Abstract
The human gastrointestinal (GI) system can be affected by various illnesses which results in the death of about two million patients globally. Endoscopy helps to detect such diseases as identifying these abnormalities in GI tract endoscopic images is crucial for therapy and follow-up decisions. However, clinicians require adequate time to examine such follow-ups that hinders manual diagnosis. As a result, the aim of the study is to detect and classify various gastric based diseases using deep transfer learning models such as DenseNet201, EfficientNetB4, Xception, InceptionResNetV2, and ResNet152V2, which have been assessed on the basis of precision, loss, accuracy, F1 score, root mean square error, and recall. In this study, Kvasir’s dataset has been used, which is divided into five categories: dyed-lifted polyps, esophagitis, normal cecum, dyed resection margins, and normal colon of endoscopic images. All the images are enhanced by removing the noise before being sent into the deep transfer learning algorithms. During experimentation, it has been analyzed that to detect dyed-lifted polyps, Inception ResNetV2 obtained the highest testing accuracy by 97.32%. On the other hand, Xception model efficiently detects dyed resection margins, esophagitis, normal cecum, and normal colon by computing the best testing accuracy of 95.88%, 96.88%, 97.16%, and 98.88%, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Healthcare is one of the most critical areas in the field of big data for its vital role in a dynamic and thriving society, this is the reason why artificial intelligence (AI) based technologies have attracted much interest in the medical field [1]. In case of gastrointestinal disorders, AI is increasingly being used, and it is becoming more helpful to identify the diseases for which it requires an understanding as how to maximize AI’s potential in diagnosing and treating such disorders [2].
Gastrointestinal diseases affect the entire gastrointestinal (GI) tract, starting from the mouth to the anus. The GI tract is a collection of hollow organs connected by a long, twisting tube that extends from the mouth to the anus and comprises of mouth, oesophagus, stomach, small intestine, large intestine, and anus as shown in Fig. 1 [3].
Gastrointestinal tract [4]
According to the National Institute of Diabetes and Digestive and Kidney Diseases, 60 million to 70 million people suffer from gastrointestinal problems each year, resulting in almost 250,000 deaths. These disorders cause over 50 million hospital visits and 21.7 million hospital admissions per year [5] and as per the World Health Organization (WHO), each year, gastrointestinal problems such as gastrointestinal polyps (Fig. 2) which are abnormal tissue that grows on the stomach and colon mucosa, and causes gastrointestinal cancer, take the lives of million people [6].Therefore, after a long research it has been observed that the increase in gastrointestinal cases are due to a variety of factors which includes unhealthy eating habits among middle and upper-class people, a hectic work schedule, a lack in exercise, an increase of stress levels, malnutrition among children from low-income families, and the unsanitary environment at rural and slum areas [7, 8].
Early identification of gastrointestinal disorders may reduce the chances of developing severe medical problems. It is because an intelligent healthcare system based on artificial intelligence (AI) technology provides fast and accurate diagnosis of GI-tract illnesses and can also be deployed to relieve the strain and assist gastroenterologists [9, 10]. In addition to this, automatic detection, recognition, and evaluation of abnormal results also aid in reducing disparities, improving quality, and making the most use of limited medical resources. Using images of gastrointestinal diseases, researchers employed a combination method of deep residual networks to identify images and achieved a good multi-class classification performance with an RK value of 0.802 and a classification speed of 46 frames per second [11].Similarly, the authors of [3] employed VGG16, ResNet, MobileNet, Inception V3, and Xception neural networks to diagnose various gastrointestinal illnesses, and discovered that VGG16 and Xception neural networks gave the most accurate findings, with up to 98% accuracy. Some researchers predicted eight-class abnormalities of digestive tract illnesses with 97% accuracy using ResNet18, DenseNet-201, and VGG-16 CNN models as feature extractors, and global average pooling (GAP) layer [12]. In a nutshell, it can be said that machine and deep learning approaches are highly beneficial in automatically extracting features and utilizing them to evaluate images for GI tract disease diagnosis.
Hence, keeping the aforementioned tracks of the details, the primary goal of the work is to create a model that efficiently predicts the performance of numerous gastrointestinal disorders using deep transfer learning methodologies for which the following contributions have been made:
-
The images have been taken from KVASIR dataset in which 4000 dyed lifted polyps images, 5000 dyed resection margins images, 4000 images of esophagitis, 5000 images of the normal cecum, and 5000 images of the normal colon are used in this investigation
-
Images have been pre-processed and then summed and presented graphically to create RGB histogram for images of every disease. Further the pre-processed images have been used to extract region of interest by applying adaptive thresholding, morphological feature extraction techniques to obtain the contour features.
-
Later, various deep transfer learning models have been applied such as DenseNet201, EfficientNetB4, Xception, InceptionResNetV2, and ResNet152V2 where it has been analyzed that to detect dyed-lifted polyps, Inception ResNetV2 obtained the highest accuracy by 97.32%. On the other hand, Xception model efficiently detects dyed resection margins, esophagitis, normal cecum, and normal colon by computing the best accuracy of 95.88%, 96.88%, 97.16%, and 98.88%, respectively.
-
All these models have been evaluated using various evaluation metrics such as accuracy, F1 score, loss, precision, recall, and root mean square error.
1.1 Organization of the Paper
After covering Sect. 1 i.e. the introduction part, the rest of the paper is presented in such a way where Sect. 2 presents the contribution of the researchers in the field of gastric disease detection, Sect. 3 provides the information about the methodology that have been used to develop the model for the classification of GI- tract diseases. Section 4 presents the results along with the discussion, and Sect. 5 wraps up the research with future scope.
2 Background
Researchers have done tremendous work to detect gastrointestinal diseases using deep learning as well as machine learning models. In [13], the authors introduced a unique way for autonomously detecting and localizing gastrointestinal illnesses in endoscopic video frame sequences using a weakly trained convolution neural network. The technique was utilized to categorize the video frames as abnormal or usual, and then an iterative cluster unification technique was employed to locate GI anomalies in them. Similarly, the researcher in [14] used video capsule endoscopy frames to provide a worldwide statistical technique for automatically detecting polyps and determining their radii. Their approach collected statistical data from available RGB channels and then loaded it into a support vector machine (SVM), which determines the presence and radius of polyps. In [15], the researchers extracted disease regions from endoscopic images of GI tract using a new method called contrast-enhanced color features while as Geometric feature techniques were used to recover features from the segmented disease region. In [16], the authors created a new CAD approach for classifying GIT disorders. Using the K-Means clustering approach, color scale-invariant characteristics are identified and isolated from all four categories of GIT disorders, including polyps, bleeding, ulcers, and healthy. A linear coding approach called saliency, and adaptive locality-constrained linear coding was presented for feature coding, which encodes the piece of features using adaptive coding. The authors of [17] used two CNN models, ResNet50 and DenseNet121, to accurately identify underlying issues in GI tract endoscopic images. In addition to this, each model had been trained for 20 epochs during training. The authors of [18] proposed a new approach for detecting polyp regions based on LSTM. To decode feature vectors, they employed the LSTM algorithm. The results of their experiments revealed that their technology could exactly determine the position of the ROI of diseased image. In continuation of the work done by researchers for identifying and diagnosing gastrointestinal illnesses, Table 1 provides a comparative study of prior work which includes performance of their models as well as their limitations.
After assaying the table, it has been discovered that the InceptionV2 technique had obtained 98.42% accuracy for the Kvasir dataset, VGG obtained 99.42% accuracy for wireless endoscopic capsule pictures (WCE), and ResNet 50 obtained 95.7% accuracy for MediaEval images. Regardless, there are several flaws in these methodologies that have been attempted to be solved in this work.
3 Materials and Methods
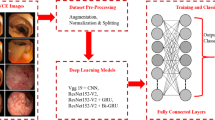
The framework of the proposed model (Fig. 3) has been shown in this section, along with the dataset description in sect. 3.1, the libraries that have been imported in sect. 3.2, the various phases used in the entire study such as data pre processing in sect. 3.3, exploratory data analysis in sect. 3.4, feature extraction in sect. 3.5, the models that have been applied in sect. 3.6, and finally the parameters that have been used to analyze the models performance.
3.1 Dataset Description
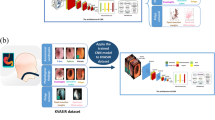
The Kvasir dataset includes photos that have been annotated and confirmed by medical professionals (experienced endoscopists). It contains hundreds of photos in each category that offer anatomical landmarks, disease abnormalities, or endoscopic operations in the GI system. Anatomical landmarks include the pylorus, cecum, and so on. Pathological findings include esophagitis, polyps, ulcerative colitis, and so on [24]. Furthermore, several image sets related to lesion removal, such as “dyed resection margins”, “dyed and lifted polyps” and so on, have been displayed.
The dataset comprises images with resolutions ranging from 720 × 576 to 1920 × 1072 pixels that are organized into different categories and labelled according to their content. 4000 images of dyed lifted polyps, 5000 images of dyed resection margins, 4000 images of esophagitis, 5000 images of the normal cecum, and 5000 images of the normal colon are used in this research (Fig. 4). The number of images is adequate for a variety of applications such as machine learning, image retrieval, transfer learning, and deep learning [24].
3.2 Libraries
Various libraries have been used such as Keras, Tensor flow, Imutlis to handle various operations of image processing which includes skeletonization, rotation, translation, scaling, recognizing edges, and sorting contours. To create and remove directories (folders) as well as to modify and identify the directory is being provided by OS module in python [25].
In addition, Matplotlib, a Python data visualization as well as graphical package has been used to see huge bulk of complex data in simple representations [26]. Python data visualization, seaborn is a toolkit which is tightly integrated with matplotlib and pandas. It is used for exploratory data analysis and visualization of data. It’s ideal for data frames with the Pandas library which isa Python computer language-based data manipulation and analysis application [27]. The main components are data structures and procedures to process numerical tables and temporal series. CV2 also uses the imread() method to load an image from a provided file. Along with this, NumPy, Scikit-learn, and the OpenCV package were employed [28].
3.3 Data Pre-processing
Before any classification algorithms can be used for an image class, the dataset must be pre-processed. The dataset used in this study is KVASIR V2, which is freely accessible to researchers participating in numerous technological research efforts. This dataset is a challenge in image pre-processing because many of the photos contain unwanted artifacts. As a result, the dataset encompassing various stomach illnesses was pre-processed using the Opencv and Imultis tools. Opencv (name, flag) method is loaded to open the new window as well as to display the images in full-screen mode. The width and height of the image are modified while resizing to uphold the aspect ratio (224,224,3).
3.4 Exploratory Data Analysis
After pre-processing the images from both datasets, the information has been summed and graphically displayed in order to generate the RGB histogram of images. It helps to provide insight into image data, such as the image size, colour space, resolution, and pixel values through which we can interpret the images more accurately and make more informed decisions regarding pre-processing steps and model selection.
In addition, it also helps us to identify relationships and patterns within the image data, such as common shapes or objects, colour distributions, and texture patterns. In fact, these insights can inform the design of features and the selection of suitable image processing techniques. In Fig. 5, the pixel intensity values of images are depicted as a histogram that have been used to map one intensity distribution to another, to enhance the image’s overall appearance and increase its visual appeal.
3.5 Feature Extraction
In this section, the features have been extracted in a sequential way. Initially, the morphological values of the images per class have been computed which are shown in Table 2. We used Eq. (1–18) to compute various parameters from input images, which includes epsilon, area, equivalent, aspect ratio, maximum and minimum value, minimum and maximum value location, extreme leftmost, topmost, rightmost, and bottommost point etc. of dataset images.
After computing the morphological values, the function findcontours() was used to generate the contour, which is a closed curve representing the boundaries of an image’s object or region. Further The function cv2.ContourArea finds the largest contour in an image, which is the contour with the largest area, i.e., the contour that encloses the largest object or region in the image (). In addition, Extreme points, also known as the convex hull, are the outermost points of a contour as defined by the cv2.ConvexHull function (). A contour's convex hull is the smallest convex polygon that contains the contour. This convex hull can be used to crop an object or region of interest from an image by utilising the bounding rectangle's coordinates. The color image is then scaled and transformed to grayscale using the cvtColor() method, translating an image from one to another color space. Later, An adaptive thresholding approach is applied to graycolored data to emphasize the target area or extract the region of interest for the isolation of item from the background for superior feature extraction outcomes.
When applied to grayscale images, the morphological processing techniques dilation and erosion produce various results. By removing pixels from object borders, erosion shrinks the size of an image's pixels, resulting in an output pixel with the lowest value possible. The output pixel from Dilation, on the other hand, has the highest value out of all the pixels in the region because it expands the image by adding pixels to the object borders (as shown in Fig. 6).
Additionally, there are some additional considerations to make regarding the impact and repercussions of these operations on the image. Although erosion removes pixels from an object, it also results in pixel loss, whereas dilation adds pixels, resulting in pixel gain. Depending on the application, this may result in the loss or addition of essential data, such as edges or texture. Erosion can thin or reduce an object, while dilation can thicken or enlarge it. This can alter the appearance of the object and influence subsequent image processing steps. Erosion and dilation can also affect an object’s connectivity. Erosion can result in the disconnection of an object into multiple parts, whereas dilation can result in the connection of multiple objects. This can affect the interpretation and further analysis of the object. Erosion and dilation can be used to increase contrast by emphasising an object's edges and borders. However, excessive erosion or dilation can lead to over smoothing or over enhancement, which can result in inaccurate results.
The most important step after feature extraction is to split the dataset in to training and testing phase. In this study, the diseases dataset has been divided into such proportion in which training dataset of esophagitis, dyed lifted polyps, normal cecum, dyed resection margins, and normal colon have 3500, 4500, 3500, 4500, and 4500 images respectively whereas for testing phase they are 500 images each (as shown in Fig. 7).
3.6 Applied Models
In this section, we have provided the description regarding various deep transfer learning models that have been used for the detection as well as classification of gastric diseases.
3.7 DenseNet201
DenseNet-201 is a 201-layer deep convolutional neural network. Densely connected blocks of convolutional layers make up the architecture of DenseNet201, where each layer is connected to every layer that comes before it. As a result, the gradient is given a clear path through the network, which can speed up training and increase accuracy. Additionally, batch normalisation and dropout are used by DenseNet201 to regularise the network and minimise overfitting. Additionally, transition layers are used, which decrease the feature maps’ spatial dimensions while increasing the number of channels, thereby lowering the overall number of parameters. [29]. The image input size on the network is 224 by 224 which is being provided to the architecture of DenseNet201 model consisting of global average pooling 2d, dense layer, batch normalization, activation function, a dropout layer, and a second dense layer to classify the image as shown in Fig. 8.
DenseNet201 has a total parameter count of 18815813: 18586245 trainable parameters and 229568 non-trainable parameters. Table 3 gives the information related to the parameters of each layer.
3.8 EfficientNetB4
The baseline network is critical to the success of model scaling. A new baseline network has been constructed to boost performance even further by implementing a neural architecture search with the AutoML MNAS framework, optimizing accuracy and efficiency (FLOPS). AutoML MNAS generates the EfficientNet-B0 baseline network, and the EfficientNet-B1 through B7 networks are obtained by scaling up the baseline network [4, 24, 30,31,32]. The picture input size for the network is 224 × 224 pixels. The initial model, a global average pooling 2d, dense layer, batch normalization, activation function, a dropout layer, and lastly, a second dense layer are used to classify the image as shown in Fig. 9.
EfficientNetB4 has 18134884, with 18009165 trainable parameters and 125719 non-trainable parameters. Table 4 gives the information related to the parameters of each layer.
3.9 Xception
Xception is a deep convolutional neural network with 71 layers. For image classification and other computer vision tasks, the architecture known as Xception is intended to be more effective and powerful. It is made up of several depth-wise separable convolutional layers where the spatial and channel-wise convolutions are carried out separately before being combined into one channel by point-wise convolutions. With no loss in accuracy, the depth-wise separable convolutional layers in Xception’s network require fewer parameters and less computation. In comparison to other convolutional neural network architectures, Xception is consequently quicker and more memory-efficient. [33]. The image input size for the network is 299 by 299 pixels which is being fed to the layers of initial model followed by a global average pooling 2d, dense layer, batch normalization, activation function, a dropout layer, a second dense layer, and lastly, an activation function is used to classify the image as shown in Fig. 10.
Xception has 21388077 parameters, with 21333037 trainable parameters and 55040 non-trainable parameters. Table 5 gives the information related to the parameters of each layer.
3.10 InceptionResNetV2
InceptionResNetV2 model has 164 layers and the architecture has global average pooling 2d, dense layer, batch normalization, activation function, a dropout layer, and lastly, an second dense layer to classify the image as shown in Fig. 11. To enhance gradient flow and network convergence, the InceptionResNetV2 architecture combines the multi-level feature extraction of the Inception network with the residual connections of the ResNet. With bottleneck layers and residual connections to lower the number of parameters and increase training efficiency, it is made up of a deep stack of convolutional and pooling layers [34,35,36,37,38,39].
The total parameters of the InceptionResNetV2 are 54732261: 54671205 for trainable parameters and 61056 for non-trainable parameters. Table 6 gives the information related to the parameters of each layer.
3.11 ResNet152V2
A Residual Network (ResNet), as shown in Fig. 12, is a CNN design with multiple convolutional layers. ResNet is incredibly quick and has a considerable number of layers. The critical distinction between ResNetV2 and the original (V1) is that V2 does batch normalization on each weight layer before applying it [40]. ResNet excels at picture identification and localization tasks, demonstrating the usefulness of a wide range of visual recognition tasks. The model's pre-trained initial weights can be used to learn the input. This strategy reduces training time while covering a vast region with high precision. The architecture of ResNet152V2 consists of a global average pooling 2d, dense layer, batch normalization, an activation function, a dropout layer, and lastly, second dense layer which is used for the classification of image [39, 41,42,43,44].
In this research, ResNet152V2 has 58858245 parameters, with 58713989 trainable parameters and 144256 non-trainable parameters. Table 7 gives the information related to the parameters of each layer.
3.12 Evaluative Parameters
3.12.1 Accuracy
It is the parameter to define the best model by identifying the relationship as well as patterns of various attributes in a given input or a dataset which is used to train the models [45]. It is calculated by using Eq. (19)
3.13 Loss
It is the parameter that identifies how bad the algorithm is predicting the data [46]. Equation (20) is used to calculate it.
3.14 Root Mean Square Error (RMSE)
It is the standard deviation of the errors which occurs when a prediction is made in a dataset [47] and is solved by using Eq. (21).
\({\widehat{y}}_{i}\) are values that have been predicted, \({y}_{i}\) are values that have been observed, and \(n\) is the total number of observations.
3.15 Precision
It is the proportion between the number of relevant items the system retrieves and the total number of items it retrieves [48]. It is calculated by using Eq. (22)
3.16 Recall
It is the measure of relevant items that the system has successfully retrieved to all relevant items in the dataset [48]. It is calculated by using Eq. (23)
3.17 F1 Score
It defines the relationship between recall and precision. In other words, it is the harmonic mean of precision as well as recall [45]. It is calculated by using Eq. (24)
4 Results and Discussion
This section covers the results of multiple deep transfer learning models such as DenseNet201, EfficientNetB4, Xception, InceptionResNetV2, and ResNet152V2 have been shown for various diseases such as dyed lifted polyps, normal colon, normal cecum, esphaigitis, and dyed resection margins. The top two models have been selected on the basis of their best performance and merged together to test their performance for different diseases [49].
Figure 13 depicts the confusion matrix of various deep transfer learning models to compute their performances in terms of various evaluative parameters as mentioned in Sect. 3.7. In addition to this, the matrix of 5 × 5 also presents the actual as well as predicted values of various classes in the form of true positive, false positive, true negative, and false negative.
From Table 8, it has been found that Xception and InceptionResNetV2 models have been the top two models who have computed the highest accuracies as well as loss value by 98.74% and 98.93% as well as 0.03 and 0.02 during training phase while as during testing phase, these models have computed again the best accuracies as well as loss value by 97.88% and 95.32% as well as 0.06 and 0.13. These two top models have been hybridized together and while training as well as validating by the same dataset, the accuracies achieved by them are 98.83% and 96.6% respectively. In addition to this, root mean square has been also computed so that we can compare it to a reference or ground truth image to determine the degree of similarity between the two. If the RMSE is small, which indicates that any of the two images are extremely similar which has been obtained by InceptionResNetV2? On the contrast, a large RMSE indicates that the two images differ significantly which has been obtained by EfficientNetB4.
The models have been also computed for the different set of performance measures such as F1 score, precision, as well as recall (Table 9) and it has been found that the highest values have been obtained by Xception model with 98.2% each. On the contrary, the lowest values of recall, precision, and F1 score has been computed by ResNet152V2 with 92.6%, DenseNet201 with 89.8%, and DenseNet201 as well as Hybrid Model with 89.6% respectively. Hence, on assaying the overall results it can be said that if a model has low precision, recall, and F1 scores, it indicates that it is not performing well and has poor classification accuracy. In such a situation, it may be necessary to re-evaluate the model or make adjustments to enhance its performance.
As shown in Table 10, ResNet152V2 computed the highest training accuracy and best testing loss for dyed lifted polyps by 99.69% and 0.14, respectively. At the same time, the hybrid model achieved the best testing accuracy, training loss, and root mean square error values by 95.6%, 0.04, and 0.20, respectively. ResNet152V2 computed the maximum training accuracy and best testing loss by 99.69% and 0.14, respectively, for Dyed Resection Margins. At the same time, the hybrid model attained the training loss, and root mean square error values by 0.04 and 0.20, respectively. The Xception model, on the other hand, had the highest testing accuracy of 95.88%. For esophagitis, InceptionResNetV2 had the best training accuracy, training loss, as well as root mean square error of 96.93%, 0.22, and 0.47, respectively, while the Xception model had the most incredible testing accuracy of 96.88% and testing loss of 0.16. Xception and the hybrid model had the same training accuracy rating of 98.76% for Normal Cecum. The hybrid model achieved 0.01, 0.12, and 0.08, respectively, superior to other methods in terms of training loss, root mean square value, and testing loss. In contrast, Xception achieved the highest testing accuracy score of 97.16%. InceptionResNetV2 obtained the best training accuracy, training loss, and root mean square error value for the Normal colon by 99.93%, 0.01, and 0.13, respectively. In comparison, Xception obtained the best testing accuracy and testing loss by 98.88% and 0.05, respectively, compared to the other algorithms.
The graphical analysis (Fig. 14) of the models such as DenseNet201, EfficientNetB4, Xception, InceptionResNetV2, and ResNet152V2 for different gastric diseases have been computed using evaluative metrics such as F1 score, recall, and precision. The algorithms computed the highest precision value by 99%, recall by 99%, and F1 score by 99% for various diseases. On the other hand, the lowest precision, recall, and F1 score value obtained by DenseNet201 is 66%, 80%, and 78%, EfficientNetB4 is 83%, 85%, and 87%, Xception is 96%, 95%, and 97%, InceptionResNetV2 is 89%, 87%, and 92%, and ResNetV2 is 81%, 80%, and 86% respectively. On comparing the performance of all these algorithms at an individual scale with the hybrid models, it has been seen that the highest value obtained is 100% recall, 99% precision, and a 100% F1 score, and the lowest values are 86%, 87%, 79% respectively. Bold denotes the best results for each parameter out of all results in Tables 8, 9, and 10.
In Table 11, a comparison has been done for various gastric diseases datasets using various techniques on the basis of their accuracies. It can be seen that the Xception model applied in our study has obtained the highest accuracy by 97.88% as compared to the others. The least accuracy has been computed by BMFA model by 92.6% for testing on 5000 images of gastrointestinal tract.
5 Conclusion
In this study, the publicly available dataset of five gastrointestinal disorders was used to build deep transfer learning models, and then evaluated using various performance metrics. Dataset was pre-processed before training the models, and its features were retrieved using several techniques to obtain morphological information. A confusion matrix was also employed to compare and calculate the various models’ performance. During testing for various classes of diseases, it was discovered that Xception has obtained 97.88% accuracy and 0.06 loss, but a hybrid model consisting of InceptionResNetV2 and Xception had computed the highest score by 100% recall, 99% precision, and a 100% F1 score. On the contrary, for the combined dataset, Xception model computed the best precision, F1 score, and recall values of 98.2%. Finally, compared to other previously published works, the new strategy outperforms current methods. The main difficulty encountered in this study was that the images were of varying sizes. The majority of the images had been bordered in black color, which reduced the performance of the classification networks. As a result, in future, the quality of an image can be improved by using advanced image processing technologies an application should be built where the patients can themselves check which gastro-intestinal diseases they are suffering from without wasting their time.
Data availability
Not applicable.
References
Paul Y, Hickok E, Sinha A, Tiwari U, Mohandas S, Ray S, Bidare PM (2018) Artificial intelligence in the healthcare industry in India. The Centre for Internet and Society, India.
Mirbabaie M, Stieglitz S, Frick NR (2021) Artificial intelligence in disease diagnostics: a critical review and classification on the current state of research guiding future direction. Heal Technol 11(4):693–731
Dheir IM, Abu-Naser SS (2022) Classification of Anomalies in Gastrointestinal Tract Using Deep Learning. International Journal of Academic Engineering Research (IJAER), 6(3).
Kaur I, Sandhu AK, Kumar Y (2022) Artificial intelligence techniques for predictive modeling of vector-borne diseases, and its pathogens: a systematic review. Arch Comput Methods Eng 29:3741–3771. https://doi.org/10.1007/s11831-022-09724-9
Hernandez M (2016) Improving Foot Care and Kidney Disease Screening Through Implementation of American Diabetes Association Standards–2016 in The Primary Care Setting.
World Health Organization. Centre for Health Development, & World Health Organization. (2010). Hidden cities: unmasking and overcoming health inequities in urban settings. World Health Organization.
Furukawa A, Sakoda M, Yamasaki M, Kono N, Tanaka T, Nitta N et al (2005) Gastrointestinal tract perforation: CT diagnosis of presence, site, and cause. Abdom Imaging 30(5):524–534
Kumar Y, Gupta S, Singla R et al (2022) A systematic review of artificial intelligence techniques in cancer prediction and diagnosis. Arch Comput Methods Eng 29:2043–2070. https://doi.org/10.1007/s11831-021-09648-w
Öztürk S, Özkaya U (2020) Gastrointestinal tract classifica-tion using improved LSTM based CNN. Multimed Tools Appl 79(39):28825–28840
Agrebi S, Larbi A (2020) Use of artificial intelligence in infectious diseases. In Artificial intelligence in precision health (pp. 415–438). Academic Press.
Pogorelov K, Riegler M, Halvorsen P, Griwodz C, de Lange T, Randel KR, et al. (2017) A Comparison of Deep Learning with Global Features for Gastrointestinal Disease Detection. In MediaEval.
Gamage C, Wijesinghe I, Chitraranjan C, Perera I (2019) GI-Net: anomalies classification in gastrointestinal tract through endoscopic imagery with deep learning. In 2019 Moratuwa Engineering Research Conference (MERCon) (pp. 66–71). IEEE.
Iakovidis DK, Georgakopoulos SV, Vasilakakis M, Koulaouzidis A, Plagianakos VP (2018) Detecting and locating gastrointestinal anomalies using deep learning and iterative cluster unification. IEEE Trans Med Imaging 37(10):2196–2210
Zhou M, Bao G, Geng Y, Alkandari B, Li X (2014) Polyp detection and radius measurement in small intestine using video capsule endoscopy. In 2014 7th International Conference on Biomedical Engineering and Informatics (pp. 237–241). IEEE.
Sharif M, Attique Khan M, Rashid M, Yasmin M, Afza F, Tanik UJ (2021) Deep CNN and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images. J Exp Theor Artif Intell 33(4):577–599
Yuan Y, Meng MQH (2017) Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys 44(4):1379–1389
KahsayGebreslassie A, Hagos MT (2019) Automated gastrointestinal disease recognition for endoscopic images. In 2019 International Conference on Computing, Communication, and Intelligent Systems (ICCCIS) (pp. 312–316). IEEE.
Zeng X, Wen L, Liu B, Qi X (2020) Deep learning for ultrasound image caption generation based on object detection. Neurocomputing 392:132–141
Igarashi S, Sasaki Y, Mikami T, Sakuraba H, Fukuda S (2020) Anatomical classification of upper gastrointestinal organs under various image capture conditions using AlexNet. Comput Biol Med 124(6):103950
Ghatwary N, Zolgharni M, Ye X (2019) Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg 14(4):611–621. https://doi.org/10.1007/s11548-019-01914-4
Cogan T, Cogan M, Tamil L (2019) MAPGI: accurate identification of anatomical landmarks and diseased tissue in gastrointestinal tract using deep learning. Comput Biol Med 111:103351
Nadeem et al. (2018) Nadeem S, Tahir MA, Naqvi SSA, Zaid M. Ensemble of texture and deep learning features for finding abnormalities in the gastro-intestinal tract. International Conference on Computational Collective Intelligence; Springer; 2018. pp. 469–478.
A Asperti, C Mastronardo, (2017) “The Effectiveness of Data Augmentation for Detection of Gastrointestinal Diseases from Endoscopical Images,” arXiv,.
Bhardwaj P, Bhandari G, Kumar Y et al (2022) An investigational approach for the prediction of gastric cancer using artificial intelligence techniques: a systematic review. Arch Comput Methods Eng 29:4379–4400. https://doi.org/10.1007/s11831-022-09737-4
Almanifi ORA, Razman MAM, Khairuddin IM, Abdullah MA, Majeed APA (2021) Automated Gastrointestinal Tract Classification Via Deep Learning and The Ensemble Method. In 2021 21st International Conference on Control, Automation and Systems (ICCAS) (pp. 602–606). IEEE.
Cario CL, Witte JS (2018) Orchid: a novel management, annotation and machine learning framework for analyzing cancer mutations. Bioinformatics 34(6):936–942
Gupta A, Koul A, Kumar Y (2022) Pancreatic Cancer Detection using Machine and Deep Learning Techniques. In: 2nd International Conference on Innovative Practices in Technology and Management (ICIPTM) (Vol. 2, pp. 151–155). IEEE.
Nadeem S, Tahir MA, Naqvi SSA, Zaid M (2018) Ensemble of texture and deep learning features for finding abnormalities in the gastro-intestinal tract. In: Nguyen NT, Pimenidis E, Khan Z, Trawiński B (eds) International Conference on Computational Collective Intelligence. Springer, Cham, pp 469–478
Gamage C, Wijesinghe I, Chitraranjan C, Perera I (2019) GI-Net: anomalies classification in gastrointestinal tract through endoscopic imagery with deep learning. In 2019 Moratuwa Engineering Research Conference (MERCon) (pp. 66–71). IEEE
Yan T, Wong PK, Choi IC, Vong CM, Yu HH (2020) Intelligent diagnosis of gastric intestinal metaplasia based on convolutional neural network and limited number of endoscopic images. Comput Biol Med 126:104026
Kumar Y, Gupta S (2023) Deep transfer learning approaches to predict glaucoma, cataract, choroidal neovascularization, diabetic macular edema, DRUSEN and healthy eyes: an experimental review. Arch Comput Methods Eng 30:521–541. https://doi.org/10.1007/s11831-022-09807-7
Gupta S, Kumar Y (2022) Cancer prognosis using artificial intelligence-based techniques. SN Comput Sci 3:77. https://doi.org/10.1007/s42979-021-00964-3
Kumar Y, Singla R (2022) Effectiveness of Machine and Deep Learning in IOT-Enabled Devices for Healthcare System. In: Ghosh U, Chakraborty C, Garg L, Srivastava G (eds) Intelligent Internet of Things for Healthcare and Industry. Springer, Cham, pp 1–19
Bansal K, Bathla RK, Kumar Y (2022) Deep transfer learning techniques with hybrid optimization in early prediction and diagnosis of different types of oral cancer. Soft Comput 26:11153–11184. https://doi.org/10.1007/s00500-022-07246-x
Koul A, Bawa RK, Kumar Y (2023) Artificial intelligence techniques to predict the airway disorders illness: a systematic review. Arch Comput Methods Eng 30:831–864. https://doi.org/10.1007/s11831-022-09818-4
Chaplot N, Pandey D, Kumar Y et al (2023) A comprehensive analysis of artificial intelligence techniques for the prediction and prognosis of genetic disorders using various gene disorders. Arch Comput Methods Eng. https://doi.org/10.1007/s11831-023-09904-1
Kanna GP, Kumar SJKJ, Parthasarathi P et al (2023) A review on prediction and prognosis of the prostate cancer and gleason grading of prostatic carcinoma using deep transfer learning based approaches. Arch Comput Methods Eng. https://doi.org/10.1007/s11831-023-09896-y
Kumar A, Kumar N, Kuriakose J et al (2023) A review of deep learning-based approaches for detection and diagnosis of diverse classes of drugs. Arch Comput Methods Eng. https://doi.org/10.1007/s11831-023-09936-7
Sisodia PS, Ameta GK, Kumar Y et al (2023) A Review of deep transfer learning approaches for class-wise prediction of alzheimer’s disease using MRI images. Arch Comput Methods Eng 30:2409–2429. https://doi.org/10.1007/s11831-022-09870-0
Musha A, Hasnat R, Al Mamun A, Ghosh T (2022). Deep Learning-Based Comparative Study to Detect Polyp Removal in Endoscopic Images. In International Conference on Emerging Smart Computing and Informatics (ESCI) (pp. 1–5). IEEE.
Kaur S, Kumar Y, Koul A et al (2023) A systematic review on metaheuristic optimization techniques for feature selections in disease diagnosis: open issues and challenges. Arch Comput Methods Eng 30:1863–1895. https://doi.org/10.1007/s11831-022-09853-1
https://my.clevelandclinic.org/health/articles/7040-gastrointestinal-diseases
https://www.niddk.nih.gov/health-information/digestive-diseases/digestive-system-how-it-works
Kaur D, Singh S, Mansoor W, Kumar Y, Verma S, Dash S, Koul A (2022) Computational intelligence and metaheuristic techniques for brain tumor detection through IoMT-enabled MRI devices. Wirel Commun Mob Computing. https://doi.org/10.1155/2022/1519198
Kumar Y, Koul A, Singla R, Ijaz MF (2022) Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda. Journal of Ambient Intelligence and Humanized Computing, 1–28.
Petscharnig S, Schöffmann K, Lux M (2017) An Inception-like CNN Architecture for GI Disease and Anatomical Landmark Classification. In MediaEval.
Ahmed A (2022) Classification of Gastrointestinal Images Based on Transfer Learning and Denoising Convolutional Neural Networks. In: M Saraswat, S Roy, C Chowdhury, AH Gandomi (Eds) Proceedings of International Conference on Data Science and Applications (pp. 631–639), Springer, Singapore.
Yang S, Lemke C, Cox BF, Newton IP, Näthke I, Cochran S (2020) A learning-based microultrasound system for the detection of inflammation of the gastrointestinal tract. IEEE Trans Med Imaging 40(1):38–47
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhardwaj, P., Kumar, S. & Kumar, Y. A Comprehensive Analysis of Deep Learning-Based Approaches for the Prediction of Gastrointestinal Diseases Using Multi-class Endoscopy Images. Arch Computat Methods Eng 30, 4499–4516 (2023). https://doi.org/10.1007/s11831-023-09951-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11831-023-09951-8