Abstract
Ginsenoside Rb1 is the main predominant component in Panax species. In this study, an eco-friendly and convenient preparation method for ginsenoside CK has been established, and five strains of β-glucosidase-producing microorganisms were screened out from the soil of a Panax notoginseng planting field using Esculin-R2A agar. Aspergillus niger XD101 showed that it has excellent biocatalytic activity for ginsenosides; one of the isolates can convert ginsenoside Rb1 to CK using extracellular enzyme from the mycelium. Mycelia of A. niger were cultivated in wheat bran media at 30 °C for 11 days. By the removal of mycelia from cultured broth, enzyme salt fractionation by ammonium sulfate (70%, v/v) precipitation, and dialysis, sequentially, crude enzyme preparations from fermentation liquid supernatant were obtained. The enzymatic transformed Rb1 as the following pathways: Rb1→Rd→F2→CK. The optimized reaction conditions are at reaction time of 72 h, in the range of pH 4–5, and temperature of 50–60 °C. Active minor ginsenosides can be obtained by a specific bioconversion via A. niger XD101 producing the ginsenoside-hydrolyzing β-glucosidase. In addition, the crude enzyme can be resulted in producing ginsenoside CK via conversion of ginsenoside Rb1 at high conversion yield (94.4%). FDA generally regarded, A.niger as safe microorganism. Therefore, these results indicate that A. niger XD10 may provide an alternative method to prepare ginsenoside CK without food safety issues in the pharmaceutical industry.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Ginseng has been used for more than 2000 years as a traditional medicine [1] in Japan, China, Korea, and other orient countries [2]. Ginsenosides are the major active ingredients of ginseng [3] that have been reported to have useful physiological and pharmacological activities including antitumor [4], anti-inflammatory, and antiaging activities [5]. However, the major ginsenosides (including Rb1, Rb2, Re, and Rg1) in hum-absorbed intestine, constituting almost 80–90% of all ginsenosides [6], is hardly absorbed by the human body due to a larger size, less permeability through the cell membrane, and lower bioavailability [7]. Interestingly, deglycosylated ginsenosides such as ginsenoside CK do not exist in natural ginseng [8].
Removal of glycosyls on the skeleton structure of dammarane tetracyclic triterpenoid branched chains using multiple technologies and biosynthesis of specific ginsenosides are an important research topic. Previous studies focused on the production of minor active ginsenosides in recent decades. Therefore, the rare deglycosylated ginsenosides can be obtained using various approaches such as heating [9], acid treatment [10], alkali treatment, and biotransformation [11, 12]. Chemical approach has poor selectivity for the hydrolysis of glucose moieties, hence causing side reactions incorporating isomerization, hydroxylation, and hydratization; however, biotransformation is the most promising method which produces minor active ginsenosides because of its high specificity, productivity, eco-friendliness, low cost, and ability to scale up for industrial-scale production. Several excellent reviews have summarized the advances on the production of deglycosylated ginsenosides using microbial methods [13,14,15].
The search for novel and effective techniques such as microbial screening for new enzymatic and microbial cloning (metagenomics and recombinant DNA) is important for enzyme expression. Screening of suitable bacteria and optimization of its conversion condition for minor ginsenoside production will benefit natural ecology bioremediation. Many studies attempted to seek promising microorganism or enzymes that can transform Rb1 into minor active ginsenoside CK, including endophytic fungus isolated from Panax ginseng, lactic acid bacteria isolated from kimchi, or preparation of enzyme by culturing Armillaria mellea [16].
Rare ginsenoside CK which is easily absorbed by the human body is the genuine active form of protopanaxadiol-type saponins. Recently, it has been paid much attention to its intriguing biological activities in vivo such as anticarcinogenic, antiangiogenesis, antiaging, and antidiabetic activities [17] .The production of CK can be achieved by using microbes or through enzymatic processes. For example, endophyte JG09 transformed Rb1 in the following sequence: Rb1→Rd→F2→CK, successively hydrolyzing the 20-O-β-D-(1-6)-glucopyranoside and 3-O-β-D-(1-2)-glucopyranoside [18]. Ginsenoside CK was rapidly produced during the 168-h culture. Yan et al. [19] reported that minor ginsenoside CK was gated by the enzyme of Paecilomyces bainier sp. 229. Quan et al. [20] first discovered aerobic bacteria that can convert ginsenoside Rb1 to CK via the gypenoside XVII and ginsenoside Rd.
Ginsenoside Rb1-converting fungus isolated and identified from the soil of a Panax notoginseng field was most commonly applied [4, 12, 21]. However, soil microorganisms can be applied to foods provided it is safe. Fungus has been used for a long time in fermented crops because of its production of various enzymes. In this study, the conversion of ginsenoside Rb1 into specific minor ginsenosides CK was evaluated using microbes or enzymes after the Esculin-R2A assay.
Materials and Methods
Materials
Standard ginsenosides Rb1, Rd, F2, and CK were supplied by Chengdu Prefa Technology Co Ltd. (Dalian, China). p-Nitrophenyl-β-d-glucopyranoside (pNPG), p-nitrophenol (pNP), and β-glucosidase were obtained from Sigma-Aldrich (St Louis, MO, USA). Galactosidase was obtained from Aspergillus oryzae; Pectinex XXL (liquid), Celluclast 1.5L (liquid), and Snailase (BR, breaking wall of 90%) were obtained from Sigma-Aldrich, Novozymes, Novozymes, and Shanghai Yuanye Biotechnology Co., Ltd. (Shanghai, China), respectively.
Isolation Fungi from Soil of P. ginseng and Phylogenetic Analysis
Esculin-R2A agar was used to isolate β-glucosidase-producing microorganisms. They were selected according to the reddish-brown to dark brown zone surrounded from β-glucosidase that hydrolyzes the surrounding substances.
Sangon Biotech (Shanghai) Co. Ltd., China, completed the sequencing of ITS rDNA gene. According to the resulting alignment, the phylogenetic tree was constructed using the neighbor-joining method of the MEGA4 program (http://www.megasoftware.net/mega4/mega.html). Confidence levels for the branches were similarly performed to obtain a bootstrap analysis with 1000 replicates.
Growth and Crude Enzyme Production
The fungus A. niger XD101 was cultured on PDA at 30 °C for 2 days. Then, we inoculated the microorganism in a medium (100mL in 500mL Erlenmeyer flask) of which the component is 3% wheat bran, 0.3% (NH4)2SO4, and 0.2% KH2PO4 with 11 days at 30 °C. For achieving homogeneous growth, a few glass beads were added to prevent mycelial clumping during shake flask culturing. After stopping the fermentation, the culture broth was centrifuged to remove the cells at 10,000×g at 4 °C for 20min; 70% (NH4)2SO4 was added to the supernatant. The mixture was stored at 4 °C overnight to obtain the protein precipitate after centrifugation. The protein was purified by dialyzing against 0.05 M and pH 5.0 acetate buffer and diluting to 1/10 volume of culture with 0.05 M and pH 5.0 acetate buffer.
Assay of Enzyme Activity and Enzyme Characterization
The effects of temperature and pH were studied under the following conditions from pH 4.0 to 9.0 and at various temperatures from 20 to 70°C. Each reaction was incubated at 50°C for 30 min. The buffers (each at 0.05 M) used include glycine-HCl buffer (pH 3.0), NaAc-HAc (pH 4.0–5.5), sodium phosphate buffer (pH 6.0–7.0), and Tris–HCl buffer (pH 8.0–9.0) .
The effects of pH and temperature on the stability of enzyme were monitored under the following conditions: The enzyme samples were stored in buffers with different pH (4.0–9.0) for 4 h at 50 °C. The enzyme samples were incubated at 20–70 °C for 4 h and then immersed in an ice bath to cool to room temperature immediately. The activity of enzyme determined under optimal conditions (optimal pH and temperature) was set as 100%; other activities were reported relative to this activity.
Biotransformation of Ginsenoside Rb1
Microbial transformation from spore suspension was conducted as follows: Spore suspension was prepared by scraping down 2-day-old culture with 0.1% Tween 80 acetate buffer (pH 7.0) on PDA slants. Initial fermentation medium was incubated 1% (v/v) of fungal spore suspension in an Erlenmeyer flask by shaking at 30 °C with 150 rpm. Ginsenoside Rb1 was added to the flasks to a final concentration of 1 mg/mL shaken for 7 days; the culture was filtered and centrifuged. The supernatant extracted with n-butanol was analyzed using HPLC.
The spores were suspended by PDB and cultured for several days till spore germination, and the mycelium suspension system used the mycelium suspension. Microbial transformation from mycelium was conducted by adding ginsenoside Rb1 to 5-mL suspensions of 2-day-old cultures to a final concentration of 1 mg/mL ~50mg/mL dissolved in 12.5 mL 25 mM acetate buffer (pH 5.0). We mixed the suspension and an equal volume of LL medium consisting of 0.5 g/L NH4Cl, 1.0 g/L K2HPO4, 0.5 g/L KH2PO4, 0.25 g/L MgSO4, and 1.0 g/L yeast extract (pH 7.0) and shaken at 30 °C for 7 days at 150 rpm.
Enzymatic transformation was conducted with the crude enzyme prepared from the cultured supernatant as characterized in the section. A total of 2.5–10 mg/mL ginsenoside Rb1 dissolved in crude enzyme was reacted at 25–70 °C for 72 h. Samples were withdrawn at suitable time intervals (12 h, 24 h, 36 h, 48 h, 60 h,72 h). Comparison of ginsenosides produced by commercial β-glucosidase, 3 mL of each enzyme solution (approximately 20 U) was added to the same volume of ginsenoside reaction mixture, and the samples were analyzed by HPLC after 72 h.
Extraction of Crude Saponin
The reaction mixtures were extracted twice with same volume of water-saturated n-butanol. The n-butanol fraction was evaporated using a rotary vacuum evaporator to generate the crude saponin fraction (Heidolph, Germany). The crude saponin was dissolved in 5mL methanol and analyzed by TLC and HPLC. The samples were then passed through a 0.45-mm PTFE syringe filter prior to injection.
Purification of β-glucosidase from A. niger
Ten milliliters of crude enzyme was eluted on a DEAE-cellulose column (2.8cm×25 cm), and the proteins were fractionated stepwise with 0.06, 0.12, 0.18, 0.24, 0.30, 0.40 and 0.50M KCl in 0.25M NaAc-HAc buffer (pH 5.0; 3.0mL/tube). The tube was examined for β-glucosidase activity. The tube with the highest activity was carried out by SDS-PAGE.
TLC and HPLC Analysis of Ginsenosides
TLC was performed on silica gel plates (GF254; Qingdao Ocean Chemical Co., Ltd) with a mixture of chloroform/methanol/water (10:5:1 v/v/v, under layer) as the developing solvent. We detected the spots by spraying with 10% (v/v) sulfuric acid in ethanol, followed by heating at 110 °C for 5 min until the spots became clearly visible.
HPLC was performed using an Agilent 1100 system (Agilent Technologies). A reverse-phase column (C18, 4.6 mm × 150 mm, 5 mm) at 30 °C was used. The mobile phase of solvents A (CH3CN) and B (H2O) was as follows: A from 80 to 55%, 0–20 min; A from 55 to 45%, 20–45 min; A from 45 to 55%, 45–55 min; A from 55 to 45%, 55–70 min; A from 45 to 0%, 70–85 min. We detected metabolites at 203 nm.
Statistical Analysis
We performed experiments in triplicate, and then the results are shown as mean standard deviation. All the data were assigned for comparison, and an analysis of variance (ANOVA) was carried out using Origin version 8.0.
Results
Identification of β-Glucosidase-Producing Strains
A total of five fungal isolates were obtained and identified from the soil of P. ginseng using the method of the standard dilution–plating technique on Esculin-R2A agar (Table S1), for production of β-glucosidase. Among them, one isolate, XD101, was found to efficiently biotransform Rb1 to ginsenoside CK. The biotransformed products by strains isolated with ginsenoside-converting activity for 7 days were determined, as corresponding TLC and HPLC chromatograms listed in Fig. S1 and Fig. S2.
The cultured conidia and sporangia images are shown in Fig. S3. The colonial morphology varied when cultured under different conditions (Fig. S4). The morphological characteristics are shown in Table S2. The ITS gene sequence of all the screened bacteria was compared with the sequences in the NCBI database, and the DC102 was aligned with other neighboring strains, confirming the taxonomic relationships. The strain XD101 was grouped with the Aspergillus species which is revealed by phylogenetic tree. By analyzing the morphological characteristics and basing on phylogenetic analysis, it indicated the isolated which can be designed as the genus Aspergillus. It is most closely related to Aspergillus niger MK203789.1, as seen in Fig. 1, named as A. niger strain XD101.
Effects of Different Biotransformation Systems on Conversion of Ginsenoside Rb1
The two biotransformation systems (spore suspension and mycelium) were mixed with ginsenoside Rb1 at a final concentration of 1 mg/mL. The bioconversion rate (w/w) of ginsenoside Rd by spore suspension was 13.3% at 24 h, then increased to 58.6 % at the terminal 144 h (Fig. 2a). By comparison, the conversion rates (w/w) of ginsenoside Rd by mycelium suspension were 83.3% at 24 h and sharply decreased to 0% in 3 days. The transformation of ginsenoside F2 and ginsenoside CK occurred one after another. A max peak area of Rd at the first day almost decreased in the subsequent 2 days as shown in Fig. 2b. Ginsenoside F2 was detected at the second day and reached the maximum conversion of 63.2% at the fifth day. The maximum conversion of ginsenoside CK reached 51.1% at the sixth day. Based on the products including ginsenosides Rd, F2, and CK, we selected the mycelium as the final biotransformation system for the production of ginsenoside CK. This result indicates that A. niger XD101 excreted abundant ginsenoside Rb1-converting enzymes.
Effect of Rb1 Concentration with Cultured Mycelium A. niger XD101
For microorganisms or enzymes identified for converting ginsenosides, a usual problem is low yield for the reason of low substrate tolerance, requirements of pure substrates, low hydrolyzed activity, and further separation from other compounds. Therefore, the effect of substrate concentration on bioconversion was evaluated in this study. Overall, the conversion of ginsenoside CK decreased with increasing concentration of ginsenoside Rb1 in the range of 1–50 mg/mL (Fig. 3). The maximum bioconversion rates of ginsenoside CK are 51.8% and 44.3% at Rb1 concentrations of 1 mg/mL and 2 mg/mL, respectively. However, high substrate concentrations would decrease the reaction rate. The bioconversion rate of ginsenoside CK sharply decreased to l1.6% in Rb1 concentration of 1–10 mg/mL and decreased less than 10% with the increase in Rb1 concentration to 50 mg/mL.
Analysis of Hydrolysis Metabolites on Bioconversion Time-Course
The advantages of isolated enzymes are reaction specificity and simple methodology. To evaluate the transformation of ginsenoside Rb1 by the crude enzyme of cultured mycelium from A. niger strain XD101, HPLC analyzed the sample withdrawn at regular intervals (Fig. 4). As it was detected after 12 h, ginsenoside Rb1 was almost biotransformed to other three metabolites ginsenosides Rd, F2, and CK in the initial stage of reaction (Fig. 4a). According to the HPLC profiles, ginsenoside Rd was gradually hydrolyzed to metabolites ginsenosides F2 and CK. More specifically, the conversation of ginsenoside F2 was 60.3%, 34.8%, and 6.4% at 12 h, 36 h, and 72 h, respectively. Correspondingly, the amount of ginsenoside CK increased from 53.9% at 12 h to 73.7% at 36 h and reached 94.4% after 72 h. Therefore, almost all ginsenoside Rb1 were hydrolyzed to the final product ginsenoside CK after 72 h (Fig. 4c).
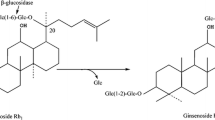
Ginsenoside Rb1 that contains C-20 and C-3 β-glucose linkages can be hydrolyzed by successive loss of glucose moiety through β-glucosidase. The HPLC examined the process that ginsenoside Rb1 was decomposed by the crude enzyme of A. niger XD101 in this study, with change in reaction time. Firstly, ginsenoside Rd is produced by hydrolyzing 20-O-β-D-(1-6)-glucopyranoside. Then, the enzyme hydrolyzed glucose moiety linkage of the C-3 position of aglycon of Rd to F2 further to CK. Therefore, the results show that the crude enzyme of A. niger XD101 utilizes the hydrolytic pathway of ginsenosides Rb1 → Rd → F2 → CK, as shown in Fig. 5.
Effect of Temperature and pH on Enzyme Activity of Crude Enzyme XD101
Industry applications require enzymes with special characteristics of different substrates and raw materials. The activity and stability of enzyme over a temperature and pH range were analyzed for the crude enzyme XD101.
In this study, the optimum temperature of β-glucosidase activity in crude enzyme is 60 °C. Relative activity was 63.6% at 50 °C, while it was 50.8% at 70 °C. The enzyme is stable at temperatures lower than 50 °C, and the thermostability sharply decreased above 60 °C (Fig. 6a). This enzyme showed that the greatest activity was at pH 5.0. At pH 4 and 6, the activity was 92.53% and 51.36% respectively. The activity was approximately 50% and remained stable in a pH range from 3 to 6. The enzyme became activated on the acid side of pH profile (above pH 7.0: Fig. 6b).
Effect of Temperature, pH, and Rb1 Concentration on the Biotransformation of Ginsenoside CK by Crude Enzyme Preparations XD101
An isolated enzyme has more advantages for one-step reaction but only works efficiently under optimum pH and temperature conditions. The ginsenoside Rb1-hydrolyzing activity of crude β-glucosidase at a reaction temperature is shown in Fig. 7a. Ginsenoside Rb1 was incubated with crude β-glucosidase for 24 h at a temperature range of 25–70 °C. Initially, at 25–30 °C, ginsenoside Rb1 hydrolyzed to ginsenosides Rd and F2. As the reaction temperature was between 40 and 60 °C, the amount of ginsenoside CK was significantly increased. When the reaction temperature was increased to 60–70 °C, the transformation of ginsenoside CK decreased.
Hydrolysis of ginsenoside Rb1 by crude enzyme XD101. Effect of a temperature and b pH on the transformation of Rb1 by crude enzyme. The reaction mixture (1 mL) containing Rb1 (6 mg) and crude enzyme (1 mL 3.5 U) in 1.0 mL of 0.05 M acetate buffer (pH 5) was incubated for 24 h at the given temperatures in the range of 25–70 °C. The optimum pH was determined by incubating the reaction mixture for 24 h at 50 °C at the given pH values (each at 0.05 M); acetate buffer (pH 4.0–5.5), sodium phosphate (pH 6.0–7.0), Tris–HCl (pH 8.0), and glycine-NaOH buffer (pH 9.0)
The results indicate that the optimum temperature for ginsenoside CK transformation by the crude enzyme of A. niger XD101 is in the range of 50–60 °C. The optimum pH range for ginsenoside CK transformation is 3.0–5.0 in this study (Fig. 7b). When the pH was increased to 5.0–8.0, the reaction rate decreased. The transformation of ginsenoside CK was not observed beyond pH 8.0. From the above result, we concluded that the formation of ginsenoside CK from ginsenoside Rb1 needed a weak acidic condition.
The effect of Rb1 concentration on the transformation of ginsenoside CK was evaluated. The conversion rate of ginsenoside CK was much higher in the concentration of Rb1 in the range of 2.5–6 mg/mL, as shown in Fig. 8. Substrate ginsenoside Rb1 almost completely converted to ginsenoside CK within 72 h. However, the yield of ginsenoside CK was 90% at 6 mg/mL of ginsenoside Rb1 in 60 h. This is slightly slower than that of 2.5 and 4 mg/mL ginsenoside Rb1. The conversion was much slower under the other two reaction conditions of 8–10mg/mL ginsenoside Rb1. We selected the conditions 6 mg/mL substrate concentration and 20 U crude enzyme.
Evaluation of Ginsenoside Rb1 Transformation Activity by Comparing with Several Commercial Glycosidases
Evaluation of ginsenoside Rb1 transformation activity by comparing with several commercial glycosidases was conducted in this study, as shown in Table 1. The generation of ginsenoside CK from ginsenosides Rb1 was catalyzed with several commercial enzymes, almost from Aspergillus. The commercial enzymes galactosidase, Pectinex XXL, and Celluclast 1.5L could not transform ginsenosides Rb1 to ginsenoside CK and instead transformed to ginsenosides F2, Rd, and Rd, respectively. As a result, comparison of the four enzymes including galactosidase, Pectinex XXL, Celluclast 1.5L, and enzyme in this work, enzyme preparation from cultured mycelia XD101 could only produce high yield of ginsenoside CK. The transformation of ginsenoside CK using the enzyme preparation obtained from the cultured mycelia XD101 reached 94.4% at pH 4.5, 50 °C, 20 U enzyme, and 6 mg/mL substrate for 3days.
Discussion
The application forms of Aspergillus strains generally depend on downstream processing and microbial inherent characteristics. Lin et al. [4] optimized the biotransformation conditions for product Rd by Aspergillus versicolor with the maximum conversion yield as high as 96% at 48 h in shake flasks. Microbial sources can biotransform the major ginsenoside Rb1 to minor ginsenosides. As such, CK can be transformed by enzymes secreted by microbes such as Caulobacter leidyi GP45, F sacchari, and A. niger [22, 23]. However, spore suspension from the fungus A. niger XD101 converted ginsenoside Rb1 to Rd, with a final conversion of 64.8% at the seventh day (Fig. 2a); mycelium from A. niger XD101–converted ginsenoside Rb1 to ginsenoside F2 reached the maximum conversion of 63.6% at the fifth day, and the maximum conversion of ginsenoside CK reached 50.4% at the sixth day (Fig. 2b).
The effect of ginsenoside Rb1 concentration on the transformation of ginsenoside CK was evaluated. The conversion of ginsenoside CK was decreased with increasing concentration of ginsenoside Rb1 in the range of 1–50 mg/mL, with maximum bioconversion rates of 50.4% at Rb1 concentrations of 1 mg/mL by mycelium of A. niger strain XD101 in 7-day culture (Fig. 3). However, studies have reported on the high-substrate-concentration bioconversion of ginsenoside by fungus via UV mutation. The mutant P. bainier 229-7 can produce ginsenoside Rd at a high bioconversion rate of 94.9% with 20 mg/mL saponin from SPNL [24]. The bioconversion ratio of a novel A. niger strain TH-10a is 60% at even 40 mg/mL ginseng root extract as the substrate after 72 h reaction under the optimal condition [25]. As a low efficiency of cultured mycelium was obtained from wild A. niger strain XD101 in our study, the crude enzyme prepared from the cultured mycelia was selected for further study on ginsenoside CK production.
Usually, Aspergillus sp. β-glucosidases have diverse molecular weight (43–138 kDa) with an optimal temperature between 45 and 70 °C and an optimal pH range between 3.5 and 7.5 [1, 26, 27]. A. versicolor excreting β-glucosidase converted ginsenoside Rb1 to Rd under optimum conditions of pH 5.0 and 37 °C [4]. The proposed optimum reaction conditions in ginsenoside F1 production by commercial enzyme Cellulase KN from A. niger are pH 5.0 and 50 °C [28]. The processing technology on biotransformation of ginsenoside Rb1 to CK using extracellular preparations from cultured mycelia in our study is as seen in Figs 9 and 10. The optimum conditions for the preparation of ginsenoside CK by crude enzyme preparations from cultured mycelium A. niger XD101 are as follows: temperature range of 50–60 °C and pH 4–5. Therefore, the results indicate that the crude enzyme XD101 is ideal for the formation of ginsenoside CK from Rb1 in a thermostable option and a weak acidic condition.
In addition, the conversion rate of ginsenoside CK was increased in the concentration of ginsenoside Rb1 in the range of 2.5–6 mg/mL by the crude enzyme of cultured mycelium from A. niger strain XD101, as shown in Fig. 8. Because of the advantages of smaller usage of enzymes and complete conversion of Rb1, the conditions—6 mg/mL substrate concentration and 20 U crude enzyme—were selected. Actually, the crude enzyme contains various glucosidases and performs the hydrolysis of Rb1 using a combination of these glucosidases [29]. After the conversion of the enzyme, the purification and characterization was estimated in further studies, the mechanism of hydrolysis will be elucidated, and the enzyme activities will be improved to shorten the production cycle and increase the yield.
For enzymes were overexpressed in Escherichia coli, with BglPC28 β-glucosidase (cloned and expressed from Pseudonocardia sp. strain Gsoil 1536), Du et al. [30] obtained 113 g of Rg2(S) by the biotransformation of 150 g of Re (20 mg/mL ginsenoside) in a 10-L jar fermenter at 30 °C for 24 h. Zhanget al. [31] reported that two thermostable glycosidases bioconverted high-substrate-concentration ginsenoside extract to ginsenoside 20(S)-Rg3. Under the optimal conditions, 6.28 g/L of Rg3 within 90 min with a corresponding molar conversion of 95.0% can be obtained by transformation of 20 g/L of ginsenoside extract. Therefore, the enzymatic conversion of ginsenoside Rb1 from wild fungus can be improved by construction and high-rate expression of genetically engineered strains for resistance to much higher concentrations of ginseng saponin used in the next scaled-up biotransformation step.
Additionally, A. niger is the most widely used genus species in food and drink production processes with more 1500 years history. Only A. oryzae have the GRAS status as listed by the U.S. Food and Drug Administration. To further improved for high-concentration substrate tolerance for ginsenoside CK transformation, fungus can be selected by mutation. In addition, many important β-glucosidases have been considered to come from Aspergillus species in recent years [32]. For example, commercial enzyme Cellulase KN obtained from food-grade A. niger was used to increase the production of ginsenoside F1 as a 10-g unit [28]. Ginsenosides Rb1, Rb2, Rc, Rd, and Rg3 were used for the bioconversion into gypenoside-XVII, compound-O, compound MC1, F2, and Rh2, respectively, by commercial β-galactosidase obtained from A. oryzae (SUMILACT LTM) [33]. Therefore, the isolated A. niger strain XD101 can be awarded for producing deglycosylated ginsenosides as a safe strain which were verified not produce mycotoxins. Then, we want to study the safety of the fungus.
The enzyme activity of fractions was examined: the 24-26 tube eluted by 0.18M and 0.24M hydrolyzed Rb1; 25 and 26 fraction exhibited the highest enzyme activity and a single band. The molecular weight the of purited enzyme was approximately 110kDa.
Change history
22 April 2021
A Correction to this paper has been published: https://doi.org/10.1007/s12010-021-03574-0
References
Park, C. S., Yoo, M. H., Noh, K. H., & Oh, D. K. (2010). Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Applied Microbiology and Biotechnology, 87, 9–19.
Yoo, M. H., Yeom, S. J., Park, C. S., Lee, K. W., & Oh, D. K. (2011). Production of aglycon protopanaxadiol via compound K by a thermostable β-glycosidase from Pyrococcus furiosus. Applied Microbiology and Biotechnology, 89, 1019–1028.
Luan, H., Qi, L. X., Hu, Y., Hao, D., Cui, Y., & Yang, L. (2006). Purification and characterization of a novel stable ginsenoside Rb1-hydrolyzing β-D-glucosidase China white jade snail. Process Biochemistry, 41(9), 1974–1980.
Duan, Z. G., Wei, B., Deng, J. J., Mi, Y., Dong, Y. F., Zhu, C. H., Fu, R. Z., Qu, L. L., & Fan, D. D. (2018). The anti-tumor effect of ginsenoside Rh4 in MCF-7 breast cancer cells in vitro and in vivo. Biochemical and Biophysical Research Communications, 499, 482–487.
Lin, F., Guo, X., & Lu, W. (2015). Efficient biotransformation of ginsenoside Rb1 to Rd by isolated Aspergillus versicolor, excreting β-glucosidase in the spore production phase of solid culture. Anton Leeuw, 108(5), 1117–1127.
Upadhyaya, J., Kim, M. J., Kim, Y. H., Ko, S. R., Park, H. W., & Kim, M. K. (2016). Enzymatic formation of compound-K from ginsenoside Rb1 by enzyme preparation from cultured mycelia of Armillaria mellea. Journal of Ginseng Research, 40, 105–112.
Lei, C., Wu, S. Q., Zhao, C. A., & Yin, C. R. (2016). Microbial conversion of major ginsenosides in ginseng total saponins by Platycodon grandiflorumendophytes. Journal of Ginseng Research, 40, 366–374.
Kim, M. J., Upadhyaya, J., Yun, M. S., Ryu, N. S., Song, Y. E., Park, H. W., Kim, Y. H., & Kim, M. K. (2017). Highly regioselective biotransformation of ginsenoside Rb2 into compound Y and compound K by β-glycosidase purified from Armillaria mellea mycelia. Journal of Ginseng Research, 42, 504–511.
Hwang, C. R., Lee, S. H., Jang, G. Y., Hwang, I. G., Kim, H. Y., Woo, K. S., Lee, J., & Jeong, H. S. (2014). Changes in ginsenoside compositions and antioxidant activities of hydroponic-cultured ginseng roots and leaves with heating temperature. Journal of Ginseng Research, 38(3), 180–186.
Sun, C., Gao, W., Zhao, B., & Cheng, L. (2013). Optimization of the selective preparation of 20(R)-ginsenoside Rg3 catalyzed by d, l-tartaric acid using response surface methodology. Fitoterapia., 84, 213–221.
Quan, L. H., Kim, Y. J., Li, G. H., Choi, K. T., & Yang, D. C. (2013). Microbial transformation of ginsenoside Rb1 to compound K by Lactobacillus paralimentarius. World Journal of Microbiology and Biotechnology, 29, 1001–1007.
Song, X., Wu, H., Piao, X., Yin, Z., & Yin, C. (2017). Microbial transformation of ginsenosides extracted from Panax ginseng adventitious roots in an airlift bioreactor. Electronic Journal of Biotechnology, 26, 20–26.
Murthy, H. N., Georgiev, M. I., Kim, Y. S., Jeong, C. S., Kim, S. J., Park, S. Y., & Paek, K. Y. (2014). Ginsenosides: prospective for sustainable biotechnological production. Applied Microbiology and Biotechnology, 98(14), 6243–6254.
Eom, S. J., Kim, K. T., & Paik, H. D. (2018). Microbial bioconversion of ginsenosides in Panax ginseng and their improved bioactivities. Food Review International, 34, 698–712.
Ku, S. (2016). Finding and producing probiotic glycosylases for the biocatalysis of ginsenosides: a mini review. Molecules, 21, 645.
Chang, K. H., Jo, M. N., Kim, K. T., & Paik, H. D. (2014). Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. Journal of Ginseng Research, 38, 47–51.
Kim, S. H., Min, J. W., Quan, L. H., Lee, S., Yang, D. U., & Yang, D. C. (2012). Enzymatic transformation of ginsenoside Rb1 by Lactobacillus pentosus strain 6105 from kimchi. Journal of Ginseng Research, 36, 291–297.
Cui, L., Wu, S. Q., Zhao, C. A., & Yin, C. R. (2016). Microbial conversion of major ginsenosides in ginseng total saponins by Platycodon grandiflorum endophytes. Journal of Ginseng Research, 40, 366–374.
Yan, Q., Zhou, W., Shi, X., Zhou, P., Ju, D., & Feng, M. (2010). Biotransformation pathways of ginsenoside Rb1 to compound K by β-glucosidases in fungus Paecilomyces Bainier sp. 229. Process Biochemistry, 45(9), 1550–1556.
Quan, L. H., Piao, J. Y., Min, J. W., Kim, H. B., Kim, S. R., Yang, D. U., & Yang, D. C. (2011). Biotransformation of ginsenoside Rb1 to prosapogenins, gypenoside XVII, ginsenoside Rd, ginsenoside F2, and compound K by Leuconostoc mesenteroides DC102. Journal of Ginseng Research, 35, 344.
Zhao, Y., Lee, H. G., Kim, S. K., Yu, H., Jin, F., & Im, W. T. (2016). Mucilaginibacter pocheonensis sp. nov. with ginsenoside converting activity isolated from soil of ginseng cultivating field. International Journal of Systematic and Evolutionary Microbiology, 66, 2862–2868.
Chi, H., & Ji, G. E. (2005). Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnology Letters, 27(11), 765–771.
Cheng, L. Q., Kim, M. K., Lee, J. W., Lee, Y. J., & Yang, D. C. (2006). Conversion of major ginsenoside Rb1 to ginsenoside F2 by Caulobacter leidyia. Biotechnology Letters, 28(14), 1121–1127.
Ye, L., Zhou, C. Q., Zhou, W., Zhou, P., Chen, D. F., Liu, X. H., Shi, X. L., & Feng, M. Q. (2010). Biotransformation of ginsenoside Rb1 to ginsenoside Rd by highly substrate-tolerant Paecilomyces bainier 229-7. Bioresource Technology, 101, 7872–7876.
Feng, L., Xu, C., Li, Z., Dai, Y., Han, H., Yu, S., & Liu, S. (2016). Microbial conversion of ginsenoside Rd from Rb1 by the fungus mutant Aspergillus niger strain TH-10a. Preparative Biochemistry, 46, 336–341.
Molina, G., Contesini, F. J., & De, R. R. (2016). β-Glucosidase from Aspergillus//GUPTA V K, Ed. New and future developments in microbial biotechnology and bioengineering. Amsterdam, 155–169.
Yang, X. D., Yang, Y. Y., Ouyang, D. S., & Yang, G. P. (2015). A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia, 100, 208–220.
Wang, Y., Choi, K. D., Yu, H., Jin, F., & Im, W. (2016). Production of ginsenoside F1 using commercial enzyme Cellulase KN. Journal of Ginseng Research, 40(2), 121–126.
Luan, H., Qi, L. X., Hu, Y., Hao, D., Cui, Y., & Yang, L. (2006). Purification and characterization of a novel stable ginsenoside Rb1-hydrolyzing β-D-glucosidase from China white jade snail. Process Biochemistry, 41, 1974–1980.
Du, J., Cui, C. H., Park, S. C., Kim, J. K., Yu, H. S., Jin, F. X., Sun, C. K., Kim, S. C., & Im, W. T. (2014). Identification and characterization of a ginsenoside-transforming β-glucosidase from Pseudonocardia sp. Gsoil 1536 and its application for enhanced production of minor ginsenoside Rg 2 (S). PLoS One, 9, e96914.
Zhang, S. S., Xie, J. C., Zhao, L. G., Pei, J. J., Su, E. Z., Xiao, W., & Wang, Z. Z. (2018). Cloning, overexpression and characterization of a thermostable β-xylosidase from Thermotoga petrophila and cooperated transformation of ginsenoside extract to ginsenoside 20(S)-Rg3 with a β-glucosidase. Bioorganic Chemistry, 85, 159–167.
Treebupachatsakul, T., Nakazawa, H., Shinbo, H., Fujikawa, H., Nagaiwa, A., Ochiai, N., Kawaguchi, T., Nikaido, M., Totani, K., Shioya, K., Shida, Y., Morikawa, Y., Ogasawara, W., & Okada, H. (2016). Heterologously expressed Aspergillus aculeatus β-glucosidase in Saccharomyces cerevisiae is a cost-effective alternative to commercial supplementation of β-glucosidase in industrial ethanol production using Trichoderma reesei cellulases. Journal of Bioscience and Bioengineering, 121, 27–35.
Kim, M. J., Upadhyaya, J., Yun, M. S., Ryu, N. S., Song, Y. E., Park, H. W., & Kim, M. K. (2017). Highly regioselective biotransformation of ginsenoside Rb2 into compound Y and compound K by β-glycosidase purified from Armillaria mellea mycelia. Journal of Ginseng Research, S1226845317300118.
Funding
This research was supported by the National Natural Science Foundation of China (21706211, 21576160, 21878246, 21676214), the National Key R&D Program of China (2019YFA0905200) and the Educational Commission of Shaanxi Province of China (16JS104).
Author information
Authors and Affiliations
Contributions
JYY, LWN, and FDD conceived and designed the study. JYY performed the experiments and analyzed the data. JYY wrote the paper. LWN and FDD reviewed and edited the manuscript. All the authors read and approved the manuscript
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
The original online version of this article was revised: Changes has been made to the authors affiliation (article note has been added) and funding.
Supplementary Information
ESM 1
(DOC 1449 kb)
Rights and permissions
About this article
Cite this article
Jiang, Y., Li, W. & Fan, D. Biotransformation of Ginsenoside Rb1 to Ginsenoside CK by Strain XD101: a Safe Bioconversion Strategy. Appl Biochem Biotechnol 193, 2110–2127 (2021). https://doi.org/10.1007/s12010-021-03485-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03485-0