients’ gender, environment or complicating disease such as hypertension or diabetes, which may modulate the relationship when discussing those confounding factors.
In summary, the current meta-analysis revealed that SNP83 displayed significant association with IS in Asian populations under the dominant model, heterozygote model and allelic model, which offering evidence that PDE4D may be involved in the pathogenesis of IS. Nevertheless, no correlation was observed in Caucasians. Additional, as with kinds of limitations, the relationships of SNP83 with IS susceptibility in Asian and Caucasian populations were worth further exploration. In future, well designed case-control studies with large sample sizes and stroke subtype analysis regarding the association of SNP83 with IS needed to be performed.
References
Bak S, Gaist D, Sindrup SH, Skytthe A, Christensen K (2002) Genetic liability in stroke: a long-term follow-up study of Danish twins. Stroke 33:769–774. https://doi.org/10.1161/hs0302.103619
Banerjee I, Gupta V, Ahmed T, Faizaan M, Agarwal P, Ganesh S (2008) Inflammatory system gene polymorphism and the risk of stroke: a case–control study in an Indian population. Brain Res Bull 75:158–165. https://doi.org/10.1016/j.brainresbull.2007.08.007
Cheng H, Jin Q, Li L, Ding X, Song X, Zeng Y (2011) Association of ALOX5AP and PDE4D with the risk of lacunar infarct in people from Jiangsu Province, China. China Nerve Regeneration Research 6:935–940
Das S, Roy S, Munshi A (2016) Association between PDE4D gene and ischemic stroke: recent advancements. Int J Neurosci 126:577–583. https://doi.org/10.3109/00207454.2015.1051621
Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM, Hawkins M, Gudmundsson G, Gudmundsdottir H, Andrason H, Gudmundsdottir AS, Sigurdardottir M, Chou TT, Nahmias J, Goss S, Sveinbjörnsdottir S, Valdimarsson EM, Jakobsson F, Agnarsson U, Gudnason V, Thorgeirsson G, Fingerle J, Gurney M, Gudbjartsson D, Frigge ML, Kong A, Stefansson K, Gulcher JR (2003) The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet 35:131–138. https://doi.org/10.1038/ng1245
Jianhua PJ, Zhang X (2012) Relationship between SNP 83 of phosphodiesterase 4D gene and ischemic cerebrovascular diseases in Xinjiang Uygur and Han population. Chinese Journal of Nervous and Mental Diseases 38:733–737
Jorgensen C, Yasmeen S, Iversen HK, Kruuse C (2015) Phosphodiesterase4D (PDE4D)--a risk factor for atrial fibrillation and stroke? J Neurol Sci 359:266–274. https://doi.org/10.1016/j.jns.2015.11.010
Kalita J, Somarajan BI, Kumar B, Kumar S, Mittal B, Misra UK (2011) Phosphodiesterase 4 D gene polymorphism in relation to intracranial and extracranial atherosclerosis in ischemic stroke. Dis Markers 31:191–197. https://doi.org/10.1155/2011/232490
Kotlęga D, Peda B, Zembroń-Łacny A, Gołąb-Janowska M, Nowacki P (2017) The role of brain-derived neurotrophic factor and its single nucleotide polymorphisms in stroke patients. Neurol Neurochir Pol 51(3):240–246. https://doi.org/10.1016/j.pjnns.2017.02.008
Kuhlenbaumer G et al (2006) Evaluation of single nucleotide polymorphisms in the phosphodiesterase 4D gene (PDE4D) and their association with ischaemic stroke in a large German cohort. J Neurol Neurosurg Psychiatry 77:521–524. https://doi.org/10.1136/jnnp.2005.073577
Li N, He Z, Xu J, Liu F, Deng S, Zhang H (2010) Association of PDE4D and IL-1 gene polymorphism with ischemic stroke in a Han Chinese population. Brain Res Bull 81:38–42. https://doi.org/10.1016/j.brainresbull.2009.09.009
Lin HF, Liao YC, Liou CW, Liu CK, Juo S (2007) The phosphodiesterase 4D gene for early onset ischemic stroke among normotensive patients. Journal of thrombosis. Haemostasis 5:436–438. https://doi.org/10.1111/j.1538-7836.2007.02350.x
Liu K, Wang JW, Yu ZP, Qin XY, Wu YQ, Li N, Kui YQ, Fang K, Wang XY, Wu T, Chen DF, Hu YH (2013) Association study between PDE4D gene polymorphism and ischemic stroke. Beijing Da Xue Xue Bao 45:359–363
Luo M et al (2014) Relation between phosphodiesterase 4D gene polymorphism and ischemic stroke in Guangdong Han population. Chinese journal of geriatric heart brain and vessel. Diseases 16:503–506
Matsushita T, Kubo M, Yonemoto K, Ninomiya T, Ashikawa K, Liang B, Hata J, Doi Y, Kitazono T, Ibayashi S, Iida M, Kiyohara Y, Nakamura Y et al (2009) Lack of association between variations of PDE4D and ischemic stroke in the Japanese population. Stroke 40:1245–1251. https://doi.org/10.1161/STROKEAHA.108.527408
Meschia JF, Brott TG, Brown RD Jr, Crook R, Worrall BB (2005) Phosphodiesterase 4D and 5-lipoxygenase activating protein in ischemic stroke. Ann Neurol 58:351–361. https://doi.org/10.1002/ana.20585
Mika D, Conti M (2016) PDE4D phosphorylation: a coincidence detector integrating multiple signaling pathways. Cell Signal 28:719–724. https://doi.org/10.1016/j.cellsig.2015.11.001
Munshi A, Kaul S (2008) Stroke genetics--focus on PDE4D gene. Int J Stroke 3:188–192. https://doi.org/10.1111/j.1747-4949.2008.00199.x
Munshi A, Babu MS, Kaul S, Shafi G, Anila AN, Alladi S, Jyothy A et al (2009) Phosphodiesterase 4D (PDE4D) gene variants and the risk of ischemic stroke in a south Indian population. J Neurol Sci 285:142–145. https://doi.org/10.1016/j.jns.2009.06.024
Quarta G, Stanzione R, Evangelista A, Zanda B, Angelantonio ED (2009) Phosphodiesterase 4D and 5-lipoxygenase activating protein genes and risk of ischemic stroke in Sardinians. Eur J Hum Genet 17:1448–1453. https://doi.org/10.1038/ejhg.2009.71
Saleheen D, Bukhari S, Haider SR, Nazir A, Khanum S, Shafqat S, Anis MK, Frossard P et al (2005) Association of Phosphodiesterase 4D gene with ischemic stroke in a Pakistani population. Stroke 36:2275–2277. https://doi.org/10.1161/01.STR.0000182242.59466.ee
Shao M, Yi X, Chi L, Lin J, Zhou Q, Huang R (2015) Ischemic stroke risk in a southeastern Chinese population: insights from 5-lipoxygenase activating protein and phosphodiesterase 4D single-nucleotide polymorphisms. J Formos Med Assoc 114:422–429. https://doi.org/10.1016/j.jfma.2013.12.004
Song Q (2006) Phosphodiesterase 4D polymorphisms and the risk of cerebral infarction in a biracial population: the stroke prevention in young women study. Hum Mol Genet 15:2468–2478. https://doi.org/10.1093/hmg/ddl169
Song HJ, Zhou XH, Guo L, Tian FL, Guo XF, Sun YX (2015) Association of phosphodiesterase 4D gene and interleukin-6 receptor gene polymorphisms with ischemic stroke in a Chinese hypertensive population. Genet Mol Res 14:19396–19403. https://doi.org/10.4238/2015.December.29.50
Song YL et al (2017) Phosphodiesterase 4D polymorphisms associate with the short-term outcome in ischemic stroke. Sci Rep 7:42914. https://doi.org/10.1038/srep42914
Staton JM, Sayer MS, Hankey GJ, Attia J, Thakkinstian A (2006) Association between phosphodiesterase 4D gene and ischaemic stroke. Journal of Neurology Neurosurgery Psychiatry 77:1067–1069. https://doi.org/10.1136/jnnp.2006.092106
Sun Y, Huang Y, Chen X, Liu Y, Lu X (2009) Association between the PDE4D gene and ischaemic stroke in the Chinese Han population. Clin Sci 117:265–272. https://doi.org/10.1042/CS20080471
van Rijn MJ et al (2005) Familial aggregation, the PDE4D gene, and ischemic stroke in a genetically isolated population. Neurology 65:1203–1209. https://doi.org/10.1212/01.wnl.0000178744.42953.b7
Wang HM, Chen XL, Ye HH, Bi Y, Pan DB, Xu LY et al (2012) Association ot phosphodiesterase 4D gene with Atherothrombosis ischemic Strok. J Med Res 41:134–137
Wang X, Sun Z, Zhang Y, Tian X, Li Q, Luo J (2017) Impact of the PDE4D gene polymorphism and additional SNP-SNP and gene-smoking interaction on ischemic stroke risk in Chinese Han population. Neurol Res 39(4):351–356. https://doi.org/10.1080/01616412.2017.1289309
Woo D, Kaushal R, Kissela B, Sekar P, Wolujewicz M, Pal P, Alwell K, Haverbusch M, Ewing I, Miller R, Kleindorfer D, Flaherty M, Chakraborty R, Deka R, Broderick J et al (2006) Association of Phosphodiesterase 4D with ischemic stroke: a population-based case-control study. Stroke 37:371–376. https://doi.org/10.1161/01.STR.0000198843.72824.0a
Xu S, Zhang Y, Lin X (2008) Relationship between phosphodiesterase 4D gene polymorphism and ischemic cerebral vascular disease. J clin Neurol 21:249–252
Xue H, Wang H, Song X, Li W, Sun K, Zhang W, Wang X, Wang Y, Hui R (2009) Phosphodiesterase 4D gene polymorphism is associated with ischaemic and haemorrhagic stroke. Clin Sci (Lond) 116(4):335–340. https://doi.org/10.1042/CS20080162
Yan Y, Luo X, Zhang J, Su L, Liang W, Huang G, Wu G, Huang G, Gu L et al (2014) Association between phosphodiesterase 4D polymorphism SNP83 and ischemic stroke. J Neurol Sci 338:3–11. https://doi.org/10.1016/j.jns.2013.12.012
Yang F, Liu S, Yu C, Wang SJ, Paganini-Hill A, Fisher MJ (2012) PDE4 regulates tissue plasminogen activator expression of human brain microvascular endothelial cells. Thromb Res 129:750–753. https://doi.org/10.1016/j.thromres.2011.12.008
Yoon D, Park SK, Kang D, Park T, Park JW (2011) Meta-analysis of homogeneous subgroups reveals association between PDE4D gene variants and ischemic stroke. Neuroepidemiology 36:213–222. https://doi.org/10.1159/000327915
Zhang HL, Wang SR, Li SM (2009) Study on association of the single nucleotide polymorphism of phosphodiesterase 4D with stroke. J Chin Microcirc 13:624–627
Zhang S, Wang XB, Han YD, Wang C, Zhou Y, Zheng F (2017) Certain polymorphisms in SP110 gene confer susceptibility to tuberculosis: a comprehensive review and updated meta-analysis. Yonsei Med J 58:165–173. https://doi.org/10.3349/ymj.2017.58.1.165
Zhao J, Wang X, Xu J, Li N, Shang X, He Z, Yang J (2012) Association of inflammatory response gene polymorphism with atherothrombotic stroke in Northern Han Chinese. Acta Biochim Biophys Sin (Shanghai) 44(12):1023–1030. https://doi.org/10.1093/abbs/gms088
Acknowledgements
This work was supported by Wu Xi Health Technology Project (YGM1125).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Wang, P., Yang, F., Liu, C.X. et al. Association between PDE4D rs966221 polymorphism and risk of ischemic stroke: a systematic review and meta-analysis. Metab Brain Dis 33, 637–645 (2018). https://doi.org/10.1007/s11011-017-0158-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-017-0158-2