Abstract
To heterologously express a Talaromyces thermophilus lipase gene in Trichoderma reesei, an efficient binary vector pChph-pCBH1sigpro-ttl which includes a newly designed cbh1 promoter and hygromycin-resistant marker was constructed. This plasmid was then transformed into T. reesei via improved Agrobacterium EHA 105-mediated transformation. After modification of co-culture conditions and enzymolysis treatment of conidia, 258 transformants were produced. A two-step screening method based on antibiotic resistance and capacity to utilize lactose and tributyrin was introduced to further select promising candidates, which would be additionally verified by PCR analysis, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and lipase activity assay. Lipase production was carried out in shaking flasks, and the activity reached 241 IU/mL (7415.4 IU/mg) after 84-h fermentation. It was found that this lipase performed high alkali and thermostable tolerance with the optimal pH 9.5 and temperature 60 °C, and it could retain more than 70 % activity after being disposed in pH 11 or 70 °C for 1 h. This study herein would benefit the genetic engineering of T. reesei and the industrial application of this important fungal lipase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipase is characterized by its ability to hydrolyze long-chain triacylglycerides into free fatty acids. In most cases, lipases possess a catalytic center, which is buried under a surface loop (lid) and could be activated upon oil–water interface [1]. Due to its versatility of catalyzing numerous reactions, lipase is widely used in bioenergy, paper making, fine chemicals and food industry, and pharmaceutical and waste management [2, 3].
With more than half a century research, the outstanding filamentous fungal host, Trichoderma reesei, is hitherto the workhorse for biomass conversion [4]. The decisive advantage of this strain for its extensive application is the salient secretory capacity, not only for the three cellulolytic enzymes (cellobiohydrolase, endo-β-1,4-glucanase, β-glucosidase) [5], but also for various heterologous proteins [6]. Additionally, excellent post-translational modifications such as glycosylation and phosphorylation together with simplified downstream purification make these versatile model fungi more suitable for heterologous expression [7].
The alkali and thermostable lipase from Talaromyces thermophilus performed remarkably in different conditions, endowing itself great potential in biodiesel production [8], paper making, and detergent ingredient [9]. Whereas, low productivity of this fungus held back its further applications. Among the solutions, heterologous expression is always counted upon for its high efficiency and practicability [6]. There are various host candidates for removing this bottleneck [10], such as the prevalent prokaryotic Escherichia coli, eukaryotic yeast, and T. reesei. According to previous studies, heterologous productivity in T. reesei could exceed its fungal counterparts like Pichia pastoris in some cases [11–13], and the produced protein could be economically attained in quantity through rough filtration. Therefore, T. reesei was selected as the expression host in this study.
To express in T. reesei, the indispensable prerequisite is establishing an efficient and convenient transformation platform [14], which seems to be more urgent for the functional genomic analysis after the sequencing of its genome [15]. There are mainly chemical (PEG or CaCl2-mediated protoplast transformation), physical (biolistic and electroporated transformation), and biological ways to transfer target genes into the Trichoderma [16], among is the Agrobacterium-mediated transformation (AMT). AMT is frequently adopted to integrate multitudinous genes randomly and mainly in single copy into genomes of various starting materials including spores, protoplasts, mycelium, and germlings [17]. Studies had manifested that AMT was an efficient tool for mutagenic manipulation of Trichoderma [18]. However, measures should be further taken to increase the transformation efficiency so as to make this methodology better qualified in T. reesei expression.
In this work, a genetic manipulation platform was set up, through which optimized lipase gene was successfully expressed in T. reesei under control of a self-designed cbh1 promoter. Optimization of AMT and a two-step screening method were applied to boost transformation efficiency. Additionally, lipase production was discussed in a lab scale, and lipase properties were further researched aiming to pave the way for industrial application of this fungal lipase.
Materials and Methods
Strains and Media
T. reesei ZU-02 was used for making recipients and chromosomal DNA preparation. E. coli DH5α and Agrobacterium tumefaciens EHA 105 were applied for plasmid propagation and transformation mediation, respectively. Other microorganisms were lab stored.
The seed, induction, and fermentation media were the same as that described by Gu et al. [19]. For transformant screening, the medium was supplemented by 1 % tributyrin and 4 % lactose on the basis of Fang et al. [20].
Preparation of Lipase Gene
The T. thermophilus lipase (TTL) gene (GenBank JF414585.1) encodes an alkali and thermostable lipase with bright prospect in industrial application. According to the codon usage preference of T. reesei [21], TTL was optimized using the Software Gene Designer [22] by reference to KAZUSA database (http://www.kazusa.or.jp/codon/). Resultantly, 184 nucleotides were changed in order to increase the GC content from 55.8 to 59.4 %. The minimum free energy of RNA secondary structure changed from −307.3 to −322.4 kcal/mol, which meant that RNA was more stable in T. reesei for translation. Modified DNA sequence was further synthesized by ABI3900 DNA high-throughput synthesizer. The corresponding primers for PCR of TTL were L1 (5′-ATGTATCAAAAGTTGGCCCTC-3′) and L2 (5′-TTACAGGAACGATGGGTTTGC-3′). For detailed sequence alignment, please refer to “Appendix S1.”
Plasmid Construction
Chromosomal DNA of T. reesei was prepared according to Wang et al. [23]. The promoter with the secretion sequence (pCBH1-sigpro, 1.4 kb) and terminator sequence (tCBH1, 0.6 kb) of cbh1 was then cloned from the genome of T. reesei with primers P1 (5′-GTAGGATCCAAGCTTCCATTTG-3′) and P2 (5′-CCGCTCGAGAGCTCGAGCAGTAGCCAAG-3′) and T1 (5′-CGCTCTAGATGAACCCTTACTACTCTCAGT-3′) and T2 (5′-ATTAAGCTTACTAGTGTCCTCGGCACGTTGTCATC-3′), respectively. They were then constructed into the PUC18 backbone to form gene expression cassette.
For hygromycin B-resistant marker, the PtrpC-hph expression cassette (1.4 kb) was cloned from pDESTR (GenBank AB218275.1) with primers H1 (5′-CGCCACCATGTTGGGACGTTAACTGATATTGAAGG-3′) and H2 (5′-GCCTCGAGCGTTAACTGGTTCCCGGTCGGC-3′). This cassette was further ligated into the binary vector pCAMBIA1300.
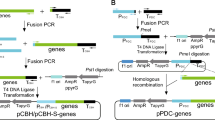
The gene expression cassette was afterwards released from PUC18 using BamHI and SpeI and cloned into the pCAMBIA1300 (containing the selection maker) to form the final expression vector pChph-pCBH1sigpro-ttl (Fig. 1).
Recombinant plasmid pChph-pCBH1sigpro-ttl used for transformation of T. reesei. Employing pCAMBIA 1300 (pC) as backbone, elements inside the expression cassette were as follows: enhanced T. reesei cbh1 promoter (pCBH1), signal sequence (sigpro), heterologous gene (ttl), and terminator of cbh1. Hygromycin B resistance marker (hph) in this plasmid was applied for selection of fungal transformants. Image was generated using DNAman 5.2.2 software
Agrobacterium-Mediated Transformation
The AMT was based upon previous publications [16, 13] with adjustments. There were three main parts during this process, namely, plasmid transformation into Agrobacterium, propagation of Agrobacterium and T. reesei, and co-cultivation. Transformation into Agrobacterium and microorganism propagation were done according to prior references [13].
For co-culture, the Agrobacterium (OD600 0.5–1.0) and spores of T. reesei (1.0E + 7 spores/mL) were mixed and transferred onto the nitrocellulose filter with 200 μmol acetosyringone for a 36–48-h cultivation on induction plate under the pH and temperature range from 4.5 to 6.3 and 20 to 30 °C, respectively. Afterwards, the nitrocellulose filter was tiled reversely on PDA at 30 °C with 150 μg/mL hygromycin B and 200 μg/mL cefotaxime. In order to increase the AMT efficiency, T. reesei spores could be treated in induction medium supplemented with 10 mg/mL snailase for 1–5 h at 24 °C in advance.
Screening of Transformants
Transformant selection was based on a two-step screening method. Antibiotic resistance was used for the first round screening. Potential transformants grown under the nitrocellulose filter were picked out by cutters (diameter 0.5 cm) and further cultivated on PDA supplemented with 200 μg/mL hygromycin B. After that, lactose-consuming ability and lipase activity were compared for further confirmation. Hygromycin-resistant candidates were additionally valued according to diameters of colonies and transparent zones after being cultivated on selective plates for 48 h.
Detection of Lipase Gene in Recombinant T. reesei
To evaluate the mitotic stability, heterologous TTL gene was detected by PCR in transformants. Candidates were subcultured on PDA without antibiotics for five generations. Subsequently, the genome of each mono-conidial was extracted for PCR analysis to verify the existence of lipase gene using primers L1 and L2. Genome of original T. reesei ZU-02 was taken as contrast.
Enzyme Production
For lipase fermentation at lab scale, operation was based upon Gu et al. [19]. T. reesei without genetic manipulation was applied as control, and experiments were replicated three times.
Analysis Method
Cellulase activity was assayed according to Jin et al. [13]. Lipase activity was determined by titration, and olive oil emulsion was used as substrate [24]. One unit lipase activity was defined as the amount of enzyme that released 1 μmol of free fatty acids per minute. All the experiments were repeated three times for integrity.
After fermentation, the UPPA-PROTEIN-ConcentrateTM kit (Sangon, Shanghai, China) was applied to increase the lipase concentration before sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) for determining its protein profile. The gel (12 % polyacrylamide) was stained by Coomassie Brilliant Blue R-250 and further destained overnight by a methanol-acetic acid–water mixture (20:10:70, v/v/v). Then, Micro Protein PAGE Recovery Kit (Sangon, Shanghai, China) was adopted to recover the recombinant lipase from the gel before the property research.
During studying of lipase characteristics, 5 mL olive oil emulsion with 4 mL buffers were preheated at 60 °C for 5 min. After that, 1 mL lipase solution (also being preheated) was added into that system for a 10-min reaction with temperature-controlled shaking. Inactivated lipase by boiling water was added to the control group. Fifteen milliliters of ethanol was then used to stop the reaction, followed by supplementing of 15 mL sterilized water before titration using 0.1 mol/L NaOH. For testing, the optimal temperature and pH and hydrolyzing reactions were proceeded within a temperature range (30–80 °C) and pH range (pH 6–12) before titration. For pH and temperature resistance assay, lipase was treated in different pH and temperatures for 1 h in advance.
For kinetic hydrolysis of olive oil, normal hydrolytic reactions were carried out expect for that the amount of produced fatty acids were measured continuously for half an hour after reaction.
Results and Discussion
Construction of the Recombinant Plasmid pChph-pCBH1sigpro-ttl
In the era of functional genomics or system biology [25], evidence is accumulating that an efficient and rapid platform for gene overexpression or downregulation at molecular level is always the cornerstone for genetic engineering in Trichoderma [14]. The promoter of cellobiohydrolase I (CBH I), which was encoded by a single cbh1 gene while occupying the majority of T. reesei secreted protein, was proved to be a strong promoter for both homologous and heterologous expression [26]. Its promotion was inhibited by glucose while induced by other substrates like lactose and avicel [27]. Regulation of this promoter mainly happened at transcription level, which involves participation of various transcriptional factors [27]. Sequence analysis found that there were three binding motifs for Hap2/3/5 [28], three binding sites for XYRI [29], and one locus for ACEII [30] in the upstream sequence (from −805 to −606) of cbh1 promoter, all of which could boost the promotion. Whereupon, this sequence was repeated four times in the newly designed promoter aiming to further increase promotion effect. Moreover, signal sequence of CBHI was introduced to stabilize mRNA of TTL and facilitate intracellular transportation [21]. Heterologous TTL was inserted between the modified cbh1 promoter and its terminator to form the expression cassette (Fig. 1), which would be further constructed into the final binary vector pChph-pCBH1sigpro-ttl.
Approaches from transcriptional level come foremost for genetic manipulation of T. reesei [14]. With promotion of the multi-duplicate motifs for positive transcriptional factors and screening easement by hygromycin-resistant marker, various proteins could be efficiently expressed in T. reesei by means of this platform.
Optimization of AMT Protocol
Efficient and convenient transformation is also an indispensable prerequisite for T. reesei engineering. During the AMT process, co-cultivation of Agrobacterium and fungi is top-drawer as sorts of parameters could affect the final efficiency [16, 20], especially temperature, pH, and concentration of Agrobacterium [19]. Based on prior studies [19, 13], a new transformation platform mediated by A. tumefaciens EHA 105 was set up. In accordance with Combier et al. [31], we found that the optimal co-cultivation temperature was 24 °C (Fig. 2), of which the transformation efficiency was 3.56 times higher than that at 30 °C, implying that high temperature would decline the T-DNA transfer rate. For the effects of pH (Fig. 2), results showed that pH 5.3 was far more preferable than other alkaline conditions. This difference with former studies in our lab [19] may be attributed to the novel species, A. tumefaciens EHA 105. Additionally, maintaining the growth balance between bacteria and fungi played a vital role [32]. The most appropriate bacteria–fungi concentration during co-cultivation in this study was OD660 0.8 versus 1.0E + 7 spores/mL. More Agrobacterium would exhaust the nutrient and space during co-cultivation, whereas additional T. reesei could make isolation of single colony troublesome.
The effects of concentration of Agrobacterium (OD600 filled squares), pH (filled circles), and temperature (filled triangles) on the efficiency of transformation during co-cultivation. After 24-h culture of Agrobacterium possessing heterologous lipase gene, it was diluted to different concentration, then mixed with T. reesei spores (1.0E + 7 spores/mL) using pipettes. Mixture was further spread on the nitrocellulose filter of induction medium (IM) supplemented with 200 μM acetosyringone for a 36–48-h dark cultivation under different pH and temperatures. Data shown are average of triplicate experiments
Further results indicated that pre-germination would remarkably augment the efficiency of AMT. Through treating the conidia of T. reesei with 10 mg/mL snailase at 30 °C for 3 h (Fig. 3), the number of transformants turned to be a magnitude higher. However, longer period of pre-germination was harmful for that extra lysing of fungal cells came into being. This germination time was the same as Jin et al. [13], while half of Curvularia lunata reported by Liu et al. [33], indicating that variance of cell wall recalcitrance may result in different treating times [19].
Using this optimized protocol, the final transformation rate was 1.05/1.0E + 3 conidia, which was 100 times higher than Abuodeh et al. [34]. However, this efficiency was only about 80 % of that mediated by A. tumefaciens AGL 1 [13], proving that AGL 1 was more effective than EHA 105 in genetic transformation of T. reesei [35]. Compared with other physical or chemical approaches [17], the optimized AMT here possesses such advantages, e. g., no preference toward target genes, randomly integration into genome with T-DNA tag [18], and highly efficient transformation in large scale mutagenesis [31], that renders itself a reliable and competent platform for T. reesei genetic transformation.
Screening of T. reesei Transformants
To efficiently screen the lipase-producing candidates, a two-step screening method was introduced. After growing 2 days on lactose-tributyrin plates, diameters of most transformants (total number 258) were 2 to 4 cm, and only a minority (about 3 %) could reach 4.5 cm (Fig. 4). Due to the correlation between growth rate and lipase-producing ability [20], final fermentative transformants exactly came from this minority. Our work demonstrated this two-step screening method that depending on antibiotic resistance and protein secretion capacity could be further applied to alleviate the laborious selection work during expression in T. reesei.
Screening of transformants on MCC plates. Transformants which had bigger colony after growing on PDA added with 150 μg/mL hygromycin B for 2 days were preliminarily selected. These candidates were then cultured on MCC plates for 36–48 h, where 20 g/L microcrystalline cellulose and 1 % tributyrin were the sole carbon source and lipase indicator, respectively. Blank bars refer to the number of defined transformants, and filled squares represent the corresponding percentage. Results are the total of three independent trials
Verification of Lipase Gene in Transformants
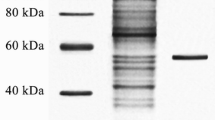
It is necessary to test the mitotic stability of recombinant transformants as some genetic traits like antibiotic resistance would lose during subculturing [13]. Candidates were cultivated on PDA without hygromycin B for five generations, and then genomes were extracted for PCR confirmation of lipase gene. Resultantly, a band about 0.8 Kb could be detected in their genomes (Fig. 5a), corresponding with the size of TTL. This meant that lipase gene had been stably integrated into the genome of recombinant transformants.
PCR verification of lipase gene (a) and SDS-PAGE analysis (b). a Primers L1 and L2 were applied to detect the TTL gene in the T. reesei genome. Lane M: the DAN molecular mass maker; Lane 1–2: using transformants genome as templates; Lane 3: control T. reesei without genetic manipulation. b Lane M: the protein molecular mass marker; Lane 1: supernatant of T. reesei fermentation without TTL integration; Lane 2: lipase from the fermentation broth of transformants
Lipase Production by T. reesei Transformants
To test the lipase-producing capacity of recombinant T. reesei, a 4-day fermentation at lab scale was performed. From Fig. 6, we could see that variation of lipase production, reducing sugar, and pH during fermentation could be divided into two stages on the whole. During the first 24 h, the reducing sugar tobogganed due to the rapid consumption of lactose by the booming mycelia (Fig. 6). After that reducing sugar stabilized, during which, induction of lipase production by avicel dominated strain growth. Following the same trend, pH decreased from 5 to around 4 took place in the first 24 h (Fig. 6), and then stabilized at 5.2 afterwards. Decrease of pH during T. reesei fermentation was previously reported in many other papers [36, 37], and this may result from the fact that the optimal pH for growth and protein production of T. reesei was 4 and 5, respectively [38]. Conclusions may be drawn that before the induction for protein expression, there would be a preliminary mycelia growth stage (lasting about 24 h), during which the reducing sugar and pH of fermentation broth would decline.
The time course of lipase production by recombinant T. reesei. Five milliliters seed broth was inoculated into 50-mL fermentation medium in 250-mL Erlenmeyer flasks with initial pH 4.8. Since then, flasks were shaken at 30 °C and 200 rpm. Lipase activity (filled squares), pH (filled triangles), and reducing sugar (filled circles) in the medium were measured every 12 h. Data shown are the means of triplicate samples
After 84-h fermentation, lipase activity increased sostenuto to its peak, where concentration of lipase in culture supernatant was 32.5 mg/L, and lipase activity was 241 IU/mL (7415.4 IU/mg) (Fig. 6). It could be further found that lipase production increased faster at strain growth stage than the later period. The lipase activity attained here was nearly four times the level that without RNAi mediated gene silencing in Qin et al. [39] and was about 60 times as high as the basic lipase production by T. reesei itself when 5 % olive oil was used as inducer in submerge fermentation [40]. SDS-PAGE analysis moreover proved that T. reesei transformants secreted the recombinant lipase of about 39 kDa, a molecular mass expected for TTL (Fig. 5b). Determined by the random integration in AMT, higher lipase production in this study may relate to the strong promoter, better location for gene insertion, and proper gene copies [41].
Further verification of lipase production by recombinant T. reesei at higher scale should be carried out, for that growth conditions turn to be much more suitable in fermentation tanks. Fed-batch technique is frequently applied during a designed fermentation cycle [42]. Similarly, new purification methodologies like the routine frame filtration-ultrafiltration-spray drying other than centrifugation in lab should be researched for industrial applications.
Kinetic Study of Lipase
Figure 7 shows the released fatty acids as a function of reaction time. It could be seen that at a fixed concentration of substrate (33.3 %, v/v) and enzyme (32.5 μg protein/mL), the lipase-catalyzed hydrolysis followed a typical hyperbolic form. After a 5-min incubation, formation of fatty acid was linearly proportional to reaction time up to 10 min. Catalytic stability plays an important part for lipase application, for that accumulation of released fatty acids at lipid–water interfaces would inactivate lipase itself. Therefore, materials accepting the free fatty acids such as bile salt and beta-cyclodextrin are always added to the reaction system [9]. The linear kinetic of T. thermophilus lipase here demonstrated that this lipase could resist the denaturation caused by inhibition of fatty acids and keep strong catalytic stability.
Effects of pH and Temperature on Lipase Activity
Performance in different pH and temperatures are one of the main properties determining lipase application. From Fig. 8a, we can see that this lipase preferred alkali conditions with the optimum pH 9.5. It also showed such a strong alkali tolerance that more than 70 % activity could be maintained after treatment in pH 11 for 1 h. This optimal pH was even higher than some bacterial lipases [43], which were usually thought to possess stronger adaptability toward varying pH than fungal lipases [44].
The effects of pH (a) and temperature (b) on lipase. a The optimum (filled squares) pH was measured at 60 °C in buffer with varying pH. For pH stability (inverted triangles), lipase was first treated in different pH buffers for one hour before titration. b The optimal temperature (filled squares) was determined at different temperatures in pH 9.5. For thermostability (inverted triangles), lipase should be deposited at various temperatures for 1 h in advance. Data corresponding to optimal condition and stability were measured three times
It could also be found that the optimal reaction temperature of this lipase was 60 °C (Fig. 8b), and it could retain more than half the activity within a temperature range from 40 to 80 °C for an hour. In the matter of thermostability, we could see TTL surpassed many other fugal lipases [45]. For instance, the lipase from Penicillium wortmannin was stable at 40 °C and only 20 % activity could be remained after preincubation at 60 °C for 1 h [46].
Though most of the industrial lipases are from fungi, it is rare to find a fungal lipase that is stable at both alkaline conditions and high temperatures [47]. Prominent alkali and thermotolerance of TTL may result from the stability of its protein sequence, which showed high similarity with the commercial basophilic Thermomyces lanuginosa lipase [48]. This advantage makes it promising for industrial applications like paper making and detergent addition [2]. Furthermore, the synergetic degradation of cellulase and lipase (secreted at specific ratio by recombinant T. reesei) makes this research meaningful in certain environmental fields such as oil-rich mill waste treatment [49], where the corresponding cellulose and triglycerides coexist.
Conclusions
In conclusion, a versatile platform was established for homologous or heterologous gene expression in T. reesei. A de novo designed cbh1 promoter with repeated locus for positive transcriptional factors was applied. Assisted by the improved Agrobacterium EHA 105-mediated transformation and an effective two-step screening method, T. thermophilus lipase gene was successfully expressed in T. reesei. After fermentation in lab scale and property study, this lipase was proved to possess high alkali and thermotolerance, which determined its future applications in industry.
References
Brzozowski, A. M., Derewenda, U., Derewenda, Z. S., Dodson, G. G., Lawson, D. M., Turkenburg, J. P., Bjorkling, F., Huge-Jensen, B., Patkar, S. A., & Thim, L. (1991). A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature, 351, 491–494.
Stergiou, P.-Y., Foukis, A., Filippou, M., Koukouritaki, M., Parapouli, M., Theodorou, L. G., Hatziloukas, E., Afendra, A., Pandey, A., & Papamichael, E. M. (2013). Advances in lipase-catalyzed esterification reactions. Biotechnology Advances, 31, 1846–1859.
Dai, D., & Xia, L. (2006). Effect of lipase immobilization on resolution of (R, S)-2-octanol in nonaqueous media using modified ultrastable-Y molecular sieve as support. Applied Biochemistry and Biotechnology, 134, 39–50.
Gusakov, A. V. (2011). Alternatives to Trichoderma reesei in biofuel production. Trends in Biotechnology, 29, 419–425.
Amore, A., & Faraco, V. (2012). Potential of fungi as category I consolidated bioprocessing organisms for cellulosic ethanol production. Renewable and Sustainable Energy Reviews, 16, 3286–3301.
Jorgensen, M., Skovlund, D., Johannesen, P., & Mortensen, U. (2014). A novel platform for heterologous gene expression in Trichoderma reesei (Teleomorph Hypocrea jecorina). Microbial Cell Factories, 13, 33.
Nevalainen, K. M. H., Te'o, V. S. J., & Bergquist, P. L. (2005). Heterologous protein expression in filamentous fungi. Trends in Biotechnology, 23, 468–474.
Romdhane, I. B.-B., Romdhane, Z. B., Gargouri, A., & Belghith, H. (2011). Esterification activity and stability of Talaromyces thermophilus lipase immobilized onto chitosan. Journal of Molecular Catalysis B: Enzymatic, 68, 230–239.
Romdhane, I. B.-B., Fendri, A., Gargouri, Y., Gargouri, A., & Belghith, H. (2010). A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochemical Engineering Journal, 53, 112–120.
Valero, F. (2012). Lipases and phospholipases, vol. 861. In G. Sandoval (Ed.), Methods in molecular biology (pp. 161–178) Humana Press.
Kontkanen, H., Reinikainen, T., & Saloheimo, M. (2006). Cloning and expression of a Melanocarpus albomyces steryl esterase gene in Pichia pastoris and Trichoderma reesei. Biotechnology and Bioengineering, 94, 407–415.
Jin, X., Meng, N., & Xia, L.-M. (2011). Expression of an endo-β-1, 4-glucanase gene from Orpinomyces PC-2 in Pichia pastoris. International Journal of Molecular Sciences, 12, 3366–3380.
Jin, X., & Xia, L. (2011). Heterologous expression of an endo-β-1,4-glucanase gene from the anaerobic fungus Orpinomyces PC-2 in Trichoderma reesei. World Journal of Microbiology and Biotechnology, 27, 2913–2920.
Meyer, V. (2008). Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnology Advances, 26, 177–185.
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., Chapman, J., Chertkov, O., Coutinho, P. M., Cullen, D., Danchin, E. G. J., Grigoriev, I. V., Harris, P., Jackson, M., Kubicek, C. P., Han, C. S., Ho, I., Larrondo, L. F., de Leon, A. L., Magnuson, J. K., Merino, S., Misra, M., Nelson, B., Putnam, N., Robbertse, B., Salamov, A. A., Schmoll, M., Terry, A., Thayer, N., Westerholm-Parvinen, A., Schoch, C. L., Yao, J., Barabote, R., Nelson, M. A., Detter, C., Bruce, D., Kuske, C. R., Xie, G., Richardson, P., Rokhsar, D. S., Lucas, S. M., Rubin, E. M., Dunn-Coleman, N., Ward, M., & Brettin, T. S. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nature Biotechnology, 26, 553–560.
Michielse, C., Hooykaas, P. J., van den Hondel, C. M. J. J., & Ram, A. J. (2005). Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Current Genetics, 48, 1–17.
Su, X., Schmitz, G., Zhang, M., Mackie, R. I., & Cann, I. K. O. (2012). Heterologous gene expression in filamentous fungi. Advances in Applied Microbiology, 81, 1–61.
Zhong, Y. H., Wang, X. L., Wang, T. H., & Jiang, Q. (2007). Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Applied Microbiology and Biotechnology, 73, 1348–1354.
Gu, B., & Xia, L. (2013). High expression of a neutral endo-β-glucanase gene from Humicola insolens in Trichoderma reesei. Journal of Industrial Microbiology & Biotechnology, 40, 773–779.
Fang, H., & Xia, L. (2013). High activity cellulase production by recombinant Trichoderma reesei ZU-02 with the enhanced cellobiohydrolase production. Bioresource Technology, 144, 693–697.
Te’o, V. S. J., Cziferszky, A. E., Bergquist, P. L., & Nevalainen, K. M. H. (2000). Codon optimization of xylanase gene xynB from the thermophilic bacterium Dictyoglomus thermophilum for expression in the filamentous fungus Trichoderma reesei. FEMS Microbiology Letters, 190, 13–19.
Villalobos, A., Ness, J., Gustafsson, C., Minshull, J., & Govindarajan, S. (2006). Gene designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics, 7, 285.
Wang, B., & Xia, L. (2011). High efficient expression of cellobiase gene from Aspergillus niger in the cells of Trichoderma reesei. Bioresource Technology, 102, 4568–4572.
Meyers, S. A., Cuppett, S. L., & Hutkins, R. W. (1996). Lipase production by lactic acid bacteria and activity on butter oil. Food Microbiology, 13, 383–389.
Weld, R. J., Plummer, K. M., Carpenter, M. A., & Ridgway, H. J. (2006). Approaches to functional genomics in filamentous fungi. Cell Research, 16, 31–44.
Peterson, R., & Nevalainen, H. (2012). Trichoderma reesei RUT-C30—thirty years of strain improvement. Microbiology, 158, 58–68.
Mach, R., & Zeilinger, S. (2003). Regulation of gene expression in industrial fungi: Trichoderma. Applied Microbiology and Biotechnology, 60, 515–522.
Ries, L., Belshaw, N. J., Ilmén, M., Penttilä, M. E., Alapuranen, M., & Archer, D. B. (2014). The role of CRE1 in nucleosome positioning within the cbh1 promoter and coding regions of Trichoderma reesei. Applied Microbiology and Biotechnology, 98, 749–762.
Stricker, A. R., Grosstessner-Hain, K., Würleitner, E., & Mach, R. L. (2006). Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryotic Cell, 5, 2128–2137.
Aro, N., Saloheimo, A., Ilmén, M., & Penttilä, M. (2001). ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. Journal of Biological Chemistry, 276, 24309–24314.
Combier, J.-P., Melayah, D., Raffier, C., Gay, G., & Marmeisse, R. (2003). Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Microbiology Letters, 220, 141–148.
Gardiner, D., & Howlett, B. (2004). Negative selection using thymidine kinase increases the efficiency of recovery of transformants with targeted genes in the filamentous fungus Leptosphaeria maculans. Current Genetics, 45, 249–255.
Liu, T., Liu, L., Jiang, X., Hou, J., Fu, K., Zhou, F., & Chen, J. (2010). Agrobacterium-mediated transformation as a useful tool for the molecular genetic study of the phytopathogen Curvularia lunata. European Journal of Plant Pathology, 126, 363–371.
Abuodeh, R. O., Orbach, M. J., Mandel, M. A., Das, A., & Galgiani, J. N. (2000). Genetic transformation of Coccidioides immitis facilitated by Agvobactevium tumefaciens. Journal of Infectious Diseases, 181, 2106–2110.
Zhang, Y., Li, G., He, D., Yu, B., Yokoyama, K., & Wang, L. (2011). Efficient insertional mutagenesis system for the dimorphic pathogenic fungus Sporothrix schenckii using Agrobacterium tumefaciens. Journal of Microbiological Methods, 84, 418–422.
Giese, H., Kruithof, P., Meier, K., Sieben, M., Antonov, E., Hommes, R. W. J., & Büchs, J. (2014). Improvement and scale-down of a Trichoderma reesei shake flask protocol to microtiter plates enables high-throughput screening. Journal of Bioscience and Bioengineering, 118, 702–709.
Li, C., Yang, Z., He Can Zhang, R., Zhang, D., Chen, S., & Ma, L. (2013). Effect of pH on cellulase production and morphology of Trichoderma reesei and the application in cellulosic material hydrolysis. Journal of Biotechnology, 168, 470–477.
Prasetyo, J., Sumita, S., Okuda, N., & Park, E. (2010). Response of cellulase activity in pH-controlled cultures of the filamentous fungus Acremonium cellulolyticus. Applied Biochemistry and Biotechnology, 162, 52–61.
Qin, L.-N., Cai, F.-R., Dong, X.-R., Huang, Z.-B., Tao, Y., Huang, J.-Z., & Dong, Z.-Y. (2012). Improved production of heterologous lipase in Trichoderma reesei by RNAi mediated gene silencing of an endogenic highly expressed gene. Bioresource Technology, 109, 116–122.
Rajesh, E. M., Arthe, R., Rajendran, R., Balakumar, C., Pradeepa, N., & Anitha, S. (2010). Investigation of lipase production by Trichoderma reesei and optimization of production parameters. Electronic Journal of Environmental, Agricultural and Food Chemistry, 9, 1177–1189.
Gouka, R. J., Punt, P. J., & van den Hondel, C. A. M. J. J. (1997). Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Applied Microbiology and Biotechnology, 47, 1–11.
Ahamed, A., & Vermette, P. (2008). Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochemical Engineering Journal, 40, 399–407.
Dutta, S., & Ray, L. (2009). Production and characterization of an alkaline thermostable crude lipase from an isolated strain of Bacillus cereus C7. Applied Biochemistry and Biotechnology, 159, 142–154.
Gupta, R., Gupta, N., & Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology, 64, 763–781.
Papaparaskevas, D., Christakopoulos, P., Kekos, D., & Macris, B. (1992). Optimizing production of extracellular lipase from Rhodotorula glutinis. Biotechnology Letters, 14, 397–402.
Costa, M. A. F., & Peralta, R. M. (1999). Production of lipase by soil fungi and partial characterization of lipase from a selected strain (Penicillium wortmannin). Journal of Basic Microbiology, 39, 11–15.
Singh, A., & Mukhopadhyay, M. (2012). Overview of fungal lipase: a review. Applied Biochemistry and Biotechnology, 166, 486–520.
Romdhane, I. B.-B., Frikha, F., Maalej-Achouri, I., Gargouri, A., & Belghith, H. (2012). Gene cloning and molecular characterization of the Talaromyces thermophilus lipase catalyzed efficient hydrolysis and synthesis of esters. Gene, 494, 112–118.
Fickers, P., Marty, A., & Nicaud, J. M. (2011). The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnology Advances, 29, 632–644.
Acknowledgments
This work was supported by the National High-tech R&D Program (2007AA05Z401) and the Program for Zhejiang Leading Team of S&T Innovation (2011R50002) of China.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
Sequence alignment of the original and optimized lipase sequence (GIF 498 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Li, X. & Xia, L. Heterologous Expression of an Alkali and Thermotolerant Lipase from Talaromyces thermophilus in Trichoderma reesei . Appl Biochem Biotechnol 176, 1722–1735 (2015). https://doi.org/10.1007/s12010-015-1673-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1673-4