Abstract
Purpose of Review
The replenishment of lost or damaged myocardium has the potential to reverse heart failure, making heart regeneration a goal for cardiovascular medicine. Unlike adult mammals, injury to the zebrafish or neonatal mouse heart induces a robust regenerative program with minimal scarring. Recent insights into the cellular and molecular mechanisms of heart regeneration suggest that the machinery for regeneration is conserved from zebrafish to mammals. Here, we will review conserved mechanisms of heart regeneration and their translational implications.
Recent Findings
Based on studies in zebrafish and neonatal mice, cardiomyocyte proliferation has emerged as a primary strategy for effecting regeneration in the adult mammalian heart. Recent work has revealed pathways for stimulating cardiomyocyte cell cycle reentry; potential developmental barriers for cardiomyocyte proliferation; and the critical role of additional cell types to support heart regeneration.
Summary
Studies in zebrafish and neonatal mice have established a template for heart regeneration. Continued comparative work has the potential to inform the translation of regenerative biology into therapeutics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Current therapeutics for heart failure largely seek to ameliorate symptoms or limit the progression of disease. For instance, loop diuretics are used for symptomatic decongestion, and β-adrenergic receptor blockers attenuate pathologic remodeling due to an activated sympathetic nervous system. Even those patients who respond to guideline-directed medical therapies are considered to be in remission and are committed to complex medical regimens indefinitely for fear of relapse [1, 2]. For the select patients with end-stage heart failure that are eligible for advanced therapies, treatment with mechanical circulatory support or transplantation carries considerable morbidity. By contrast, strategies that could replace lost or dysfunctional myocardium through tissue regeneration offer the potential to fundamentally reverse heart failure.
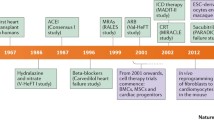
A remarkable ability of adult zebrafish is their high capacity for tissue regeneration. Scarless regeneration after resection of the ventricular apex in zebrafish was first described by Poss et al. in 2002 [3]. Zebrafish heart regeneration has since proved to be robust through a variety of injuries, including apical resection, cryoinjury, and genetic ablation of cardiomyocytes (CMs) [3,4,5,6]. Although the zebrafish heart is two-chambered, studies in zebrafish are highly applicable to the mammalian heart, having greatly influenced our understanding of heart development [7, 8]. Indeed, one decade after the description of zebrafish heart regeneration, a similar regenerative response to ventricular resection was described in neonatal mice [9]. The murine capacity for regeneration is stage-specific, limited to the early neonatal period and absent in adulthood. Heart regeneration has now been described in neonatal pigs and even suggested for the neonatal human heart, pointing to a conserved regenerative program across fish and mammals [10,11,12] (Fig. 1a). Redeploying this regenerative program in the adult mammalian heart is considered a primary approach for achieving therapeutic heart regeneration. Here, we will review mechanisms for heart regeneration that are conserved from zebrafish to mammals and discuss potential implications for translation (Fig. 1b).
A roadmap to heart regeneration. a Capacity for heart regeneration by species. Zebrafish and neonatal mice have a high capacity for regeneration while adult mammals have little-to-no ability for regeneration after injury. b Conserved mechanisms for cardiomyocyte proliferation in zebrafish and neonatal mice. Following injury, mature CMs reenter the cell cycle and undergo sarcomere remodeling before completing mitosis to generate new cardiomyocytes. Factors that promote CM proliferation at each stage are shown in green and inhibitory factors are shown in red
Cellular Mechanisms of Heart Regeneration
After the first descriptions of zebrafish heart regeneration, questions about the cellular sources for regenerating myocardium were in the forefront. Over the past two decades, these cellular mechanisms have been largely clarified in zebrafish and neonatal mice, establishing important themes for heart regeneration.
Cardiomyocytes Beget New Cardiomyocytes
Proliferating CMs can be identified in the border zone of the injured zebrafish heart within a few days of injury [3]. Genetic fate-mapping experiments in zebrafish, during which cells and their progeny are indelibly labeled with a genetic marker, have shown new CMs to be derived from preexisting CMs, without evidence for a major stem cell contribution [13,14,15]. This finding was surprising at the time, because multiple groups had suggested a progenitor cell, resident within the myocardium or within the bone marrow, to effect cardiomyogenesis in the mammalian heart [16,17,18]. In 2011, Porrello et al. demonstrated that neonatal mice, in the first few days of life, can regenerate their hearts after resection of the left ventricular apex [9]. Like zebrafish, CM proliferation was noted as a part of the injury response. Fate-mapping experiments in neonatal mice also suggested that preexisting CMs were the source of newly formed myocardium rather than a progenitor cell population. Shortly after the description of heart regeneration in the neonatal mouse, fate-mapping technologies for the adult mouse heart evolved with the conclusion that low numbers of new CMs in the adult heart are primarily derived from preexisting CMs [19]. As a result, methods to stimulate proliferation of CMs have emerged as a dominant strategy to effect therapeutic heart regeneration, making a better understanding of the mechanisms that regulate CM cell cycle reentry of significant importance [20].
CM Cell Cycle Reentry Is Modulated by Physiological, Genetic, and Structural Factors
Heart regeneration in zebrafish is remarkably robust. Loss of up to 60% of the CMs in the adult zebrafish heart can be overcome by innate regenerative responses [4]. By contrast, the adult mammalian heart has little-to-no ability for CM proliferation and endogenous regeneration. Comparative studies of the zebrafish and the mammalian heart have provided important insights into factors that might affect the proliferative capacity of adult mammalian CMs.
Tissue hypoxia is an important modifier of CM cell cycle reentry. After injury to the zebrafish heart, CMs adjacent to the wound become relatively hypoxic [21]. Experimental hypoxia can boost CM proliferation during zebrafish heart regeneration by almost 50%. Because the zebrafish and fetal mouse heart have ~ 70% less arterial oxygen content than the adult mammalian heart, Puente and colleagues reasoned that oxygen tension could mediate cell cycle arrest of cardiomyocytes [22]. Consistent with their hypothesis, experimental hypoxia more than doubles CM proliferation, and hyperoxia suppresses CM proliferation by nearly 7-fold in neonatal mice. Mechanistically, oxygen tension regulates the cell cycle in CMs by increasing reactive oxygen species and inducing a DNA damage response. Impressively, hypoxia induced 1 week after experimental myocardial infarction in adult mice results in increased CM cycling and functional recovery [23•]. While prolonged hypoxia may not be feasible for patients with heart failure, manipulation of the pathways that regulate hypoxia-induced CM proliferation could be an attractive target for therapeutics.
A second potential barrier to mammalian heart regeneration is polyploidization. Unlike proliferating zebrafish CMs which complete cytokinesis, proliferating CMs in the adult mammalian heart preferentially undergo endomitosis (DNA synthesis without cytokinesis). In fact, CMs in the zebrafish heart are mononuclear and diploid while many adult mammalian CMs are multinucleated or polyploid [24, 25]. The tendency for polyploidization makes regenerative responses after injury less efficient in the adult mammalian heart, because ~ 97% of the rare cycling CMs in the adult mammalian heart fail to undergo cytokinesis [19]. Several functional experiments point to polyploidization as a significant impediment to heart regeneration. Using a combination of genetic fate-mapping and genetic methods to induce polyploidization, González-Rosa et al. found diploid CMs to preferentially regenerate the zebrafish heart after ventricular resection [26•]. Importantly, zebrafish hearts with > 45% polypoid CMs formed large scars after ventricular resection, establishing a threshold for polyploidization beyond which regeneration is impaired. Similarly, genetic deletion of the nuclear filament protein Lmnb2 from CMs of neonatal mice induces polyploidization and subsequently impairs regeneration [27••]. Conversely, viral overexpression of Lmnb2 increases CM proliferation and enhances regeneration after cryoinjury. Thus, approaches that can overcome the polyploidy barrier are likely to result in more efficient therapeutics.
Sarcomere density may be among the array of factors that limit the mitotic potential of adult mammalian CMs. A cardinal feature of proliferating CMs in zebrafish and neonatal mice is loss of sarcomeric structures [14, 15]. Mechanisms for sarcomere dissolution in the zebrafish heart include increased autophagy and epigenetic repression of sarcomeric genes. Interference with either of these pathways compromises the regenerative ability of zebrafish [28, 29••, 30•]. While analogous studies have yet to be done in the adult mammalian heart, recent work has suggested that mechanical unloading of the human heart with a left ventricular assist device (LVAD) is associated with reversal of hypertrophy and increased markers of CM DNA synthesis and cytokinesis [31].
Developmental Programs Are Reactivated During Heart Regeneration
A distinct feature of zebrafish heart regeneration is the re-expression of factors typically found in the fetal heart. Genetic fate-mapping of CMs that upregulate the developmental transcription factor gata4 demonstrate that these dedifferentiated CMs are primarily responsible for the newly formed muscle after apical resection [14]. More recently, genetic fate-mapping of cells marked by the neural crest marker sox10, suggest that less developmentally mature CMs in general more efficiently contribute to regeneration [32, 33]. Dedifferentiation of adult CMs is likely to be required for zebrafish heart regeneration. Inhibition of BMP signaling or myocardial NF-κβ, which has been associated with reactivation of developmental factors, limits CM proliferation and regeneration [34, 35]. Similarly, inhibition of gata4 signaling by expression of a dominant negative version of gata4 attenuates CM proliferation and regeneration in zebrafish [36].
The concept of dedifferentiation has informed approaches to rescue CM proliferation defects in adult mammals. Several groups have used knowledge of developmental pathways regulating CM hyperplasia to develop potential methods for adult heart regeneration. For instance, Tbx20 and YAP are transcription factors with key roles in CM growth during development that have been redeployed in the adult mammalian heart to promote CM proliferation [37,38,39,40,41,42]. Repression of the Hippo pathway, and activation of YAP in particular, is a promising method for effecting heart regeneration as multiple embryonic growth pathways, including Erbb2 signaling and IGF signaling, appear to have YAP activation in common [43, 44].
Non-Cardiomyocyte Populations Are Critical to Heart Regeneration
Despite a focus on CMs, zebrafish heart regeneration is an intricately orchestrated program involving multiple tissue types. Interference with cardiac endothelial cells, epicardial cells, inflammatory cells, or nerves can suppress CM proliferation and limit regeneration [45,46,47,48]. How non-cardiomyocyte populations influence and shape regeneration of the mammalian heart is the subject of ongoing research.
Cardiac endothelial cells are among the first cells to infiltrate the wound, forming nascent vessels within hours of injury [46]. Interestingly, inhibition of several signaling pathways that mediate revascularization, such as vegfaa, retinoic acid, and Fgf signaling also result in CM proliferation defects [46, 49, 50]. Recent work using cell-type specific tools have directly linked cardiac endothelial responses to CM proliferation. For example, inhibition of endocardial Notch signaling directly leads to reduced CM proliferation after injury by modulating the Wnt rheostat around regenerating myocardium [51•]. In addition to supporting CM proliferation, revascularization of the injury site may shape regeneration by providing a scaffold for cardiomyogenesis [52••]. Thus, revascularization strategies may be an important adjunct to therapeutics that stimulate CM proliferation.
The epicardium comprises the outer mesothelial covering of the heart. Injury to the zebrafish heart activates the epicardium, as evidenced by rapid molecular and morphologic changes [49, 53]. Activated epicardial cells migrate to cover and infiltrate the injured area, ultimately giving rise to mural cells, fibroblasts, and additional epicardial cells [54, 55]. Like development, the epicardium and epicardial-derived cells serve important paracrine functions during zebrafish heart regeneration [56]. Ablation of the epicardium in the context of injury blunts revascularization and CM proliferation, demonstrating its importance to a regenerative program in zebrafish [45]. Mechanistically, epicardial-derived cells are a source for growth factors, such as nrg1 and neuropilins, and extracellular matrix [57,58,59,60]. In the postnatal mammalian heart, epicardial responses to injury appear to be less dynamic than in zebrafish, raising the possibility that augmenting epicardial responses to injury might enhance the regenerative potential of the mammalian heart [50, 61]. Along these lines, conditioned media from epicardial cultures has been used to screen for mitogens of mammalian CMs, leading to the identification of FSTL1 as a factor able to induce CM proliferation in adult mice and pigs [62].
After injury to the zebrafish heart, the inflammatory response is initially dominated by neutrophils but gives way to macrophages and leukocytes 1 week after cryoinjury [63••]. This transition to a macrophage response is critical to regeneration. For example, medaka, a non-regenerative fish, have delayed recruitment of macrophages to the injury site compared to zebrafish. Accelerating macrophage recruitment by chemical stimulation of TLR signaling can partially rescue regeneration defects in medaka [63••]. Analogously, chemical inhibition of the macrophage response in neonatal mice results in regeneration defects with reduced neovascularization [48]. The quality of the macrophage response is likely to modulate regeneration, as distinct sets of pro-regenerative and pro-fibrotic populations have been identified in both zebrafish and neonatal mice [64,65,66]. Along with the macrophage response, the T cell response also regulates regenerative capacity, with ablation of Treg-like cells compromising heart regeneration [67]. Of great interest, the Treg-like response appears to be dynamic and capable of enhancing regenerative programs through upregulation of tissue-specific mitogens. Together, these data raise the possibility of manipulating the inflammatory response to injury in order to influence the capacity for tissue repair.
Conserved Signaling Pathways for Reactivating Endogenous Cardiac Regeneration
As the cellular contributions to heart regeneration are understood, key pathways that underlie regeneration are emerging. Because manipulation of these pathways can lead to methods for therapeutic heart regeneration, knowledge of how these pathways affect cardiac growth and homeostasis across developmental stages in the mammalian heart is essential. Here, we will discuss several pathways that are conserved from zebrafish to mammals and their potential for translation.
Neuregulin 1/Erbb2
Nrg1 was originally identified from conditioned media as a growth factor capable of inducing phosphorylation of the proto-oncogene Erbb2 [68]. Nrg1 and Erbb2 knockout mice were found to have hypoplastic hearts with impaired trabeculation, suggesting that Nrg1/Erbb2 signaling regulates CM proliferation during development [69, 70]. Subsequent studies have demonstrated that recombinant NRG1 can stimulate proliferation of adult rat CMs in vitro and enhance functional recovery of mice after experimental infarction [71]. However, mice treated with NRG1 after infarction had only ~ 0.2% of CMs with evidence of cell cycle reentry, suggesting that the beneficial effects of NRG1 on cardiac function were not entirely due to regeneration. To better define the effect of nrg1 on CM hyperplasia, Gemberling et al. generated transgenic zebrafish to conditionally overexpress nrg1 from CMs [57]. They found that nrg1 overexpression resulted in an ectopic regeneration program with massive CM proliferation. To better understand why NRG1 effects on CM proliferation are not as potent in adult mice compared to zebrafish, several groups have examined the kinetics of Erbb2 expression, finding Erbb2 to be developmentally downregulated in adult mice [43, 72]. Accordingly, NRG1 is able to more potently stimulate regeneration in neonatal mice and has been proposed as a potential therapeutic for pediatric patients [73]. Indeed, recombinant NRG1 is able to stimulate proliferation of pediatric human CMs [73]. However, the absence of Erbb2 in the adult heart limits the use of NRG1 as a primary strategy for adult heart regeneration and may limit other strategies that depend on Erbb2 for their mitogenic effects, such as the use of vitamin D [74]. Instead, re-expression of an activated Erbb2 could be a more tenable therapeutic option [43, 75, 76].
Insulin-Like Growth Factors
Insulin-like growth factors (IGFs) are structurally related to insulin and influence organ growth. During zebrafish heart regeneration, IGFs are dynamically induced [77]. igf2b transcripts are upregulated by 2–3-fold in the wound within 3 days of injury. Functional inhibition of Igf signaling with a pharmacologic inhibitor or by expression of a dominant negative igf1r decreases CM proliferation by nearly 50%, highlighting the important contributions of Igf signaling to zebrafish heart regeneration [77].
Recent work has demonstrated an analogous role for IGF signaling during neonatal heart regeneration. Genetic deletion of Igf2 from the neonatal mouse heart decreases CM proliferation by nearly 60% and results in larger scars after injury [78]. Unlike the developing heart in which Igf2 is an epicardial-derived CM mitogen, cardiac endothelial cells are the probable source for Igf2 during murine heart regeneration. Interestingly, in mice, IGF2 effects are thought to be mediated through its interaction with the INSR rather than IGF1R, which differs from zebrafish [77, 78]. Recent work has suggested that stabilizing Igf2 transcripts by overexpressing Igf2bp3 can augment regenerative responses in neonatal mice outside of the regenerative window [79]. Therefore, enhancing IGF2 signaling is a potential mechanism to stimulate heart regeneration. Indeed, exogenous IGF2 enhances cardiac function and reduces scarring following experimental infarction in adult mice [80]. However, additional work is needed to determine whether these effects are attributable to heart regeneration.
VEGF
Therapeutic inhibition of VEGF signaling has been leveraged for the treatment of various malignancies. However, efforts to use VEGF and other angiogenic factors for the treatment of ischemic disease have not been as successful, probably related to ineffective delivery mechanisms and narrow therapeutic indices [81]. Indeed, subtle changes to Vegfa levels in mice lead to developmental defects, including cardiomegaly and aberrant coronary vascularization [82]. In the regenerating zebrafish heart, vegfaa is rapidly upregulated by cardiac endothelial cells at the site of injury and leads to rapid revascularization of the wound [46, 83•]. Transgenic overexpression of a dominant negative vegfaa inhibits revascularization, CM proliferation, and regeneration, highlighting the importance of sentinel revascularization to the regenerative program [46]. We recently demonstrated that overexpression of vegfaa from CMs results in both hypervascularization and ectopic cardiomyogenesis with hallmarks of a regenerative program [83•]. To determine how vegfaa might influence regeneration, we resected the ventricular apex of vegfaa-overexpressing hearts. These hearts misexpressed vegfaa throughout the heart compared to the local expression of vegfaa at the site of injury under endogenous conditions. Despite increased global CM proliferation and tissue growth adjacent to the wound, we observed diminished tissue regeneration at the resection site with global vegfaa overexpression. Our work indicates a regulatory role of vegfaa to instruct revascularization and tissue morphogenesis within the injured zebrafish heart.
Prior work has shown that Vegfa can improve cardiac function after experimental infarction in mice and pigs [81, 84]. However, formal linkage to an innate regenerative program is lacking. Nevertheless, Vegfa gain of function has a renewed interest for the treatment of ischemic heart disease, and new, more efficient delivery methods are being developed [85, 86]. Based on our work in zebrafish, the timing and domain of Vegfa treatment are likely to be critical determinants of efficacy.
CXCL12/CXCR4
In addition to Vegfa, alternate approaches for manipulating revascularization responses have emerged through studies of heart development and regeneration. CXCL12 is a chemokine that interacts with the G protein–coupled receptor CXCR4 to regulate the formation of coronary arteries in zebrafish and mice [87, 88]. During regeneration, cxcr4a+ arterial coronary endothelial cells follow cxcl12 expressing mural cells to revascularize the injured area [83•]. Functionally, cxcr4a mutant fish fail to regenerate their hearts after injury with defects in revascularization [83•]. In the neonatal mouse heart, CXCL12/CXCR4 signaling mediates collateral artery formation from coronary arteries, a process that is required for intact regeneration [89•]. Importantly, this collateral artery formation does not occur in the adult mammalian heart and exogenous CXCL12 can rescue collateralization after experimental infarction in adult mice [89, 90]. Thus, CXCL12 supplementation may be a therapeutic revascularization strategy, potentially with regenerative implications.
Extracellular Matrix
Extracellular matrix (ECM) has long been known to facilitate microenvironments by sequestering growth factors in niches. During zebrafish heart regeneration, the ECM is dynamic. For example, treatment of mice with decellularized extracts from regenerating zebrafish hearts, but not uninjured hearts, has been reported to increase cellular proliferation after infarction [91]. Analogously, treatment of CMs with decellularized extracts from early neonatal mice is able to stimulate CM proliferation [92•]. Remarkably, much of the effect from neonatal mouse extracts can be recapitulated with the extracellular proteoglycan Agrin. Agrin is critical to regeneration in vivo as genetic deletion of Agrn results in reduced heart function, increased fibrosis, and decreased CM proliferation after resection of the neonatal mouse heart [92•]. After myocardial infarction, recombinant Agrin is able to stimulate CM cell cycle reentry and reduce scarring in both juvenile and adult mice, suggesting that Agrin can enhance regeneration independently of developmental stage [92•]. Mechanistically, Agrin exerts its effects on CM proliferation through YAP signaling [92•]. Of translational interest, Agrin can induce the proliferation of human iPSC-CMs and improve cardiac function of the infarcted porcine heart [92•, 93•]. Overall, these results indicate the therapeutic potential of Agrin in human cardiac disease and the need for further research on the role of other ECM components in cardiac regeneration.
Tissue Regeneration Enhancer Elements
Among the most potent preclinical stage methods for inducing heart regeneration are suppression of Hippo signaling with activation of YAP, ectopic expression of an activated version of the Nrg1 signaling intermediary Erbb2, and expression of transcription factor combinations for coaxing adult CMs to reenter the cell cycle [41, 43, 94,95,96]. Such approaches are not easily “druggable” but could be deployed via gene therapy. However, gene therapy for the heart requires careful spatiotemporal control to minimize potential off-target effects caused by stimulating growth pathways in distal tissues. Recent work in zebrafish has inspired methods for tight spatiotemporal control of factor expression through the identification of Tissue Regeneration Enhancer Elements or TREEs [97, 98]. TREEs are discrete DNA regulatory sequences originally identified in the zebrafish heart by epigenetic profiling and histone replacement profiling of regenerating tissue. TREEs can be used to spatiotemporally target expression to injured tissues. In proof of concept studies, a TREE for the injury-induced gene, lepb, was able to drive expression of nrg1 after injury in zebrafish, leading to increases in CM proliferation. Importantly, TREEs in zebrafish have activity in the neonatal mouse heart, but analogous work in the adult mouse heart has yet to be reported. Injury response elements for other cardiac tissues have also been reported and thus a catalog of elements may be one approach for spatiotemporal expression of multiple factors across multiple cell types to recapitulate a heart regeneration program [99].
Conclusions
Once considered to be a post-mitotic organ, the adult mammalian heart is now known to harbor a limited potential for self-renewal. Such low-grade responses are insufficient to result in meaningful recovery after injury. However, mechanisms for zebrafish and neonatal murine heart regeneration have provided key insights for enhancing the regenerative capacity of the adult mammalian heart including (1) a renewed focus on stimulating CM proliferation; (2) the identification of barriers that limit the efficiency of CM proliferation; and (3) potential targets for achieving therapeutic heart regeneration.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Wilcox JE, Fang JC, Margulies KB, Mann DL. Heart failure with recovered left ventricular ejection fraction: JACC Scientific Expert Panel. J Am Coll Cardiol. 2020;76(6):719–34.
Mann DL, Barger PM, Burkhoff D. Myocardial recovery and the failing heart: myth, magic, or molecular target? J Am Coll Cardiol. 2012;60(24):2465–72. https://doi.org/10.1016/j.jacc.2012.06.062.
Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. https://doi.org/10.1126/science.1077857.
Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138(16):3421–30. https://doi.org/10.1242/dev.068601.
Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138(9):1663–74. https://doi.org/10.1242/dev.060897.
Gonzalez-Rosa JM, Mercader N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat Protoc. 2012;7(4):782–8. https://doi.org/10.1038/nprot.2012.025.
Gut P, Reischauer S, Stainier DYR, Arnaout R. Little fish, big data: zebrafish as a model for cardiovascular and metabolic disease. Physiol Rev. 2017;97(3):889–938. https://doi.org/10.1152/physrev.00038.2016.
Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2(1):39–48. https://doi.org/10.1038/35047564.
Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–80. https://doi.org/10.1126/science.1200708.
Zhu W, Zhang E, Zhao M, Chong Z, Fan C, Tang Y, et al. Regenerative potential of neonatal porcine hearts. Circulation. 2018;138(24):2809–16.
Ye L, D’Agostino G, Loo SJ, Wang CX, Su LP, Tan SH, et al. Early regenerative capacity in the porcine heart. Circulation. 2018;138(24):2798–808.
Haubner BJ, Schneider J, Schweigmann U, Schuetz T, Dichtl W, Velik-Salchner C, et al. Functional recovery of a human neonatal heart after severe myocardial infarction. Circ Res. 2016;118(2):216–21. https://doi.org/10.1161/CIRCRESAHA.115.307017.
Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484(7395):479–84. https://doi.org/10.1038/nature11045.
Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–5. https://doi.org/10.1038/nature08804.
Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–9. https://doi.org/10.1038/nature08899.
Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410(6829):701–5.
Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, et al. Human cardiac stem cells. Proc Natl Acad Sci. 2007;104(35):14068–73.
Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8(4):389–98.
Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–6. https://doi.org/10.1038/nature11682.
Eschenhagen T, Bolli R, Braun T, Field LJ, Fleischmann BK, Frisen J, et al. Cardiomyocyte regeneration: a consensus statement. Circulation. 2017;136(7):680–6. https://doi.org/10.1161/CIRCULATIONAHA.117.029343.
Jopling C, Suñé G, Faucherre A, Fabregat C, Izpisua Belmonte JC. Hypoxia induces myocardial regeneration in zebrafish. Circulation. 2012;126(25):3017–27.
Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157(3):565–79.
• Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541(7636):222–7. https://doi.org/10.1038/nature20173This work demonstrated that experimental hypoxia can promote heart regeneration in adult mice.
Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2008;135(1):183–92. https://doi.org/10.1242/dev.010363.
Adler C-P, Friedburg H, Herget GW, Neuburger M, Schwalb H. Variability of cardiomyocyte DNA content, ploidy level and nuclear number in mammalian hearts. Virchows Arch. 1996;429(2-3):159–64.
• González-Rosa JM, Sharpe M, Field D, Soonpaa MH, Field LJ, Burns CE, et al. Myocardial polyploidization creates a barrier to heart regeneration in zebrafish. Dev Cell. 2018;44(4):433–46. e7 This work demonstrated that polyplodization is a barrier to innate heart regeneration and established a threshold for polyploidization beyond which regeneration is impaired.
•• Han L, Choudhury S, Mich-Basso JD, Ammanamanchi N, Ganapathy B, Suresh S, et al. Lamin B2 levels regulate polyploidization of cardiomyocyte nuclei and myocardial regeneration. Dev Cell. 2020;53(1):42–59.e11. https://doi.org/10.1016/j.devcel.2020.01.030This work demonstrated that experimental modification of polyploidization can enhance regeneration of the mammalian heart.
Chávez MN, Morales RA, López-Crisosto C, Roa JC, Allende ML, Lavandero S. Autophagy activation in zebrafish heart regeneration. Sci Rep. 2020;10(1):1–11.
•• Beisaw A, Kuenne C, Günther S, Dallmann J, Wu C-C, Bentsen M, et al. AP-1 contributes to chromatin accessibility to promote sarcomere disassembly and cardiomyocyte protrusion during zebrafish heart regeneration. Circ Res. 2020;126:1760–78 This report demonstrated the need for sarcomere disassembly for a successful heart regeneration program in zebrafish.
• Ben-Yair R, Butty VL, Busby M, Qiu Y, Levine SS, Goren A, et al. H3K27me3-mediated silencing of structural genes is required for zebrafish heart regeneration. Development. 2019;146(19):dev.178632 This report demonstrated the need for sarcomere disassembly for a successful heart regeneration program in zebrafish.
Canseco DC, Kimura W, Garg S, Mukherjee S, Bhattacharya S, Abdisalaam S, et al. Human ventricular unloading induces cardiomyocyte proliferation. J Am Coll Cardiol. 2015;65(9):892–900. https://doi.org/10.1016/j.jacc.2014.12.027.
Sande-Melon M, Marques IJ, Galardi-Castilla M, Langa X, Perez-Lopez M, Botos MA, et al. Adult sox10(+) cardiomyocytes contribute to myocardial regeneration in the zebrafish. Cell Rep. 2019;29(4):1041–54 e5. https://doi.org/10.1016/j.celrep.2019.09.041.
Tang W, Martik ML, Li Y, Bronner ME. Cardiac neural crest contributes to cardiomyocytes in amniotes and heart regeneration in zebrafish. Elife. 2019;8:e47929. https://doi.org/10.7554/eLife.47929.
Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, et al. Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell. 2016;36(1):36–49. https://doi.org/10.1016/j.devcel.2015.12.010.
Karra R, Knecht AK, Kikuchi K, Poss KD. Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc Natl Acad Sci U S A. 2015;112(43):13255–60. https://doi.org/10.1073/pnas.1511209112.
Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, Poss KD. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol. 2013;23(13):1221–7. https://doi.org/10.1016/j.cub.2013.05.028.
Chakraborty S, Yutzey KE. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev Biol. 2012;363(1):234–46.
Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–61.
von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci. 2012;109(7):2394–9.
Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, et al. Hippo signaling impedes adult heart regeneration. Development. 2013;140(23):4683–90.
Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, et al. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature. 2017;550(7675):260–4.
F-l X, Guo M, Yutzey KE. Overexpression of Tbx20 in adult cardiomyocytes promotes proliferation and improves cardiac function after myocardial infarction. Circulation. 2016;133(11):1081–92.
D’Uva G, Aharonov A, Lauriola M, Kain D, Yahalom-Ronen Y, Carvalho S, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol. 2015;17(5):627–38. https://doi.org/10.1038/ncb3149.
Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4(196):ra70.
Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature. 2015;522(7555):226–30. https://doi.org/10.1038/nature14325.
Marin-Juez R, Marass M, Gauvrit S, Rossi A, Lai SL, Materna SC, et al. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc Natl Acad Sci U S A. 2016;113(40):11237–42. https://doi.org/10.1073/pnas.1605431113.
Mahmoud AI, O’Meara CC, Gemberling M, Zhao L, Bryant DM, Zheng R, et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev Cell. 2015;34(4):387–99. https://doi.org/10.1016/j.devcel.2015.06.017.
Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, et al. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124(3):1382–92. https://doi.org/10.1172/JCI72181.
Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–19. https://doi.org/10.1016/j.cell.2006.08.052.
Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20(3):397–404. https://doi.org/10.1016/j.devcel.2011.01.010.
• Zhao L, Ben-Yair R, Burns CE, Burns CG. Endocardial notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through Wnt pathway antagonism. Cell Rep. 2019;26(3):546–54 e5. https://doi.org/10.1016/j.celrep.2018.12.048This work demonstrated a paracrine role of the cardiac endothelium to promote heart regeneration in zebrafish.
•• Marin-Juez R, El-Sammak H, Helker CSM, Kamezaki A, Mullapuli ST, Bibli SI, et al. Coronary revascularization during heart regeneration is regulated by epicardial and endocardial cues and forms a scaffold for cardiomyocyte repopulation. Dev Cell. 2019;51(4):503–15 e4. https://doi.org/10.1016/j.devcel.2019.10.019This report suggests a mechanism for how revascularization shapes regenerative growth in zebrafish.
Cao J, Wang J, Jackman CP, Cox AH, Trembley MA, Balowski JJ, et al. Tension creates an endoreplication wavefront that leads regeneration of epicardial tissue. Dev Cell. 2017;42(6):600–15. e4.
Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138(14):2895–902. https://doi.org/10.1242/dev.067041.
Sanchez-Iranzo H, Galardi-Castilla M, Sanz-Morejon A, Gonzalez-Rosa JM, Costa R, Ernst A, et al. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc Natl Acad Sci U S A. 2018;115(16):4188–93. https://doi.org/10.1073/pnas.1716713115.
Cao Y, Duca S, Cao J. Epicardium in heart development. Cold Spring Harb Perspect Biol. 2020;12(2):a037192.
Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4:e05871. https://doi.org/10.7554/eLife.05871.
Lowe V, Wisniewski L, Sayers J, Evans I, Frankel P, Mercader-Huber N, et al. Neuropilin 1 mediates epicardial activation and revascularization in the regenerating zebrafish heart. Development. 2019;146(13):dev174482.
Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol. 2013;382(2):427–35.
Sánchez-Iranzo H, Galardi-Castilla M, Sanz-Morejón A, González-Rosa JM, Costa R, Ernst A, et al. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc Natl Acad Sci. 2018;115(16):4188–93.
Cai W, Tan J, Yan J, Zhang L, Cai X, Wang H, et al. Limited regeneration potential with minimal epicardial progenitor conversions in the neonatal mouse heart after injury. Cell Rep. 2019;28(1):190–201 e3. https://doi.org/10.1016/j.celrep.2019.06.003.
Wei K, Serpooshan V, Hurtado C, Diez-Cunado M, Zhao M, Maruyama S, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525(7570):479–85. https://doi.org/10.1038/nature15372.
• Lai S-L, Marín-Juez R, Moura PL, Kuenne C, Lai JKH, Tsedeke AT, et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife. 2017;6:e25605 This report demonstrated that manipulation of the inflammatory response to injury in a non-regenerative fish could rescue regenerative capacity.
Bevan L, Lim ZW, Venkatesh B, Riley PR, Martin P, Richardson RJ. Specific macrophage populations promote both cardiac scar deposition and subsequent resolution in adult zebrafish. Cardiovasc Res. 2020;116(7):1357–71.
Sanz-Morejón A, García-Redondo AB, Reuter H, Marques IJ, Bates T, Galardi-Castilla M, et al. Wilms tumor 1b expression defines a pro-regenerative macrophage subtype and is required for organ regeneration in the zebrafish. Cell Rep. 2019;28(5):1296–306. e6.
Lavine KJ, Epelman S, Uchida K, Weber KJ, Nichols CG, Schilling JD, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci. 2014;111(45):16029–34.
Hui SP, Sheng DZ, Sugimoto K, Gonzalez-Rajal A, Nakagawa S, Hesselson D, et al. Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev Cell. 2017;43(6):659–72 e5. https://doi.org/10.1016/j.devcel.2017.11.010.
Holmes WE, Sliwkowski MX, Akita RW, Henzel WJ, Lee J, Park JW, et al. Identification of heregulin, a specific activator of p185erbB2. Science. 1992;256(5060):1205–10. https://doi.org/10.1126/science.256.5060.1205.
Lee K-F, Simon H, Chen H, Bates B, Hung M-C, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378(6555):394–8.
Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378(6555):386–90. https://doi.org/10.1038/378386a0.
Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–70. https://doi.org/10.1016/j.cell.2009.04.060.
Ma H, Yin C, Zhang Y, Qian L, Liu J. ErbB2 is required for cardiomyocyte proliferation in murine neonatal hearts. Gene. 2016;592(2):325–30.
Polizzotti BD, Ganapathy B, Walsh S, Choudhury S, Ammanamanchi N, Bennett DG, et al. Neuregulin stimulation of cardiomyocyte regeneration in mice and human myocardium reveals a therapeutic window. Sci Transl Med. 2015;7(281)):281ra45. https://doi.org/10.1126/scitranslmed.aaa5171.
Han Y, Chen A, Umansky K-B, Oonk KA, Choi W-Y, Dickson AL, et al. Vitamin D stimulates cardiomyocyte proliferation and controls organ size and regeneration in zebrafish. Dev Cell. 2019;48(6):853–63. e5.
Honkoop H, de Bakker DE, Aharonov A, Kruse F, Shakked A, Nguyen PD, et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife. 2019;8:e50163.
Aharonov A, Shakked A, Umansky KB, Savidor A, Genzelinakh A, Kain D, et al. ERBB2 drives YAP activation and EMT-like processes during cardiac regeneration. Nat Cell Biol. 2020;22:1346–56 1-11.
Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, et al. Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS One. 2013;8(6):e67266. https://doi.org/10.1371/journal.pone.0067266.
Shen H, Gan P, Wang K, Darehzereshki A, Wang K, Kumar SR, et al. Mononuclear diploid cardiomyocytes support neonatal mouse heart regeneration in response to paracrine IGF2 signaling. Elife. 2020;9:e53071.
Wang Z, Cui M, Shah AM, Ye W, Tan W, Min Y-L, et al. Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc Natl Acad Sci - PNAS. 2019;116(37):18455–65. https://doi.org/10.1073/pnas.1905824116.
Sui Y, Zhang W, Tang T, Gao L, Cao T, Zhu H, et al. Insulin-like growth factor II overexpression accelerates parthenogenetic stem cell differentiation into cardiomyocytes and improves cardiac function after acute myocardial infarction in mice. Stem Cell Res Ther. 2020;11(1):86. https://doi.org/10.1186/s13287-020-1575-4.
Taimeh Z, Loughran J, Birks EJ, Bolli R. Vascular endothelial growth factor in heart failure. Nat Rev Cardiol. 2013;10(9):519–30. https://doi.org/10.1038/nrcardio.2013.94.
Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127(18):3941–6.
• Karra R, Foglia MJ, Choi WY, Belliveau C, DeBenedittis P, Poss KD. Vegfaa instructs cardiac muscle hyperplasia in adult zebrafish. Proc Natl Acad Sci U S A. 2018;115(35):8805–10. https://doi.org/10.1073/pnas.1722594115This report suggested that activation of the cardiac endothelium could promote heart regeneration.
Giacca M, Zacchigna S. VEGF gene therapy: therapeutic angiogenesis in the clinic and beyond. Gene Ther. 2012;19(6):622–9. https://doi.org/10.1038/gt.2012.17.
Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31(10):898–907. https://doi.org/10.1038/nbt.2682.
Lin YD, Luo CY, Hu YN, Yeh ML, Hsueh YC, Chang MY, et al. Instructive nanofiber scaffolds with VEGF create a microenvironment for arteriogenesis and cardiac repair. Sci Transl Med. 2012;4(146):146ra09. https://doi.org/10.1126/scitranslmed.3003841.
Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, et al. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev Cell. 2015;33(4):442–54. https://doi.org/10.1016/j.devcel.2015.04.001.
Ivins S, Chappell J, Vernay B, Suntharalingham J, Martineau A, Mohun TJ, et al. The CXCL12/CXCR4 axis plays a critical role in coronary artery development. Dev Cell. 2015;33(4):455–68.
• Das S, Goldstone AB, Wang H, Farry J, D’Amato G, Paulsen MJ, et al. A unique collateral artery development program promotes neonatal heart regeneration. Cell. 2019;176(5):1128–42 e18. https://doi.org/10.1016/j.cell.2018.12.023This work demonstrated that enhancing revascularization through collaterals could enhance regeneration in mice.
Goldstone AB, Burnett CE, Cohen JE, Paulsen MJ, Eskandari A, Edwards BE, et al. SDF 1-alpha attenuates myocardial injury without altering the direct contribution of circulating cells. J Cardiovasc Transl Res. 2018;11(4):274–84.
Chen WC, Wang Z, Missinato MA, Park DW, Long DW, Liu H-J, et al. Decellularized zebrafish cardiac extracellular matrix induces mammalian heart regeneration. Sci Adv. 2016;2(11):e1600844.
• Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Umansky KB, Yifa O, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547(7662):179–84 This report demonstrated that the extracellular matrix protein agrin can promote regeneration in mice.
• Baehr A, Umansky KB, Bassat E, Jurisch V, Klett K, Bozoglu T, et al. Agrin promotes coordinated therapeutic processes leading to improved cardiac repair in pigs. Circulation. 2020;142(9):868–81. https://doi.org/10.1161/CIRCULATIONAHA.119.045116This report demonstrated that the extracellular matrix protein agrin can promote regeneration in pigs.
Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, et al. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173(1):104–16 e12. https://doi.org/10.1016/j.cell.2018.02.014.
Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci. 2013;110(34):13839–44.
Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115(3):354–63.
Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532(7598):201–6. https://doi.org/10.1038/nature17644.
Goldman JA, Kuzu G, Lee N, Karasik J, Gemberling M, Foglia MJ, et al. Resolving heart regeneration by replacement histone profiling. Dev Cell. 2017;40(4):392–404 e5. https://doi.org/10.1016/j.devcel.2017.01.013.
Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338(6114):1599–603.
Acknowledgements
We apologize to colleagues whose work we could not include due to space considerations.
Funding
This work was funded by a Duke Undergraduate Independent Study Grant (AWG), R03 HL144812 (RK), a Duke University Strong Start Physician Scientist Award (RK), and a Mandel Foundation Seed Grant (RK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Ravi Karra has a patent 62/309,649 issued. The other authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Regenerative Medicine
Rights and permissions
About this article
Cite this article
Brezitski, K.D., Goff, A.W., DeBenedittis, P. et al. A Roadmap to Heart Regeneration Through Conserved Mechanisms in Zebrafish and Mammals. Curr Cardiol Rep 23, 29 (2021). https://doi.org/10.1007/s11886-021-01459-6
Accepted:
Published:
DOI: https://doi.org/10.1007/s11886-021-01459-6