Abstract
Heart failure caused by cardiomyocyte loss after ischemic tissue damage is a leading cause of death worldwide, since adult mammals cannot regenerate heart injuries. While some new cardiomyocytes are produced in adult mammals during normal ageing and after infarction, this occurs at insufficient rates for effective cardiac regeneration. Zebrafish, on the contrary, are able to regenerate multiple organs including the heart. Injuries induce complex cellular and molecular responses in endocardium, epicardium and myocardium, which robustly regenerate in a coordinated manner, resulting in full morphological and functional recovery. In particular, differentiated cardiomyocytes re-enter the cell cycle and proliferate to regenerate the myocardium. Thus, the zebrafish has emerged as an important model to study mechanisms of naturally occurring cardiac regeneration. Here, we describe zebrafish heart injury techniques and review current data on mammalian cardiomyocyte turnover and production in response to injury as well as our current knowledge of the cellular and molecular mechanisms of zebrafish heart regeneration.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
10.1 Introduction
10.1.1 Cardiomyocytes Are Insufficiently Replaced After Injury of the Adult Mammalian Heart
Myocardial infarction (MI) is a leading cause of death worldwide. Compared to other organs in mammals (e.g. lung, liver, bone, skin), the adult heart displays very little regenerative capacity in response to injury. In particular, lost adult mammalian cardiomyocytes (CMs ) are largely not replaced after MI or other disorders resulting in CM death, leading to muscle loss, irreversible scarring and eventually heart failure (Laflamme and Murry 2011). In contrast, adult zebrafish can completely regenerate their heart via CM replacement , resulting in scar-free recovery (Poss et al. 2002; Schnabel et al. 2011; Gonzalez-Rosa et al. 2011; Chablais et al. 2011; Wang et al. 2011). Intriguingly, neonatal mice can regenerate the myocardium after partial surgical resection as well, but this ability is lost by 7 days after birth (Porrello et al. 2011). While complete functional heart regeneration requires not only restoration of lost muscle tissue, but also coordinated replacement of the other tissues of the heart, in particular of endocardium, epicardium and coronary vasculature, a central question in heart regeneration research has been how CMs can be restored. Genetic lineage-tracing experiments have indicated that during neonatal mouse and adult zebrafish heart regeneration CMs are replaced via proliferation of pre-existing CMs, and not from stem or progenitor cells (Kikuchi et al. 2010; Jopling et al. 2010; Porrello et al. 2011). In adult mammals , CMs have long been believed to be terminally differentiated and thus unable to proliferate (Zebrowski and Engel 2013). Instead, much of the growth of the postnatal mammalian heart occurs via enlargement of pre-existing CMs, i.e. hypertrophy (Li et al. 1996; Yoshizumi et al. 1995). Thus, one explanation for the differential ability of the adult zebrafish and mammalian heart to regenerate would be the idea that CMs retain the ability to re-enter the cell cycle and proliferate in the zebrafish, while such capacity is lost during ontogeny in mammals.
However, there is now good evidence that new CMs do form during adult mammalian life, including humans. Taking advantage of the integration of atmospheric 14C, which was generated by nuclear bomb tests during the Cold War, into cells born during that time, Frisén and colleagues estimated the age of CMs in humans. They showed that human CMs are being renewed during adult life, but at a very low rate of 1 % per year at the age of 25 and 0.45 % at the age of 75 (Bergmann et al. 2009). Thus, approximately 45 % of all CMs are exchanged during a human lifetime while 55 % remain from neonatal growth (Bergmann et al. 2009).
While it is not possible to identify the cellular source of newly forming CMs in the adult human heart, several elegant studies in mice have addressed whether CM renewal during aging and the low-rate CM replacement after cardiac injury are due to proliferation of existing CMs or due to CM differentiation from progenitor cells (Ali et al. 2014; Ellison et al. 2013; Hsieh et al. 2007; Malliaras et al. 2013; Senyo et al. 2013, 2014; van Berlo et al. 2014). Unfortunately, the conclusions drawn by several studies based on genetic lineage-tracing of CMs or cardiac progenitor cells differ considerably. While most reports using different means to track the fate of differentiated CMs found that renewal of CMs during homeostasis is due to proliferation of existing CMs (Senyo et al. 2014), one study concluded that progenitor cells contribute to CM formation after cardiac injury, since the progeny of differentiated CMs got diluted with other cells in injured hearts (Hsieh et al. 2007). In contrast, other studies using lineage-tracing of differentiated CMs and multi-isotope imaging mass spectrometry concluded that the limited CM replacement after injury is due to CM proliferation (Senyo et al. 2013; Ali et al. 2014). However, c-kit positive cardiac progenitor cells have been reported to contribute significantly to CM production after cardiac injury as well (Ellison et al. 2013). In contrast, another study found that c-kit positive cells form negligible numbers of CMs both during homeostasis and after injury (van Berlo et al. 2014).
Thus, while further studies are needed to clarify how CMs are formed in the adult mammalian heart, there is a growing consensus that the rate of CM formation under homeostatic conditions and in response to heart injury is low, too low for effective myocardial regeneration after heart injury (Senyo et al. 2014).
10.1.2 Mammalian Cardiomyocyte Proliferation Can Be Experimentally Induced
Despite the uncertainties about the cellular basis of CM turnover in adult mammals, the above mentioned studies suggest that adult mammalian CMs are able to proliferate and could potentially be augmented therapeutically. In support of this idea, injection of the growth factor Neuregulin 1 (Nrg1) has been shown to induce proliferation of CMs in both healthy and injured adult mouse hearts, to reduce scar size and to improve functional recovery after MI (Bersell et al. 2009). Likewise, induction of constitutively active ERBB2, a tyrosine kinase receptor of Nrg1, in neonatal, juvenile or adult mouse CMs was found to lead to cardiomegaly caused by CM hypertrophy and proliferation, while ERBB2 induction after MI improved functional recovery (D’Uva et al. 2015).
Interestingly, several studies indicated that proliferation might be actively inhibited in adult mammalian CMs. First, overexpression of several miRNAs was found to stimulate both DNA synthesis and cytokinesis in neonatal mouse and rat CMs in culture and in adult animals (Eulalio et al. 2012). Second, deletion of components of the Hippo signaling pathway , which controls organ size and suppresses cell proliferation, has been shown to induce adult CM renewal via cell cycle re-entry and mitosis (Heallen et al. 2013). In addition, removal of Hippo components in P7 mice, a time at which natural heart regeneration is compromised (Porrello et al. 2011), followed by apical resection at P8 resulted in significant muscle regeneration from spared CMs and improved functional recovery (Heallen et al. 2013). Together, these studies suggest the potential to augment cardiac regeneration by stimulating adult mammalian CM proliferation in vivo through supplying pro-proliferation factors and/or removing natural inhibitors.
10.2 Zebrafish Heart Regeneration – Injury Models
A promising way to uncover factors that are able to stimulate CM proliferation after heart injury would be to elucidate the molecular basis underlying naturally occurring adult heart regeneration in non-mammalian vertebrates. Cardiac regeneration after injury has predominantly been studied in zebrafish, but also in salamanders (Oberpriller and Oberpriller 1974; Piatkowski et al. 2013). Interestingly however, it has been reported that heart regeneration does not occur in medaka, a teleost fish model organism that otherwise shares many features with zebrafish (Ito et al. 2014). Thus, it is unclear whether heart regeneration is a common feature amongst teleost fish and other lower vertebrates, and more data will be needed to address the intriguing question of how heart regeneration evolved or whether cardiac regenerative capacity was lost during evolution in certain lineages.
Since the discovery of heart regeneration in zebrafish, this model has developed rapidly over the past years, owing to its robust regenerative capacity and its propensity for genetic manipulation (Gemberling et al. 2013). In this chapter, we summarize the current knowledge about the cellular basis and molecular regulation of zebrafish heart regeneration.
10.2.1 Ventricular Resection
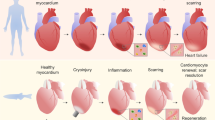
Zebrafish hearts consist of one atrium, one ventricle and a prominent non-muscular extension of the aorta, termed the bulbus arteriosus. The ability of zebrafish to regenerate the heart was first reported by Keating and colleagues (Poss et al. 2002). After surgical removal of around 20 % of the apex of the ventricle, the injury was found to be rapidly sealed by a fibrin clot. This fibrinous wound tissue increased in size till 7–9 days post injury (dpi). Evidence for CM cell cycle activity was obtained, and a contiguous wall of muscle was restored by 30 dpi (see Sect. 10.3 below). By 60 dpi, the fibrin clot disappeared and the size and shape of the ventricle appeared to be normal, with little or no collagen deposition, indicating that, in contrast to adult mammals, permanent scarring does not occur (Fig. 10.1a) (Poss et al. 2002). To date, ventricular resection remains a frequently used injury model, but several studies have developed additional ways to damage the heart, which better mimic some aspects of mammalian cardiac injury.
Regeneration of ventricular myocardium in the resected and cryoinjured zebrafish heart
(a) Acid fuchsin orange G (AFOG) staining on longitudinal sections of uninjured and resected hearts at different times after injury. At 7 days post injury (dpi), the injured area is sealed by a fibrin clot (red). The wound tissue progressively reduces in size from 7 to 21 dpi. At 30 dpi, the resected apex is filled with muscle (orange/brown) and no obvious wound tissue and collagen deposition (blue) is observed, indicative of complete morphological regeneration.
(b) Acid fuchsin orange G (AFOG) staining on longitudinal sections of cryoinjured hearts at different time points after injury. At 4 dpi, the injured area is characterized by fibrin deposition (red). The wound tissue progressively reduces in size from 4 to 60 dpi where little or no collagen deposition is observed. Dashed lines, wound boundary
10.2.2 Cryoinjury
Acute loss of myocardial tissue after MI is typically caused by ischemia and subsequent CM death throughout the affected area. After clearance of dead cells and matrix debris from the infarct zone, damaged tissue is replaced by scar tissue characterized by collagen deposition. While zebrafish effectively regenerate the resected heart, the correlation between ventricular resection in zebrafish and MI in mammals is limited. For instance, ventricular resection does not cause massive cell death and thus no cell debris removal before muscle replacement is required. In addition, scar tissue formed after resection mainly comprises fibrin with little or no collagen deposition. Therefore, the value of ventricular resection as a translationally relevant cardiac injury model has been questioned and cryoinjury has been employed in zebrafish as an alternative to better model certain aspects of MI (Chablais et al. 2011; Gonzalez-Rosa et al. 2011; Schnabel et al. 2011).
In these models, either a small piece of dry ice (Schnabel et al. 2011) or a thin metal filament cooled by liquid nitrogen is applied to the surface of the ventricle (Chablais et al. 2011; Gonzalez-Rosa et al. 2011). This treatment was found to cause extensive cell death and to result in an injury affecting approximately 25 % of the ventricle size. Infiltration of immune cells was observed as early as 1 dpi (Schnabel et al. 2011), while morphological regeneration (muscle restoration and wound tissue resolution) took longer than that after ventricular resection, around 60–130 days. Cellular responses including elevated CM proliferation and reactivation of developmental gene programs in the epicardium and endocardium were found to be largely similar to those seen in the resection model (see Sect. 10.3 below; Gonzalez-Rosa et al. 2011; Schnabel et al. 2011). In contrast to ventricular resection, scar formation with extensive collagen deposition was observed as early as 3 days after cryoinjury (Gonzalez-Rosa et al. 2011). Intriguingly however, the collagen-scar was progressively resolved and scar-free regeneration occurred (Fig. 10.1b) (Gonzalez-Rosa et al. 2011; Schnabel et al. 2011; Chablais et al. 2011). Transient scar formation has been suggested to be functionally important for regeneration in zebrafish, implying that scarring and regeneration are not necessarily mutually exclusive (Chablais and Jazwinska 2012). However, whether the molecular composition of the transient scar in zebrafish differs from that of permanent mammalian scars is unknown, and the molecular basis underlying scar resolution remains to be elucidated.
10.2.3 Genetic Ablation
10.2.3.1 Cardiomyocyte Ablation
In addition to mechanical and cryoinjury, in which several cardiac cell types are affected, myocardial-specific injury has also been achieved. Poss and colleagues generated an ablation system which induces expression of the diphtheria toxin A chain (DTA) in CreERT2-expressing CMs upon 4-hydroxytamoxifen (4-HT)-mediated recombination (Wang et al. 2011). This system caused massive cell death and depleted > 60 % of the ventricular CMs, resulting in lethargy, severe stress hypersensitivity, gasping phenotypes and reduced swimming performance, which are classical signs of heart failure and are not seen in other zebrafish heart injury models. Surprisingly, complete muscle regeneration occurred rapidly within 30 days and full functional recovery was observed at 45 dpi. Such rapid recovery could be explained by the fact that proliferation levels of spared CMs were found to be two to four times higher than those seen in other injury models. Intriguingly, robust cellular responses in the epicardium and the endocardium similar to other injury models were also observed, suggesting that CM cell death is sufficient to initiate an organ-wide regenerative program (Wang et al. 2011).
10.2.3.2 Epicardial Ablation
Poss and colleagues also employed an inducible cell ablation system using tcf21-driven expression of bacterial nitroreductase, which converts the drug metronidazole into a cytotoxin, to inducibly kill epicardial cells in the adult zebrafish heart (Wang et al. 2015). While this technique revealed that the epicardium is required for myocardial regeneration (see Sect. 10.4.2), it also demonstrated that the epicardium itself can rapidly and efficiently regenerate from spared epicardial cells. Epicardial regeneration was found to involve directed migration of proliferating epicardial cells from the base of the ventricle towards its apex (Wang et al. 2015). The bulbus arteriosus, which is located at the base of the ventricle, appears to provide signals that direct this migration by repelling epicardial cells, as shown in a series of elegant tissue recombination experiments in cultures of explanted hearts (Wang et al. 2015). Sonic hedgehog (Shh) is a strong candidate for such a bulbus arteriosus-derived signal, since Hedgehog signaling was shown to be required for epicardial regeneration and Shh-soaked beads could restore epicardial migration in hearts whose bulbus arteriosus had been removed (Wang et al. 2015). It will be interesting to test to which extent Shh-mediated directed cell migration is also involved in epicardial development.
10.3 Functional Recovery
Assays for assessing heart function, in particular quantification of cardiac hemodynamics using ultrasound, are difficult to perform in zebrafish due to the small size of the adult heart (Huang et al. 2015; Kang et al. 2015). Nevertheless, there is increasing evidence for functional recovery during zebrafish heart regeneration. Myocardium at the apex of the ventricle, which presumably has formed anew during regeneration in response to ventricular resection, was found to be electrically coupled to the rest of the ventricle, indicating functional recovery (Kikuchi et al. 2010). Likewise, coupling measured by optical mapping and injury-induced lengthening of action potential duration was found to be restored to pre-injury levels 45 days post genetic ablation of CMs (Wang et al. 2011). Electrocardiogram recordings showed prolonged QT intervals after cryoinjury, which recovered by 30 dpi (Chablais et al. 2011). However, another report found that regenerated hearts retained a prolonged QT interval after ventricular resection (Yu et al. 2010). Side-by-side comparison of different injury models as well as measurements of the electrophysiological properties of individual CMs (Nemtsas et al. 2010) will be instrumental in clarifying whether zebrafish can completely recover the electrophysiological properties of ventricular myocardium. Measurements of cardiac performance based on echocardiography indicated that heart function does recover; yet this might take longer than morphological regeneration. One study reported that heart function after cryoinjury recovered only within 180 days (Hein et al. 2015), while another showed that pumping function based on relative fractional volume shortening recovered within 60 days in cryoinjured hearts, while ventricular wall motion remained altered even after 140 days (Gonzalez-Rosa et al. 2014). Overall, these data indicate that zebrafish cannot only regenerate the architecture of the heart but also its function, albeit it remains unclear to what extent.
10.4 Regeneration of Non-myocardial Tissues
Heart injury in zebrafish triggers regenerative responses in all three primary layers of the heart, namely endocardium, epicardium and myocardium, plus regeneration of coronary vasculature. In this section, we summarize the current knowledge of cellular responses and their molecular regulation in the non-myocardial layers.
10.4.1 Endocardium
Endocardial cells were found to be the first to respond to heart injury after ventricular resection (Kikuchi et al. 2011b). In the uninjured ventricle, endocardial cells typically possess elongated nuclei and thin cell bodies that tightly adhere to myofibers, as revealed by transmission electron microscopy (TEM). As early as 1 h post ventricular resection (hpi), endocardial cells both near and distant from the amputation plane appear to round up and to detach from the underlying myofibers (Kikuchi et al. 2011b). Endocardial cells close to the amputation plane were reported to retain a disorganized appearance until 7 dpi, coinciding with rapid CM proliferation. There is also evidence that the endocardium is a source of retinoic acid, which promotes CM proliferation. As early as 3 and 6 hpi, expression of the retinoic acid-synthesizing enzyme aldh1a2 (raldh2) was found to be upregulated in the entire endocardium, representing an intriguing organ-wide response. From 1 dpi onwards, aldh1a2 expression became localized to endocardial cells at the injury site, where it persisted throughout the early stages of regeneration (until 14 dpi). Intriguingly, endocardial upregulation of aldh1a2 occurred also in response to spontaneous infarcts observed in explanted adult zebrafish hearts and upon lipopolysaccharide (LPS) injection , which triggers a systemic inflammatory response. These data suggest that endocardial upregulation of aldh1a2 is a fundamental response to cardiac injury and inflammation (Fig. 10.2) (Kikuchi et al. 2011b). To test the role of RA signaling in heart regeneration, Poss and colleagues inhibited the signal by systemic overexpression of dominant-negative retinoid acid receptors or the RA-degrading Cyp26 enzyme. Both treatments inhibited CM proliferation at 7 dpi, while RA injection was not sufficient to induce CM proliferation. These results suggest that RA is a permissive but not instructive signal for CM proliferation and regeneration (Kikuchi et al. 2011b). Since the epicardium was also found to upregulate aldh1a2 expression in response to heart injury (albeit later than the endocardium) (Kikuchi et al. 2011b), tissue-specific interference with RA production will be required to clarify whether endocardium or epicardium-derived RA regulates CM proliferation. However, injured mammalian hearts were found to display epicardial, but not endocardial aldh1a2 expression, suggesting that endocardial RA production supports myocardial regeneration (Kikuchi et al. 2011b).Platelet derived growth factor (PDGF) and Fibroblast growth factor (FGF) signaling appear to be required for revascularization of the wound, and thus likely also for restoration of the endocardial layer.
Regeneration of the endocardium following injury
In the uninjured heart, endocardial cells tightly adhere to myofibers of the trabeculated myocardium . Starting very early after injury, endocardial cells show signs of morphological and transcriptional alterations. In particular, at 3 h post injury (hpi), the retinoic acid -synthesizing enzyme aldh1a2 is upregulated in endocardial cells in the entire heart, while expression is later restricted to endocardium at the wound border (1 dpi). PDGF and FGF signaling might be required for endocardial regeneration. Lineage tracing indicates that pre-existing endocardial cells contribute to the regenerated endocardium upon complete regeneration.
10.4.2 Coronary Vasculature
Vascular supply of the myocardium varies widely among teleost fish, but the compact wall of the zebrafish ventricle is supported by coronary vessels which are located in the subepicardial space (Hu et al. 2001). No molecular marker that is expressed only in the coronary endothelium but not in endocardial cells has been described. Nevertheless, several studies have argued that in addition to endocardial cells, which express a transgenic endothelial marker, fli:EGFP, at low levels, a population of cells displaying stronger fli:EGFP could be identified in the wound tissue during heart regeneration, and these cells have been addressed as regenerating coronary vessels (Lepilina et al. 2006; Kim et al. 2010; Zhao et al. 2014). Genetic lineage-tracing experiments indicated that the endothelial cells of this presumptive regenerating coronary vasculature are derived from pre-existing endothelial cells, while perivascular cells are derived from the epicardium (Fig. 10.3) (Zhao et al. 2014; Kikuchi et al. 2011a). Systemic inhibition of FGF signaling via overexpression of a dominant-negative FGF receptor or pharmacological inhibition of PDGF signaling interfered with re-vascularization of the wound tissue (Lepilina et al. 2006; Kim et al. 2010). While FGF inhibition also resulted in defects in myocardial regeneration (Lepilina et al. 2006), neither FGF nor PDGF signaling are thought to regulate CM proliferation (Lepilina et al. 2006; Kim et al. 2010), indicating that re-vascularization is important for myocardial regeneration.
Regeneration of coronary vasculature
In the uninjured heart, coronary vasculature is observed in the compact layer myocardium . At 7 dpi, coronary vasculature beings to appear in the wound. At 14 dpi, the regenerating myocardium is heavily vascularized, which persists after complete regeneration. Regeneration of the coronary vasculature appears to be regulated by PDGF and FGF signaling.
10.4.3 Epicardium
The epicardium , a mesothelial cell layer covering the outer surface of the ventricle, has been found to respond to heart injury by re-expression of genes associated with epicardial development, including tbx18, wt1b and aldh1a2 and also by becoming proliferative at 3 dpi (Gonzalez-Rosa et al. 2011; Kikuchi et al. 2011b; Lepilina et al. 2006; Schnabel et al. 2011). By 7 dpi, epicardial cells were reported to enclose the wound and to integrate into the fibrous wound tissue. For instance, a large number of cells retaining the expression of tbx18 was found in the wound by 14 dpi and in the regenerate at 30 dpi (Lepilina et al. 2006). During embryonic heart development epicardial cells are thought to undergo epithelial-to-mesenchymal transition (EMT) and to invade the subepicardial space and the myocardium to form smooth muscle cells of the coronary vasculature and myocardial fibroblasts (Dettman et al. 1998). In addition, some lineage tracing studies have found evidence for formation of CMs from epicardial cells during development and after heart injury, although the specificity of the Cre lines used for some of these studies has been questioned (Zhou et al. 2008; Cai et al. 2008; Smart et al. 2011; Rudat and Kispert 2012). To address the potential of epicardial cells during zebrafish heart regeneration , Poss and colleagues used inducible Cre lines driven by tcf21 regulatory sequences and found that epicardial cells give rise to perivascular cells but not CMs after injury (Kikuchi et al. 2011a). This finding was substantiated by Mercader and colleagues using transplantation experiments, which in addition indicated that myofibroblasts are derived from epicardial cells during heart regeneration as well (Gonzalez-Rosa et al. 2012). Thus, during naturally occurring heart regeneration, the epicardium does not adopt CM fate (Fig. 10.4).
Regeneration of the epicardium
In the uninjured heart, the epicardium consists of a single layer of cells at the outline of the compact myocardium . At 3 dpi, epicardial activation, which is characterized by upregulation of tbx18, wt1b, fn1 and nrg1, is observed in the entire heart. From 7 to 14 dpi, epicardial cells cover and invade the wound tissue. Lineage-tracing experiments demonstrate that epicardial cells give rise to the regenerated epicardium, perivascular cells of the coronary vasculature and myofibroblasts. Regeneration of the epicardium appears to be regulated by PDGF, FGF and Shh signaling.
During embryonic development, the epicardium is thought to provide signals regulating myocardial development, including CM proliferation (Lavine and Ornitz 2008; Riley 2012). To test the requirement of the epicardium for zebrafish myocardial regeneration, Poss and colleagues ablated epicardial cells using tc21-driven expression of nitroreducatase (Wang et al. 2015). Epicardial ablation induced early after ventricular resection (from 2 to 5 dpi) reduced CM proliferation at 7 dpi and resulted in defects in myocardial restoration, as evidenced by presence of scar tissue by 30 dpi. Thus, the epicardium is essential for myocardial regeneration in zebrafish. The molecular signals mediating its effects on CMs during regeneration are however unknown.
Epicardial EMT during heart regeneration appears to be controlled by FGF and PDGF signaling (Lepilina et al. 2006; Kim et al. 2010). Inhibition of FGF signaling by systemic overexpression of a dominant-negative FGF receptor compromised integration of tbx18-positive cells into the regenerating myocardium in addition to its effects on re-vascularization (see Sect. 10.4.2) (Lepilina et al. 2006). Using an adult zebrafish epicardium explant culture, Lien and colleagues showed that PDGF signaling triggers epicardial cells to detach and form mesenchymal-like cell types. PDGF signaling was also required for epicardial cell proliferation both in vitro and in vivo, and treatment with a PDGF receptor inhibitor reduced the expression of the EMT marker snail2 in regenerating hearts (Kim et al. 2010).
The epicardium also appears to be an important source of extracellular matrix (ECM), which influences the cellular behavior of regenerating CMs. Poss and colleagues showed that expression of fibronectin, a major ECM component, is strongly induced upon heart injury and mainly deposited by injury-activated epicardial cells (Wang et al. 2013). Myocardial regeneration was significantly inhibited in fish overexpressing a dominant-negative human fibronectin fragment as well as in fish homozygous for a fibronectin1 mutation, resulting in severe fibrosis. Interestingly, such myocardial regeneration defects were not caused by defective CM proliferation. Instead, at 30 dpi, a larger number of CMs was present in the area flanking the injury in homozygous fibronectin1 mutants, suggesting that fibronectin might be required for CM mobilization and integration into the injury site (Wang et al. 2013). Altogether, these data suggest that the epicardium regulates zebrafish heart regeneration in a number of ways.
10.5 Regeneration of the Myocardium
10.5.1 Cellular Sources of Regenerating Cardiomyocytes
Central to the ability of zebrafish to recover from heart injuries is the fact that they can replace lost CMs. So far, research into the cellular and molecular mechanisms of heart regeneration thus has largely focused on CM restoration. This is usually studied in the adult heart, where homeostatic myocardial cell cycle activity is very low, which allows for the identification of injury-induced regenerative mechanisms of CM production. Nevertheless, injuries to the rapidly developing embryonic zebrafish heart at stages where CMs are normally produced have also been proposed to represent a useful model of regeneration (Zhang et al. 2013). Using a nitroreductase-based CM-specific ablation system, Chi and colleagues reported that lost embryonic ventricular CMs are rapidly replaced (Zhang et al. 2013). Interestingly, they suggested that this is not only due to elevated proliferation of spared CMs, but also due to transdifferentiation of atrial into ventricular CMs as revealed by genetic lineage-tracing experiments. This process is at least partly mediated by Notch signaling since treatment of ablated hearts with the γ-secretase inhibitor DAPT reduced the appearance of atrially-derived CMs in the injured ventricle. Though intriguing, such atrial contribution to the regenerating ventricle was found to be minimal in adult CM-ablated hearts, suggesting that adult zebrafish rely on other sources of CMs for ventricular regeneration (Zhang et al. 2013).
For a period of time, it remained unclear whether the newly forming CMs in adult regenerating hearts are derived from existing differentiated CMs or other sources like stem cells. Initial findings from Poss and colleagues suggested that CMs regenerate from progenitors, but not from differentiated spared CMs (Lepilina et al. 2006). However, subsequent genetic lineage-tracing experiments from two independent studies provided strong evidence for the latter mechanism (Jopling et al. 2010; Kikuchi et al. 2010), indicating that the earlier data which were based on differential maturation and stability of fluorescent proteins were artefacts (Kikuchi et al. 2010). Using an inducible CM-specific Cre line (myl7:CreERT2), Poss and colleagues demonstrated that differentiated CMs labeled 5 days before ventricular resection in adult fish contribute to the vast majority of the regenerated myocardium (Fig. 10.5) (Kikuchi et al. 2010). Izpisua-Belmonte and colleagues genetically lineage-traced embryonic CMs and similarly concluded that adult regenerating CMs are derived from cells that once had CM character (Jopling et al. 2010). Thus, the current consensus in the field is that zebrafish myocardial regeneration is achieved via cell cycle re-entry and proliferation of spared differentiated CMs (Figs. 10.6 and 10.7a). Similarly, neonatal mouse heart regeneration is thought to occur via spared CM proliferation as well (Porrello et al. 2011). Whether all CMs can equally contribute to regeneration or whether a particular subset of potentially less differentiated cells has higher regenerative potential is currently unknown. It is also worth noting that quantitative data showing that heart regeneration involves the full restoration of CM numbers is missing. In addition, whether CM hypertrophy, which is prominent in injured mammalian hearts (Zebrowski and Engel 2013), also occurs in the zebrafish and if so, to which extent injury-induced CM cell cycle activity could be due to CM polyploidization has not been explored.
Cardiomyocyte lineage tracing experiments
To test whether differentiated cardiomyocytes are the precursors of regenerating cardiomyocytes, a dual transgenic system has been used. Tamoxifen-inducible CreERT2 recombinase is expressed specifically in differentiated cardiomyocytes under control of the myl7 promoter. Three days before injury, Cre activity is induced by treating the fish with 4-hydroxytamoxifen. Consequently, CreERT2 removes a floxed DsRed-STOP cassette from a second transgene expressed under control of a β−actin2 promoter, which is active in cardiomyocytes in the adult heart. Removal of the STOP cassette allows transcription of GFP, thus cardiomyocytes are genetically labeled with GFP. At 30 dpi, all of the regenerated cardiomyocytes are GFP positive, indicating that cardiomyocytes regenerate from pre-existing cardiomyocytes but not from stem or progenitor cells, which are GFP negative.
Regeneration of the myocardium
At 7 dpi, cardiomyocytes close to the wound are activated, as indicated by dedifferentiation, activation of the regulatory sequences of gata4 and proliferation. Cardiomyocyte proliferation is regulated positively by RA, Tgfß, Shh, Igf, Jak1/Stat3, Hypoxia, H2O2, Notch, Nrg1, Fn1, Gata4, NF-κB and BMP and negatively by p38αMAPK, miR-99/100, miR-133 and miR-101a. Lineage tracing indicates that regenerated cardiomyocytes are derived from spared, "activated" CMs at the wound border.
Cellular responses of cardiomyocytes to injury
(a) At 7 dpi, cardiomyocytes (marked by myl7:GFP) at the wound border re-enter the cell cycle and express the cell-cycle marker PCNA.
(b) At 7 dpi, cardiomyocytes (marked by myosin heavy chain (MHC) expression, red) at the wound border activate the regulatory sequence of gata4 in gata4:GFP transgenic fish.
10.5.2 Cardiomyocyte Dedifferentiation
Cardiomyocytes do not only re-enter the cell cycle, but also appear to dedifferentiate during regeneration, which is both suggested by reported loss of characteristics of the differentiated state and by upregulation of embryonic genes. After injury, CMs close to the wound were found to be characterized by reduced expression of GFP driven by the myl7 promoter, which is active in differentiated CMs, by partial loss of sarcomeric structures revealed by transmission electron microscopy and by alpha-actinin disorganization (Jopling et al. 2010; Kikuchi et al. 2010; Sallin et al. 2015; Schnabel et al. 2011; Wang et al. 2011; Wu et al. 2016). These changes, which are suggestive of CM dedifferentiation in response to injury, were found to be accompanied by reactivated expression of genes that are highly expressed in embryonic hearts, including nppa, nppb, embryonic myosin chains, and likely also cardiogenic transcription factors like gata4 (Kikuchi et al. 2010; Sallin et al. 2015; Wang et al. 2013; Wu et al. 2016). Poss and colleagues found that a transgenic line driving GFP from regulatory sequences of gata4, a cardiac transcription factor expressed during embryonic heart development, is re-expressed in a subpopulation of CMs in the compact layer myocardium near the wound from three to seven dpi in injured hearts (Fig. 10.7b). By 14 dpi, GFP-positive cells were found in many cells around and within the injury site. A sub-set of these gata4:GFP-expressing CMs expressed markers of cell cycle activity, and their descendants, genetically marked by inducible gata4:CreERT2 from five to seven dpi, contributed significantly to the regenerated myocardium (Kikuchi et al. 2010). While strong evidence for the re-expression of endogenous gata4 or other cardiogenic factors in CMs in injured hearts is still missing, overexpression of a dominant-negative form of Gata4 reduced CM proliferation in the compact layer myocardium, compromised myocardial regeneration at 30 dpi and resulted in extensive scarring (Gupta et al. 2013). Thus, reactivation of gata4 in the compact layer myocardium appears to be crucial for successful heart regeneration (Gupta et al. 2013).
Izpisua-Belmonte and colleagues also suggested that dedifferentiation is of functional importance for CMs to proliferate since no mitotic CMs displayed highly organized sarcomeric structures (Jopling et al. 2010). However, mammalian CMs also reactivate expression of embryonic genes and show partial loss of sarcomeric structure after heart injury (D’Uva et al. 2015; Kubin et al. 2011). Therefore, CM dedifferentiation could be a shared injury response between the non-regenerating mammalian and the regenerating zebrafish heart (Szibor et al. 2014). Dedifferentiation thus might represent a prerequisite for proliferation, but likely is not sufficient or causal for CM proliferation, considering that mammalian CMs do not proliferate in response to heart injury. More detailed systematic analyses of molecular changes in CMs induced by injury in both models and the identification of experimental interventions that specifically block dedifferentiation will be essential to understand the functional role of CM dedifferentiation during heart regeneration.
Migration of CMs into the injured area, regulated by the Cxcl12-Cxcr4 system, has also been suggested to be an important event during regeneration (Itou et al. 2012). Pharmacological inhibition of Cxcr4 reduced the number of CMs in the injury site at 14 dpi and compromised myocardial regeneration at 60 dpi without affecting CM proliferation. While these data have been interpreted as indicative of the importance of CM migration during regeneration, more direct evidence for active CM migration is lacking.
10.5.3 Molecular Regulation of Cardiomyocyte Regeneration
Several studies have identified factors and signaling pathways regulating CM proliferation during zebrafish heart regeneration. Obvious candidates for such factors are those that also regulate embryonic CM proliferation during heart development. To screen for molecular regulators of embryonic CM proliferation, Poss and colleagues employed the fluorescent ubiquitination-based cell cycle indicator (FUCCI ) system, which comprises two fusion proteins, mCherry-zCdt1 and Venus-hGeminin, which are expressed cyclically in the G1 and S/G2/M phases of the cell cycle, respectively (Choi et al. 2013). By combining transgenic expression of these proteins specifically in CMs (myl7:FUCCI) with a small-scale chemical screen targeting common developmental signaling pathways, Hedgehog (Hh), Insulin-like growth factor (Igf), and Transforming growth factor ß (Tgfß) signaling were identified as positive regulators of CM proliferation during development (Choi et al. 2013).
10.5.3.1 Hedgehog Signaling
Hh signaling has been implicated in driving cardiac specification in the zebrafish embryo and reduction of Hh signaling causes cardiac morphogenesis defects in mice (Washington Smoak et al. 2005). Treatment of myl7:FUCCI embryos with the Smoothened agonist SAG, which activates Hh signaling, increased CM proliferation by 60 % as well as CM numbers by 10 % (Choi et al. 2013). On the other hand, treatment with cyclopamine (a Smoothened antagonist) reduced CM proliferation and numbers by 27 % and 19 %, respectively. These data suggest that Hh signaling is required for embryonic CM proliferation.
Interestingly, Hh signaling was also activated in CMs in the adult zebrafish heart 7 days after ventricular resection, as indicated by the expression of the Hh target gene ptch2 in a transgenic reporter line (Choi et al. 2013). Treatment with cyclopamine 6 days after ventricular resection or genetic CM ablation reduced expression of the cell cycle maker PCNA in CMs. SAG treatment, on the other hand, increased CM cell cycle activity remarkably by 65 % after ventricular resection while it had no effect on adult uninjured zebrafish hearts. Together, these data suggest that zebrafish CM proliferation requires Hh signaling, which on its own is not sufficient to trigger regenerative proliferation (Choi et al. 2013). Hh ligands might be provided by the epicardium, which was reported to express a transgenic sonic hedgehog reporter line (Choi et al. 2013). Whether Hh signaling acts directly on CMs and is indeed activated by epicardium-derived ligands will have to be clarified using tissue-specific experimental manipulations.
Hh signaling was also found to be activated in the mouse myocardium after myocardial infarction (Kusano et al. 2005). Injection of Sonic Hedgehog (Shh) plasmid into infarcted hearts was sufficient to promote neovascularization, to protect CMs from apoptosis, to reduce post-MI remodeling and to improve functional recovery (Kusano et al. 2005). Therefore, although the beneficiary effects of Hh signaling on cardiac repair are conserved between zebrafish and mammals, the mode of actions seems to be different. It would be interesting to characterize the interaction partners and downstream targets of Hh signaling in both models to understand the molecular basis behind these differential responses to the activation of the same signaling pathway.
10.5.3.2 Insulin-Like Growth Factor Signaling
Igf signaling has been described to have positive effects on CM proliferation during mouse development (Li et al. 2011). In myl7:FUCCI zebrafish embryos, treatment with the Igf receptor antagonist NVP AEW541 reduced CM proliferation and number by 33 % and 14 %, respectively (Choi et al. 2013). This proliferation defect was verified by Lien and colleagues in an independent study (Huang et al. 2013). On the other hand, treatment with an Igf signaling agonist increased embryonic CM proliferation and number by 41 % and 17 %, respectively (Choi et al. 2013). These data suggest that Igf signaling is required for CM proliferation in the developing mammalian and zebrafish heart. Components of the Igf signaling pathway were also found to be upregulated in the adult zebrafish heart in response to injury. RT-PCR analysis showed that expression of the ligand igf2b is upregulated by three dpi and peaks at seven dpi, in a temporal profile similar to that of CM proliferation, which peaks between seven and 14 dpi. igf2b expression appeared to be restricted to endocardial cells in the injury site as detected by in situ hybridization while Igf receptor 1 was expressed in CMs close to the amputation plane (Choi et al. 2013; Huang et al. 2013). Systemic Igf signaling inhibition in adult fish either by treatment with the NVP AEW541 antagonist or by overexpression of a dominant-negative form of Igf receptor 1 (hspl70:dnigf1ra) reduced PCNA expression in CMs after injury. Lien and colleagues found that activation of gata4:EGFP in a subset of CMs at the wound border was not affected by Igf receptor antagonist treatment, while Igf signaling inhibition blocked proliferation of this population and appeared to reduce their accumulation in the injured area by 14 dpi (Huang et al. 2013). Prolonged treatment with antagonist until 30 dpi inhibited regeneration, resulting in excessive fibrin and collagen deposition (Huang et al. 2013). On the other hand, overactivation of Igf signaling was sufficient to increase CM proliferation by 65 % and 36 % after ventricular resection and genetic CM ablation, respectively (Choi et al. 2013; Huang et al. 2013). Together, these data suggest that endocardium-derived Igf ligands regulate regenerative CM proliferation and successful regeneration. Again, whether proliferation is cell-autonomously regulated by Igf signaling activation in CMs and whether it is indeed activated by endocardially produced ligands will have to be clarified using tissue-specific manipulations.
10.5.3.3 Transforming Growth Factor ß Signaling
Treatment of myl7:FUCCI embryos with the Tgfß receptor inhibitor SB431542 reduced CM proliferation and number by 30 %, suggesting that Tgfß signaling promotes CM proliferation during development (Choi et al. 2013). The role of Tgfß signaling during zebrafish adult heart regeneration was further studied by Chablais and Jaźwińska (Chablais and Jazwinska 2012). Following cryoinjury, expression of three Tgfß isoforms (tgfß1, tgfß2 and tgfß3) was upregulated in the wound at four and 14 dpi as detected by in situ hybridization. These Tgfß isoforms appeared to be expressed in several cell types including fibroblasts, epithelial cells of the wound, as well as some CMs at the wound border. Tgfß receptors, on the other hand, appeared to be expressed in the wound (both alk4 and alk5a) and in the entire heart possibly in Vimentin-positive fibroblasts (alk5a), suggesting that both CMs and non-CMs can be responsive to Tgfß ligands. This idea was verified by immunostaining against phosphorylated Smad3, a readout for active Tgfß signaling, which was expressed in a large number of cells in the wound and in a sub-set of CMs at the wound border (Chablais and Jazwinska 2012). To understand the functional role of Tgfß signaling, cryoinjured fish were treated with the Tgfß receptor antagonist SB431542 and analyzed by histological staining at different time points. At four dpi, in both control and treated fish, fibrin deposition was evident in the wound. Interestingly, at 14 dpi, while scar tissue containing fibrin and collagen was evident in control hearts (Chablais et al. 2011; Gonzalez-Rosa et al. 2011), Tgfß signaling-inhibited fish failed to deposit collagen in the wound (Chablais and Jazwinska 2012). At 30 dpi, while little scar tissue could be detected in control hearts, implying that the fibrin and collagen depositions present at 14 dpi had been resolved and replaced by new muscle, Tgfß inhibitor-treated fish still contained fibrin-rich wound tissue, which showed signs of mechanical deformation (Chablais and Jazwinska 2012). These data suggest an unexpected positive correlation between transient collagen deposition and successful heart regeneration in zebrafish. Based on drug-shift experiments, Chablais and Jaźwińska suggested that Tgfß signaling-mediated transient collagen-rich scar formation is required to stabilize the injured myocardium (Chablais and Jazwinska 2012). In addition to collagen deposition, Tgfß also appears to be required for the production of other ECM proteins including the tissue remodeling protein Tenascin C and Fibronectin, which is crucial for zebrafish heart regeneration (Chablais and Jazwinska 2012; Wang et al. 2013). Lastly, consistent with its role during embryonic development, Tgfß signaling inhibition reduced CM proliferation detected by either PCNA expression or BrdU incorporation (Chablais and Jazwinska 2012; Choi et al. 2013). Altogether, these studies suggest that Tgfß signaling is required for both CM proliferation and collagen deposition in the wound after injury. Chablais and Jaźwińska’s study is the first to provide evidence for the functional importance of transient scarring during zebrafish heart regeneration (Chablais et al. 2011; Gonzalez-Rosa et al. 2011). This observation challenges the prevalent concept in the heart regeneration field that scarring and regeneration is mutually exclusive, and also opens up a new area of research into the molecular mechanisms of transient scar resolution. Similar to the pathways discussed above, future studies using cell-type specific manipulations will be necessary to clarify whether the effects of Tgfß signaling on CM proliferation are direct.
10.5.3.4 Jak1/Stat3 Signaling
Several studies have performed transcriptional profiling of regenerating zebrafish hearts using whole ventricle samples (Lien et al. 2006; Sleep et al. 2010). Poss and colleagues performed profiling of RNAs that are specifically translated in zebrafish CMs using translating ribosome affinity purification (TRAP) technology (Fang et al. 2013). To do this, an EGFP reporter gene was fused to the N-terminus of the ribosomal protein L10a, whose expression was driven specifically in CMs under the regulatory sequence of myl7 (myl7:TRAP). Translating mRNAs were isolated by immunoprecipitation with an antibody against EGFP, which were then processed for microarray analysis. Among the differentially regulated mRNAs, several members of the Janus kinases1/Signal transducer and activator of transcription 3 (Jak1/Stat3) pathway were found to be upregulated in 1 dpi samples, including interleukin 6 signal transducer (il6st), jak1, stat3 and the Jak1/Stat3 target gene suppressor of cytokine signaling 3b (socs3b) (Fang et al. 2013). In situ hybridization showed that these Jak1/Stat3 pathway genes are induced in an organ-wide manner at 1 dpi but later expression appeared to localize to CMs at the amputation plane. To investigate the role of Jak1/Stat3 signaling during heart regeneration, Poss and colleagues used a Cre/lox-based system for inducible overexpression of a dominant-negative Stat3 (dnStat3) in CMs upon 4-hydroxytamoxifen induction, which was able to reduce expression of the target gene socs3b by 80 % following injury. Continuous dnStat3 expression from prior to heart injury compromised myocardial regeneration by 30 dpi and resulted in extensive scarring compared to control fish. In line with this, CM proliferation was also strongly reduced (~80 %) in dnStat3-expressing fish (Fang et al. 2013). Expression of relaxin 3a (rln3a), which codes for a peptide hormone, was found to be directly regulated by Stat3 in the regenerating heart. Daily retro-orbital injection of Rln3a protein in dnStat3-expressing fish partially rescued CM proliferation defects at seven dpi, suggesting that the effect of Jak1/Stat3 signaling on heart regeneration is at least partly mediated by Rln3a (Fang et al. 2013).
In contrast to the signaling pathways described above, the requirement of Jak1/Stat3 signaling for CM proliferation appears to be regeneration-specific. Larvae subjected to dnStat3 overexpression from 4 days post fertilization survived to adulthood, suggesting that Jak1/Stat3 signaling is not required for heart development (Fang et al. 2013). Furthermore, CM proliferation in juvenile and young adult zebrafish stimulated by low density growing conditions (Wills et al. 2008) was not affected by dnStat3 expression (Fang et al. 2013). Altogether, these data identify Jak1/Stat3 signaling as an injury-induced pathway that directly regulates regenerative CM proliferation.
10.5.3.5 Notch Signaling
Treatment of myl7:FUCCI embryos with a Notch signaling inhibitor (γ-secretase inhibitor) had no effect on CM proliferation, suggesting that Notch signaling is dispensable for CM proliferation in the embryonic heart (Choi et al. 2013). In contrast, Burns and colleagues found that Notch signaling is required for adult CM regeneration (Zhao et al. 2014). In adult zebrafish, three out of four Notch receptors (notch1a, notch1b and notch2) were upregulated in both endocardial and epicardial cells but not in CMs 7 days after ventricular resection as revealed by in situ hybridization (Zhao et al. 2014). To understand the role of Notch signaling during regeneration, Burns and colleagues inhibited Notch signaling by ubiquitously overexpressing a dominant negative isoform of the murine mastermind-like protein (dnMAML), which cannot recruit essential cofactors to the Notch transcriptional complex, rendering the complex inert. Continuous systemic Notch inhibition compromised myocardial regeneration in the resected ventricular apex by 30 dpi and resulted in extensive scarring with collagen deposition, suggesting that Notch signaling is required for heart regeneration (Zhao et al. 2014). Although Notch receptors were predominantly upregulated in endocardial and epicardial cells after injury, Notch signaling inhibition did not interfere with the early injury responses reported previously in both cell types (Kikuchi et al. 2011b; Lepilina et al. 2006), namely the formation of epicardium-derived cells and activation of aldh1a2 expression in both epicardium and endocardium at seven dpi (Zhao et al. 2014). Interestingly, while myocardial regeneration was compromised in Notch-inhibited hearts at 30 dpi, coronary endothelium regeneration as detected by kdrl:mCherry transgenics was not inhibited, indicating that Notch signaling is dispensable for neovascularization after injury. Cardiomyocyte proliferation, on the other hand, was found to be regulated by Notch signaling since continuous overexpression of dnMAML from one to seven dpi significantly reduced PCNA expression in CMs by 53 %. Intriguingly, Notch hyperactivation by systemic overexpression of the intracellular domain of the Notch receptor (NICD) also significantly reduced CM proliferation at seven dpi as well as myocardial regeneration at 30 dpi, suggesting that the right balance of Notch signaling activity is essential for successful heart regeneration (Zhao et al. 2014).
10.5.3.6 Neuregulin1 Signaling
As discussed above, while a number of pathways/factors have been identified to regulate CM regeneration, none of them are able to stimulate CM hyperplasia in the absence of injury. In contrast, a recent study by Poss and colleagues showed that Neuregulin1 (Nrg1) is a potent injury-induced mitogen for zebrafish heart regeneration and that its activation in the healthy adult heart is sufficient to induce rapid regenerative programs, resulting in excessive myocardial hyperplasia and cardiomegaly (Gemberling et al. 2015). Neuregulin1 is an extracellular factor of the EGF family that might act as a transmembrane protein, but can also give rise to secreted peptides due to ectodomain shedding (Mei and Nave 2014).
After genetic ablation of CMs, nrg1 expression was rapidly induced in the heart at three and seven dpi, coinciding with induction of CM proliferation. nrg1 was predominantly expressed in perivascular regions in the ventricular wall after genetic ablation and in the region surrounding the injury site after ventricular resection, as revealed by in situ hybridization. Co-localization studies with transgenic reporter lines marking different cardiac cell types showed that the dominant source of nrg1 in the ventricular wall is tcf21-expressing epicardial cells. After ventricular resection, treatment with a small molecule inhibitor of Erbb receptors, which are required for Nrg1 signaling, reduced CM proliferation at seven dpi by 54 %. On the other hand, inducible overexpression of Nrg1 specifically in CMs using the Cre-Lox system increased proliferation at seven dpi by 84 %. These data suggest that Nrg1 is a potent mitogenic factor for CMs after injury (Gemberling et al. 2015).
Using the same CM-specific overexpression system, Poss and colleagues tested the effect of Nrg1 on the uninjured adult heart. Nrg1 overexpression for 7 days significantly induced CM proliferation, resulting in a remarkable thickening of the ventricular wall by 460 % after 30 days of overexpression (Gemberling et al. 2015). These changes are likely the result of CM hyperplasia instead of hypertrophy since CM cell size was not affected by continuous Nrg1 overexpression. However, prolonged Nrg1-induced hyperplasia compromised cardiac function as revealed by echocardiography and a swimming test which is an assay for cardiac function that is sensitive for heart injury and failure (Wang et al. 2011).
Nrg1 overexpression in the uninjured heart was sufficient to elicit responses similar to those seen during heart regeneration. During heart development, cortical muscle in the ventricular wall was reported to be typically formed from a small number of CMs, which expand laterally on the ventricular surface to form large clones, as revealed by CM clonal analysis (Gupta and Poss 2012). In contrast, after heart injury, CM regeneration appears to occur through proliferation near the injury site, which results in a number of small CM clones in the regenerated ventricular wall (Gupta et al. 2013). Overexpression of Nrg1 in juvenile fish 5 weeks post fertilization (wpf), a time at which cortical muscle first appears, resulted in ectopic ventricular wall thickening by 10 wpf and clonal analysis showed obvious clone mixing, which is more reminiscent of injury-induced regeneration (Gemberling et al. 2015). Furthermore, Nrg1 overexpression also activated the regulatory sequence of gata4, as well as the expression of tgfß2, aldh1a2 and fibronectin 1 which have all been functionally implicated in zebrafish heart regeneration (Gupta et al. 2013; Kikuchi et al. 2010, 2011b; Chablais and Jazwinska 2012; Wang et al. 2013). Finally, CM dedifferentiation, epicardial activation and myocardial vascularization were also rapidly induced by Nrg1 overexpression in uninjured hearts (Gemberling et al. 2015). Together, these data suggest that Nrg1 signaling is a potent inducer of regenerative programs that involve different cardiac cell types. Such a mitogenic and pro-regenerative role of Nrg1 on CMs appears to be conserved between zebrafish and mammals since both injection of Nrg1 or expression of a constitutively active Erbb2 has been reported to induce CM proliferation both in vitro and in vivo and to improve functional recovery after MI (Bersell et al. 2009; D’Uva et al. 2015).
10.5.3.7 Bone Morphogenetic Protein signaling
To identify regulators of zebrafish CM regeneration, Weidinger, Bakkers and colleagues employed Tomo-Seq, a recently developed method providing genome-wide transcriptional expression data with spatial resolution (Wu et al. 2016). This resulted in the identification of several hundred genes whose expression is enriched at the wound border, where CMs re-enter the cell cycle. Ligands, receptors and target genes of Bone Morphogenetic Protein (BMP) signaling were found to be expressed at the wound border, and nuclear accumulation of phosphorylated Smad1/5/8, a readout of active BMP signaling, was detected in dedifferentiating CMs at the wound border. Systemic interference with BMP signaling showed that the pathway is required for CM proliferation and muscle regeneration, while overexpression of the ligand Bmp2b was sufficient to augment CM proliferation and to speed up muscle regeneration (Wu et al. 2016). Weidinger, Bakkers and colleagues found that BMP signaling is not required for physiological CM proliferation in juvenile fish, indicating that it represents an injury-specific regulator of CM regeneration. In part, this might be due to BMP signaling being required for CM dedifferentiation (Wu et al. 2016). Interestingly, BMP signaling has also been reported to be upregulated in injured mammalian hearts, but there it appears to play the opposite role than in zebrafish. Interference with BMP signaling protects mammalian CMs from apoptosis and improves the outcome after MI (Pachori et al. 2010). These results indicate that the differential ability of zebrafish and mammalian hearts to regenerate depends in part on opposing responses of CMs to BMP signaling.
10.5.3.8 NF-κB signaling
NF-κB signaling has also been identified as important regulator of zebrafish heart regeneration by Poss and colleagues (Karra et al. 2015). Using a transgenic reporter line, NF-κB activity was detected in CMs adjacent to the amputation plane throughout the first two weeks after injury. Inhibition of NF-κB activity specifically in CMs inhibited CM proliferation and myocardial regeneration at 7 and 30 dpi, respectively. In addition, NF-κB transcriptional complexes also appear to contribute to CM dedifferentiation by directly activating transcription of gata4. Interestingly, myocardial inhibition of NF-κB activity also resulted in defects in infiltration of epicardial cells into the wound at 14 dpi, suggesting a potential cross-talk of CMs and epicardial cells during regeneration (Karra et al. 2015).
10.5.3.9 p38α MAPK
The activity of p38α MAPK is inversely correlated with CM proliferation in mammals; inhibition of p38α MAPK induces proliferation of cultured neonatal and adult CMs, which are normally non-proliferative (Engel et al. 2005). Moreover, co-treatment of adult rats with FGF1 and p38α MAPK inhibitor after MI reduced scarring and markedly improved functional recovery (Engel et al. 2006). A study from Izpisua-Belmonte and colleagues revealed a similar role of p38α MAPK on CM proliferation in zebrafish (Jopling et al. 2012b). To manipulate p38α MAPK activity, Izpisua-Belmonte and colleagues targeted expression of either a dominant negative form of p38α MAPK (dnp38α MAPK) or a constitutive active (ca) MKK6, an upstream activator of p38α MAPK, to CreERT2-expressing CMs upon 4-hydroxytamoxifen (4-HT)-mediated induction (Jopling et al. 2012b). While p38α MAPK inhibition had no effect on myocardial regeneration after ventricular resection, caMKK6 overexpression significantly inhibited myocardial regeneration at 30 dpi. Moreover, mosaic expression of caMKK6 using a partially silenced CM-specific Cre line showed that caMKK6-expressing CMs were unable to incorporate BrdU, suggesting that p38α MAPK activity inhibits CM proliferation and regeneration (Jopling et al. 2012b). These results suggest that p38α MAPK activity in adult zebrafish CMs must be low for heart regeneration to occur.
10.5.3.10 Hypoxia
Hypoxia-inducible factors (HIFs) are the direct effectors of the hypoxic response. In mammals, it has been reported that HIF1α overexpression in the myocardium reduces infarct size and improves functional recovery after MI (Kido et al. 2005). Izpisua-Belmonte and colleagues reported that at 7 days post ventricular resection in zebrafish, the wound tissue and a subset of CMs close to the amputation plane showed hypoxia induction as evidenced by Hypoxyprobe staining (Jopling et al. 2012a). Cardiomyocyte BrdU incorporation and regeneration in adult zebrafish preconditioned with phenylhyrazine, an anemia/hypoxia-inducing drug, was enhanced, while zebrafish overexpressing a dominant-negative form of HIF1α in CMs showed inhibition of these processes. Together, these data suggest that hypoxia is required for myocardial regeneration in zebrafish (Jopling et al. 2012a). In cultured zebrafish CMs hypoxic treatment appeared to increase the number of mitotic and dedifferentiated CMs (Jopling et al. 2012a). Whether or not these effects could also be observed in vivo is, however, unclear.
10.5.3.11 Hydrogen Peroxide
Reactive oxygen species (ROS) , despite their toxic potential, are crucial for regeneration of the tadpole tail in Xenopus and of the caudal fin in zebrafish (Gauron et al. 2013; Love et al. 2013). A study by Xiong and colleagues revealed a positive role of hydrogen peroxide (H2O2) in zebrafish heart regeneration as well (Han et al. 2014). After ventricular resection, duox, a member of the NADPH-oxidase and related dual oxidase family of proteins, which is responsible for the formation of superoxide anions and subsequently H2O2, was upregulated in epicardial cells at the site of injury from three to 14 dpi (Han et al. 2014). In line with the temporal expression profile of duox, the level of H2O2 in CMs was also found to be elevated between three and 14 dpi, as detected by expression of Hyper, a fluorescent protein-based H2O2 sensor (Belousov et al. 2006), in CMs in myl7:Hyper transgenics. Pharmacological inhibition of Duox activity by diphenyleneiodonium (DPI) or apocynin reduced H2O2 production and CM proliferation at seven dpi. Prolonged treatment with DPI also compromised myocardial regeneration and resulted in scarring. These data suggest that H2O2 is required for CM proliferation and regeneration (Han et al. 2014).
Intriguingly, H2O2 was found to regulate CM proliferation and regeneration through its interaction with Erk1/2 signaling and its feedback regulator Dusp6 (Han et al. 2014). Dusp6 expression is stimulated by phosphorylated Erk1/2 (pErk1/2) but reciprocally deactivates pErk1/2 by dephosphorylation, hence forming a negative feedback loop that limits Erk1/2 activity (Nichols et al. 2000). H2O2, on the other hand, directly oxidizes Dusp6, which renders the protein prone for degradation, and hence leads to Erk1/2 overactivation (Chan et al. 2008). In zebrafish, both phosphorylation of Erk1/2 and dusp6 expression were found to be activated after ventricular resection (Han et al. 2014). DPI treatment in regenerating hearts induced Dusp6 stability and therefore reduced phosphorylation of Erk1/2. The effect of DPI on CM proliferation could be counteracted by co-treatment with BCI, a small molecule that inhibits phosphatase activity of Dusp6. In addition, systemic overexpression of Dusp6 in hspl70:dusp6 transgenics reduced pErk1/2 expression, and compromised CM proliferation, gata4 activation and myocardial regeneration. These data strongly suggest that endogenous H2O2 regulates CM proliferation and regeneration by promoting Erk1/2 signaling through repressing Dusp6 activity (Han et al. 2014).
10.5.3.12 miRNAs
miRNAs have been implicated in various responses to MI in mammals, including cell death, proliferation, and metabolism (reviewed in Wang and Martin 2014). In zebrafish, several studies have demonstrated the importance of regulation of miRNAs for successful heart regeneration (Yin et al. 2012; Aguirre et al. 2014).
Using microarrays and real-time quantitative PCR (qPCR), Poss and colleagues found that a number of miRNAs are differentially regulated 7 days after ventricular resection, a large number of which have been shown to be modulated in injured mammalian heart (Yin et al. 2012). One of the identified miRNAs was the CM-specific miR-133, which had been found to negatively regulate zebrafish fin regeneration (Yin et al. 2008). In situ hybridization and qPCR analyses demonstrated that miR-133 levels decrease by seven and 14 dpi compared to uninjured hearts, and later return to baseline level at the completion of heart regeneration by 30 and 60 dpi, suggesting that downregulation of miR-133 levels could be important for heart regeneration (Yin et al. 2012). Indeed, loss- and gain-of-function experiments demonstrated that miR-133 negatively regulates CM proliferation and regeneration. Systemic inducible overexpression of the miR-133 precursor sequence increased the levels of mature miR-133 and reduced CM proliferation and regeneration at seven and 30 dpi, respectively (Yin et al. 2012). On the other hand, inducible overexpression of a miR-133 sponge RNA, which contained triplicate perfect binding sites for miR-133, reduced the levels of miR-133 and induced CM proliferation at seven and 30 dpi. Poss and colleagues further identified the gap junctional protein connexin-43 (cx43) and the mitotic check point kinase mps1 as targets of miR-133. While Mps1 mutants showed heart regeneration defects (Poss et al. 2002), treatment with carbenoxolone (CBX), an inhibitor of Cx43, reduced CM proliferation at seven dpi after ventricular resection (Yin et al. 2012). Together, these data demonstrate the importance of downregulation of miR-133 levels during heart regeneration, whose effect is mediated through, at least partly, Mps1 and Cx43 (Yin et al. 2012).
Yin and colleagues recently also described that downregulation of miR-101a after zebrafish heart injury is essential for CM proliferation (Beauchemin et al. 2015). In addition, this miR also seems to regulate transient scar removal. While its expression is downregulated shortly after injury, expression was found to actually increase above levels in uninjured hearts between seven and 14 dpi. Depletion of miR-101a during this time using transgenic overexpression of a sponge construct resulted in increased scarring by 30 dpi (Beauchemin et al. 2015). Thus, miR-101a appears to play a dual role during heart regeneration, where its downregulation early allows CM proliferation, while increased levels later are required for scar removal.
The miRNAs miR-99/100 and Let-7a/c were also identified as important regulators of zebrafish heart regeneration and differential regulation of their expression was suggested as part of the reason why zebrafish can but adult mammals cannot regenerate their heart. Izpisua-Belmonte and colleagues showed that expression of these miRNAs is downregulated during zebrafish heart regeneration (Aguirre et al. 2014). Injection of miR-99/100 mimics into regenerating hearts reduced CM proliferation and compromised myocardial regeneration, which was suggested to be mediated through their targets beta subunit of farnesyl-transferase (Fnt) and SWI/SNF-related matrix associated actin-dependent regulator of chromatin subfamily a, member 5 (Smarca5). Interestingly, downregulation of miR-99/100 and Let-7a/c and subsequent upregulation of Fnt and Smarca5 was not observed in mouse hearts after MI (Aguirre et al. 2014). Blockage of both miRNAs in mouse after MI resulted in CM dedifferentiation and improved functional recovery (Aguirre et al. 2014). These data suggest that downregulation of miR-99/100 and Let-7a/c is an essential regenerative response, which is conserved but dormant in the mammalian heart. This study demonstrates the importance of zebrafish as a model for deciphering molecular mechanisms underlying natural heart regeneration which could be applied to heal the non-regenerating mammalian heart.
10.6 Conclusions
The zebrafish has rapidly developed over the last decade as a model for studying cardiac regeneration. Recent discoveries have greatly improved our understanding of how natural heart regeneration is regulated. After injury, developmental gene programs are activated globally in the endocardium and epicardium, which proliferate and contribute to the regenerated endocardial (including new vasculature) and epicardial cells in a lineage-restricted manner, respectively. Activation of these two layers precedes that of CMs and is likely to provide endocardial scaffold and epicardial covering of the wound to support muscle regeneration. In addition, these two layers also act as a source of pro-regenerative factors which affect CM proliferation and regeneration in a paracrine manner, likely including ligands of FGF, RA, Igf, Tgfß and Notch signaling , and Nrg1.
In response to injury, CMs have been shown to partially dedifferentiate, re-enter the cell cycle and proliferate to regenerate the lost muscle. Several signaling pathways have been identified as being required for regenerative CM proliferation, most of which appear to be also involved in regulating CM proliferation during embryonic development (RA, Hh, Igf, Tgfß), while injury-specific regulators have been identified as well (Jak1/Stat3, H2O2, Notch, BMP). Since most pathways have been studied using systemic interference, the strongest evidence for a direct role on CMs exists for Jak1/Stat3 and BMP signaling; cell type-specific pathway manipulation will be required in the future to clarify cellular interactions during heart regeneration. Currently, heart regeneration studies in adult zebrafish are also limited by the lack of inducible genetic loss-of-function systems. Hopefully, in the near future, genome-editing tools like CRISPR/Cas9 will enable zebrafish researchers to induce tissue-specific genetic deletion of genes to further dissect the requirement of various factors or signaling pathway components during heart regeneration. Together with rapidly developing transcriptomic, proteomic and epigenomic tools, future studies promise to provide insights to important outstanding questions such as: (1) Why do zebrafish but not mammalian CMs retain the ability to proliferate throughout life? (2) How are regenerative responses initiated in the zebrafish heart? (3) What are the underlying mechanisms of scar removal in zebrafish? We believe the zebrafish will continue to thrive as an important model to understand natural heart regeneration, which might provide valuable insights to refine strategies for improving mammalian cardiac repair.
References
Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, Kumar S, Moresco JJ, Yates JR 3rd, Campistol JM, Sancho-Martinez I, Giacca M, Izpisua Belmonte JC (2014) In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 15(5):589–604. doi:10.1016/j.stem.2014.10.003
Ali SR, Hippenmeyer S, Saadat LV, Luo L, Weissman IL, Ardehali R (2014) Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci U S A 111(24):8850–8855. doi:10.1073/pnas.1408233111
Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S (2006) Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 3(4):281–286. doi:10.1038/nmeth866
Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J (2009) Evidence for cardiomyocyte renewal in humans. Science 324(5923):98–102. doi:10.1126/science.1164680
Bersell K, Arab S, Haring B, Kuhn B (2009) Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138(2):257–270. doi:10.1016/j.cell.2009.04.060
Beauchemin M, Smith A, Yin VP (2015) Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration. Development 142(23):4026–4037. doi:10.1242/dev.126649
Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM (2008) A myocardial lineage derives from Tbx18 epicardial cells. Nature 454(7200):104–108. doi:10.1038/nature06969
Chablais F, Jazwinska A (2012) The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 139(11):1921–1930. doi:10.1242/dev.078543
Chablais F, Veit J, Rainer G, Jazwinska A (2011) The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol 11:21. doi:10.1186/1471-213X-11-21
Chan DW, Liu VW, Tsao GS, Yao KM, Furukawa T, Chan KK, Ngan HY (2008) Loss of MKP3 mediated by oxidative stress enhances tumorigenicity and chemoresistance of ovarian cancer cells. Carcinogenesis 29(9):1742–1750. doi:10.1093/carcin/bgn167
Choi WY, Gemberling M, Wang J, Holdway JE, Shen MC, Karlstrom RO, Poss KD (2013) In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140(3):660–666. doi:10.1242/dev.088526
D’Uva G, Aharonov A, Lauriola M, Kain D, Yahalom-Ronen Y, Carvalho S, Weisinger K, Bassat E, Rajchman D, Yifa O, Lysenko M, Konfino T, Hegesh J, Brenner O, Neeman M, Yarden Y, Leor J, Sarig R, Harvey RP, Tzahor E (2015) ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol. doi:10.1038/ncb3149
Dettman RW, Denetclaw W Jr, Ordahl CP, Bristow J (1998) Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol 193(2):169–181. doi:10.1006/dbio.1997.8801
Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, Waring CD, Henning BJ, Stirparo GG, Papait R, Scarfo M, Agosti V, Viglietto G, Condorelli G, Indolfi C, Ottolenghi S, Torella D, Nadal-Ginard B (2013) Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 154(4):827–842. doi:10.1016/j.cell.2013.07.039
Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT (2005) p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev 19(10):1175–1187. doi:10.1101/gad.1306705
Engel FB, Hsieh PC, Lee RT, Keating MT (2006) FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A 103(42):15546–15551. doi:10.1073/pnas.0607382103
Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M (2012) Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492(7429):376–381. doi:10.1038/nature11739
Fang Y, Gupta V, Karra R, Holdway JE, Kikuchi K, Poss KD (2013) Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci U S A 110(33):13416–13421. doi:10.1073/pnas.1309810110
Gauron C, Rampon C, Bouzaffour M, Ipendey E, Teillon J, Volovitch M, Vriz S (2013) Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci Rep 3:2084. doi:10.1038/srep02084
Gemberling M, Bailey TJ, Hyde DR, Poss KD (2013) The zebrafish as a model for complex tissue regeneration. Trends Genet 29(11):611–620. doi:10.1016/j.tig.2013.07.003
Gemberling M, Karra R, Dickson AL, Poss KD (2015) Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife 4:e05871. doi:10.7554/eLife.05871
Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N (2011) Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 138(9):1663–1674. doi:10.1242/dev.060897
Gonzalez-Rosa JM, Peralta M, Mercader N (2012) Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev Biol 370(2):173–186. doi:10.1016/j.ydbio.2012.07.007
Gonzalez-Rosa JM, Guzman-Martinez G, Marques IJ, Sanchez-Iranzo H, Jimenez-Borreguero LJ, Mercader N (2014) Use of echocardiography reveals reestablishment of ventricular pumping efficiency and partial ventricular wall motion recovery upon ventricular cryoinjury in the zebrafish. PLoS One 9(12):e115604. doi:10.1371/journal.pone.0115604
Gupta V, Poss KD (2012) Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 484(7395):479–484. doi:10.1038/nature11045
Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, Poss KD (2013) An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol 23(13):1221–1227. doi:10.1016/j.cub.2013.05.028
Han P, Zhou XH, Chang N, Xiao CL, Yan S, Ren H, Yang XZ, Zhang ML, Wu Q, Tang B, Diao JP, Zhu X, Zhang C, Li CY, Cheng H, Xiong JW (2014) Hydrogen peroxide primes heart regeneration with a derepression mechanism. Cell Res 24(9):1091–1107. doi:10.1038/cr.2014.108
Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF (2013) Hippo signaling impedes adult heart regeneration. Development 140(23):4683–4690. doi:10.1242/dev.102798
Hein SJ, Lehmann LH, Kossack M, Juergensen L, Fuchs D, Katus HA, Hassel D (2015) Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS One 10(4):e0122665. doi:10.1371/journal.pone.0122665
Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT (2007) Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 13(8):970–974. doi:10.1038/nm1618
Hu N, Yost HJ, Clark EB (2001) Cardiac morphology and blood pressure in the adult zebrafish. Anat Rec 264(1):1–12
Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, Sucov HM, Lien CL (2013) Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS One 8(6):e67266. doi:10.1371/journal.pone.0067266
Huang CC, Su TH, Shih CC (2015) High-resolution tissue Doppler imaging of the zebrafish heart during its regeneration. Zebrafish 12(1):48–57. doi:10.1089/zeb.2014.1026
Ito K, Morioka M, Kimura S, Tasaki M, Inohaya K, Kudo A (2014) Differential reparative phenotypes between zebrafish and medaka after cardiac injury. Dev Dyn 243(9):1106–1115. doi:10.1002/dvdy.24154
Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y (2012) Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 139(22):4133–4142. doi:10.1242/dev.079756
Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC (2010) Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464(7288):606–609. doi:10.1038/nature08899
Jopling C, Sune G, Faucherre A, Fabregat C, Izpisua Belmonte JC (2012a) Hypoxia induces myocardial regeneration in zebrafish. Circulation 126(25):3017–3027. doi:10.1161/CIRCULATIONAHA.112.107888
Jopling C, Sune G, Morera C, Izpisua Belmonte JC (2012b) p38alpha MAPK regulates myocardial regeneration in zebrafish. Cell Cycle 11(6):1195–1201. doi:10.4161/cc.11.6.19637
Kang BJ, Park J, Kim J, Kim HH, Lee C, Hwang JY, Lien CL, Shung KK (2015) High-frequency dual mode pulsed wave Doppler imaging for monitoring the functional regeneration of adult zebrafish hearts. J R Soc Interface 12(103). doi:10.1098/rsif.2014.1154
Karra R, Knecht AK, Kikuchi K, Poss KD (2015) Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc Natl Acad Sci U S A 112(43):13255–13260. doi:10.1073/pnas.1511209112
Kido M, Du L, Sullivan CC, Li X, Deutsch R, Jamieson SW, Thistlethwaite PA (2005) Hypoxia-inducible factor 1-alpha reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J Am Coll Cardiol 46(11):2116–2124. doi:10.1016/j.jacc.2005.08.045
Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD (2010) Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464(7288):601–605. doi:10.1038/nature08804
Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD (2011a) tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 138(14):2895–2902. doi:10.1242/dev.067041
Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD (2011b) Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 20(3):397–404. doi:10.1016/j.devcel.2011.01.010
Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, Starnes VA, Lien CL (2010) PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci U S A 107(40):17206–17210. doi:10.1073/pnas.0915016107
Kubin T, Poling J, Kostin S, Gajawada P, Hein S, Rees W, Wietelmann A, Tanaka M, Lorchner H, Schimanski S, Szibor M, Warnecke H, Braun T (2011) Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell 9(5):420–432. doi:10.1016/j.stem.2011.08.013
Kusano KF, Pola R, Murayama T, Curry C, Kawamoto A, Iwakura A, Shintani S, Ii M, Asai J, Tkebuchava T, Thorne T, Takenaka H, Aikawa R, Goukassian D, von Samson P, Hamada H, Yoon YS, Silver M, Eaton E, Ma H, Heyd L, Kearney M, Munger W, Porter JA, Kishore R, Losordo DW (2005) Sonic hedgehog myocardial gene therapy: tissue repair through transient reconstitution of embryonic signaling. Nat Med 11(11):1197–1204. doi:10.1038/nm1313
Laflamme MA, Murry CE (2011) Heart regeneration. Nature 473(7347):326–335. doi:10.1038/nature10147
Lavine KJ, Ornitz DM (2008) Fibroblast growth factors and Hedgehogs: at the heart of the epicardial signaling center. Trends Genet 24(1):33–40. doi:10.1016/j.tig.2007.10.007
Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD (2006) A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127(3):607–619. doi:10.1016/j.cell.2006.08.052
Li F, Wang X, Capasso JM, Gerdes AM (1996) Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 28(8):1737–1746. doi:10.1006/jmcc.1996.0163
Li P, Cavallero S, Gu Y, Chen TH, Hughes J, Hassan AB, Bruning JC, Pashmforoush M, Sucov HM (2011) IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 138(9):1795–1805. doi:10.1242/dev.054338
Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT (2006) Gene expression analysis of zebrafish heart regeneration. PLoS Biol 4(8):e260. doi:10.1371/journal.pbio.0040260
Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E (2013) Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol 15(2):222–228. doi:10.1038/ncb2659
Malliaras K, Zhang Y, Seinfeld J, Galang G, Tseliou E, Cheng K, Sun B, Aminzadeh M, Marban E (2013) Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol Med 5(2):191–209. doi:10.1002/emmm.201201737
Mei L, Nave KA (2014) Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron 83(1):27–49. doi:10.1016/j.neuron.2014.06.007
Nemtsas P, Wettwer E, Christ T, Weidinger G, Ravens U (2010) Adult zebrafish heart as a model for human heart? An electrophysiological study. J Mol Cell Cardiol 48(1):161–171. doi:10.1016/j.yjmcc.2009.08.034
Nichols A, Camps M, Gillieron C, Chabert C, Brunet A, Wilsbacher J, Cobb M, Pouyssegur J, Shaw JP, Arkinstall S (2000) Substrate recognition domains within extracellular signal-regulated kinase mediate binding and catalytic activation of mitogen-activated protein kinase phosphatase-3. J Biol Chem 275(32):24613–24621. doi:10.1074/jbc.M001515200
Oberpriller JO, Oberpriller JC (1974) Response of the adult newt ventricle to injury. J Exp Zool 187(2):249–253. doi:10.1002/jez.1401870208
Pachori AS, Custer L, Hansen D, Clapp S, Kemppa E, Klingensmith J (2010) Bone morphogenetic protein 4 mediates myocardial ischemic injury through JNK-dependent signaling pathway. J Mol Cell Cardiol 48(6):1255–1265. doi:10.1016/j.yjmcc.2010.01.010
Piatkowski T, Muhlfeld C, Borchardt T, Braun T (2013) Reconstitution of the myocardium in regenerating newt hearts is preceded by transient deposition of extracellular matrix components. Stem Cells Dev 22(13):1921–1931. doi:10.1089/scd.2012.0575
Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA (2011) Transient regenerative potential of the neonatal mouse heart. Science 331(6020):1078–1080. doi:10.1126/science.1200708
Poss KD, Wilson LG, Keating MT (2002) Heart regeneration in zebrafish. Science 298(5601):2188–2190. doi:10.1126/science.1077857
Riley PR (2012) An epicardial floor plan for building and rebuilding the mammalian heart. Curr Top Dev Biol 100:233–251. doi:10.1016/B978-0-12-387786-4.00007-5
Rudat C, Kispert A (2012) Wt1 and epicardial fate mapping. Circ Res 111(2):165–169. doi:10.1161/CIRCRESAHA.112.273946
Sallin P, de Preux Charles AS, Duruz V, Pfefferli C, Jazwinska A (2015) A dual epimorphic and compensatory mode of heart regeneration in zebrafish. Dev Biol 399(1):27–40. doi:10.1016/j.ydbio.2014.12.002
Schnabel K, Wu CC, Kurth T, Weidinger G (2011) Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One 6(4):e18503. doi:10.1371/journal.pone.0018503
Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT (2013) Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493(7432):433–436. doi:10.1038/nature11682
Senyo SE, Lee RT, Kuhn B (2014) Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res 13(3 Pt B):532–541. doi:10.1016/j.scr.2014.09.003
Sleep E, Boue S, Jopling C, Raya M, Raya A, Izpisua Belmonte JC (2010) Transcriptomics approach to investigate zebrafish heart regeneration. J Cardiovasc Med 11(5):369–380. doi:10.2459/JCM.0b013e3283375900
Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR (2011) De novo cardiomyocytes from within the activated adult heart after injury. Nature 474(7353):640–644. doi:10.1038/nature10188
Szibor M, Poling J, Warnecke H, Kubin T, Braun T (2014) Remodeling and dedifferentiation of adult cardiomyocytes during disease and regeneration. Cell Mol Life Sci: CMLS 71(10):1907–1916. doi:10.1007/s00018-013-1535-6
van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E, Molkentin JD (2014) c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509(7500):337–341. doi:10.1038/nature13309
Wang J, Martin JF (2014) Macro advances in microRNAs and myocardial regeneration. Curr Opin Cardiol 29(3):207–213. doi:10.1097/HCO.0000000000000050
Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD (2011) The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138(16):3421–3430. doi:10.1242/dev.068601
Wang J, Karra R, Dickson AL, Poss KD (2013) Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol 382(2):427–435. doi:10.1016/j.ydbio.2013.08.012
Wang J, Cao J, Dickson AL, Poss KD (2015) Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature. doi:10.1038/nature14325
Washington Smoak I, Byrd NA, Abu-Issa R, Goddeeris MM, Anderson R, Morris J, Yamamura K, Klingensmith J, Meyers EN (2005) Sonic hedgehog is required for cardiac outflow tract and neural crest cell development. Dev Biol 283(2):357–372. doi:10.1016/j.ydbio.2005.04.029
Wills AA, Holdway JE, Major RJ, Poss KD (2008) Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 135(1):183–192. doi:10.1242/dev.010363
Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noel ES, Grun D, Berezikov E, Engel FB, van Oudenaarden A, Weidinger G, Bakkers J (2016) Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell 36:36–49. doi:10.1016/j.devcel.2015.12.010
Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD (2008) Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev 22(6):728–733. doi:10.1101/gad.1641808
Yin VP, Lepilina A, Smith A, Poss KD (2012) Regulation of zebrafish heart regeneration by miR-133. Dev Biol 365(2):319–327. doi:10.1016/j.ydbio.2012.02.018
Yoshizumi M, Lee WS, Hsieh CM, Tsai JC, Li J, Perrella MA, Patterson C, Endege WO, Schlegel R, Lee ME (1995) Disappearance of cyclin A correlates with permanent withdrawal of cardiomyocytes from the cell cycle in human and rat hearts. J Clin Invest 95(5):2275–2280. doi:10.1172/JCI117918
Yu F, Li R, Parks E, Takabe W, Hsiai TK (2010) Electrocardiogram signals to assess zebrafish heart regeneration: implication of long QT intervals. Ann Biomed Eng 38(7):2346–2357. doi:10.1007/s10439-010-9993-6
Zebrowski DC, Engel FB (2013) The cardiomyocyte cell cycle in hypertrophy, tissue homeostasis, and regeneration. Rev Physiol Biochem Pharmacol 165:67–96. doi:10.1007/112_2013_12
Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin YF, Ocorr K, Kang G, Chen J, Stainier DY, Yelon D, Chi NC (2013) In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498(7455):497–501. doi:10.1038/nature12322
Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, MacRae CA, Lee RT, Burns CG, Burns CE (2014) Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci U S A 111(4):1403–1408. doi:10.1073/pnas.1311705111
Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT (2008) Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454(7200):109–113. doi:10.1038/nature07060
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Wu, CC., Weidinger, G. (2016). Cardiac Regeneration in Zebrafish. In: Steinhoff, G. (eds) Regenerative Medicine - from Protocol to Patient. Springer, Cham. https://doi.org/10.1007/978-3-319-27583-3_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-27583-3_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27581-9
Online ISBN: 978-3-319-27583-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)