Abstract
The study aimed to investigate organic waste-degrading microbial communities in compost and sugarcane rhizospheric soil from Kodinar, Gujarat, India and to assess their potential for producing novel bioactive compounds. Compost contained higher total culturable heterotrophic bacteria (1.3 × 108) and cellulolytic bacterial (4 × 106) populations compared to rhizospheric soil. A total of 96 isolates were obtained using various cultivation strategies with 35 isolates obtained through enrichment techniques, and the rest through direct plating on nutrient agar. Remarkably, four isolates CP2, CP4, CP9 and CP11 showed significant lignin hydrolysis. Further, morphological characterization revealed 26 Gram-positive isolates. Based on microscopic, cultural, and biochemical assessments, as well as 16 S rRNA gene sequencing, selective isolates with lignocellulolytic potential were identified as Bacillus licheniformis (SRS 11), Priestia megaterium (SRS 8, CP 5), Bacillus subtilis (CP11), and Priestia flexa (CP 2 & CP 9). Multiple enzyme secretion potential of the isolates was also investigated. Around 16.66% of isolates secreted at least one lytic enzyme such as cellulase, ligninase, amylase, protease, and pectinase, with variable percentages. The salt significantly affected growth and CMCase activity, with strain SRS8 being less tolerant (3% NaCl) compared to CP2, CP9, and CP11, which exhibited activity between 0 and 7% NaCl (w/v). Compost isolates preferred higher temperatures (40–45 °C), while most rhizospheric isolates thrived at lower temperatures (35 °C). Rhizospheric isolates preferred pH 7. Most compost isolates grew at pH 7, except CP9 and CP11, which showed best growth at pH 8. Overall, the study highlights microbial waste-degrading potential, enzyme production, and environmental preferences.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Solid waste substances or materials containing organic constituents generated by human activities include municipal organic solid wastes such as kitchen waste, green waste, slaughterhouse wastes and sludge. Similarly, agricultural organic solid waste includes livestock, manure, and crop straw. These types of wastes are normally incinerated, disposed to landfill sites or utilised to feed animals and dung cake preparation. Unsafe disposal of organic fractions of municipal solid wastes (MSW) causes health issues, environmental pollution in terms of methane emissions, and leachate issues thereby contaminating groundwater, etc. According to the data displayed by CPCB (Central Pollution Control Board), India, 117,644 MT of total solid wastes were collected in 2018 and out of these only 49,401 MT of solid wastes were treated. Major constituents of organic agricultural waste are materials rich in lignocellulose, minerals, proteins, and other sugars. Organic solid waste (OSW) could be productively utilized as a substrate or raw materials for various processes like composting, anaerobic digestion, ex-situ green manuring etc.
Organic solid waste is a source of underutilized, renewable biomass on the earth. An enormous quantity of organic waste is generated annually which can be managed by composting. Heterotrophic bacterial and fungal species have the potential to convert this organic heteropolymer to a simple monomeric substance which has a potential application. The major fraction of agricultural waste is lignocellulose (35-50% cellulose and 11 -15% lignin) (Jiang et al. 2020). Other domestic food wastes like bread, savoury, fruits, vegetables, onion & potato pill wastes, restaurant wastes and slaughterhouse wastes contain starch, pectin and proteins.
Endoglucanase (EC 3.2.1.4), exoglucanase (EC 3.2.1.74), and β-glucosidase (EC 3.2.1.21) are the cellulases responsible for the conversion of cellulose to glucose with a different mechanism of action and substrate specificity. Lignin is the most degradation-resistant constituent found in plant residue. Lignin degradation is facilitated by three major complex enzymes namely laccases (EC 1.10.3.2), lignin peroxidases (EC 1.11.1.14), and manganese peroxidases (EC1.11.1.13) (Tao et al. 2022). Hydrolysis of pectin is catalyzed by pectin methylesterase (EC3.1.1.11) which acts as a de-methoxylating enzyme that removes the methyl group from pectin while pectin lyase (EC 4.2.2.10) breaks pectin in exo action pattern generating oligomers (Datta et al. 2020). Amylolytic enzymes are widely distributed in bacterial and fungal genera. Endo-acting α amylase (EC 3.2.1.1) and exo-acting β amylase (EC 3.2.1.2) hydrolyse polymeric starch to monomeric glucose (Paul et al. 2021). Extracellular proteases (EC 3.4) hydrolyze proteinaceous wastes by breaking peptide bonds present in the polypeptide chain of amino acids.
Trichoderma, Penicillium, Chaetomium, Aspergillus, Phanerochaete, and other white-rot basidiomycetes have been widely studied for lignocellulases production (Li et al. 2019; Xia et al. 2019; Machado et al. 2020). Fungus has a higher potential for lignocellulose degradation than bacteria but in contrast, bacteria have the expression of multiple enzyme complexes, rapid growth rate, and extreme environmental tolerance. Due to this versatility bacterial enzyme secretion has become the emphasis of production research (Chuanjiao et al. 2021). Bacterial lignocellulases production has been extensively reported in the literature. Actinomycetes genera like Streptomyces, Arthrobacter, Micromonospora, Nocardia and non-filamentous bacterial genera like Acinetobacter, Flavobacterium, Cellulomonas, Micrococcus, Pseudomonas, and Xanthomonas have strong lignocellulose degrading ability (Wu et al. 2023; Dube et al. 2023; Zhang et al. 2021). Pectin methylestrase and pectin lyase production have been reported in many microbes, including, Pseudomonas solanacearum, Aspergillus niger, Neurospora crassa, Aspergillus sp., Bacillus sp. Lactobacillus lactis, Penicillium occitanis, A. japonicus (Datta et al. 2020; Murad et al. 2011), Erwinia carotovora, Pseudomonas syringae, and Bacillus sp. (Murad et al. 2011). Bacterial amylolytic enzymes are widely distributed in bacterial and fungal genera including, Bacillus, Proteus, Pseudomonas, Escherichia coli, Lactobacillus, Streptomyces, Talaromycesemersonii, Aspergillus ficuum, and Thermomyceslanuginosus (Wu et al. 2018; Pokhrel et al. 2013; Shishtawy et al. 2014). Extracellular protease production was reported in bacterial strains including Bacillus safensis, Bacillus lentus, Pseudomonas fuorescens, Geobacillus, Bacillus alkaloophilus, Bacillus pumilus, Bacillus halodurans, Pseudomonas aeruginosa, Bacillus licheniformis and Bacillus amyloliquefaciens (Ahmad et al. 2020; Contesini et al. 2018; Majithiya and Gohel 2020). Fungus is the next microbes in protease production. Aspergillus, Cephalosporium, Rhizopus, Fusarium, Penicillium, Mucor, and Trichoderma are the fungal protease producers (Abu-Tahon et al. 2020; Naveed et al. 2021).
Metabolites from bacteria/actinobacteria are exceptional, novel, and occasionally intricate (Gohel and Singh 2018; Chauhan and Gohel 2020). They possess excellent enzymatic and antibacterial properties, while also showing low toxicity (Chauhan et al. 2021; Majithiya and Gohel 2022a, b; Vaghela and Gohel 2023). Therefore, the study aimed to cultivate lignocellulolytic bacteria from rhizospheric soil and compost samples using an enrichment and serial dilution process with diverse, nutrient-rich growth media. Moreover, the study covers morphological, cultural, biochemical, and molecular characterization, as well as growth and enzymatic profile analysis of potent rhizospheric and compost isolates.
Materials and methods
Sample collection
Sampling sites were selected such that sufficient lignocellulose can be available naturally, whereby the inhabitant microbial population could primarily be lignocellulolytic by nature which could be easily isolated in large numbers. Samples collected from compost and rhizospheric soil of sugarcane plantations in different areas of Kodinar Taluka, Gujarat, India were screened for the isolation of organic waste-degrading bacteria. The isolation process was initiated within 8 h of collection to minimize saprophytic developments.
Enrichment of lignocellulolytic bacteria
A nutrient enrichment technique was used to enrich and isolate lignocellulolytic bacteria. A total of 05 g sample was added in 100 mL of minimal salt media (MSM-L), which contained, NaNO3 2.5 g; KH2PO4 2 g; MgSO4 0.2 g; NaCl 0.2 g; CaCl2.6H2O 0.1 g; and lignin (TCI) 100 mg in a litre. Due to the higher molecular weight of lignin, bacterial strains could not grow by consuming lignin as a single carbon source. Therefore, glucose (w/v) 10 g; peptone (w/v) 5 g; and trace elements solution (1 mL L− 1) were added in an MSM- L broth as suggested earlier (Pfenning and Lippert 1966; Zainith et al. 2019). The medium pH was adjusted to 7.5 ± 0.1. The flasks were incubated at shaking conditions (120 RPM) for seven days. After 7 days, the 10 mL sample was retransferred in fresh 90 mL of the same culture medium and incubated for 72 h. At every 72 h, three more successive transfers were done to enrich lignolytic bacteria (Chandra et al. 2012). Similarly, cellulolytic bacteria were enriched by using minimal media (MSM-CMC) supplemented with 1% Na salt of carboxymethylcellulose (medium viscosity).
Isolation of bacteria
For each sample, dilution ratios of 10− 4, 10− 5 and 10− 6 were spread in duplicate in petri dishes containing L-MSM, CMC-MSM and nutrient agar. An isolation medium was prepared, having a similar composition to the enrichment medium. The abundance of bacteria was calculated from the counting data, as colony-forming units (CFU g− 1) of the sample. Representative colonies were purified by repeated streaking on NA plates. Purified colonies from L-MSM agar plates were further studied for utilisation of sole carbon source lignin (300 mg L− 1) by nutrient enrichment plate assay (Chandra et al. 2012).
Morphological and cultural characterization
Isolates recovered on L-MSM and CMC-MSM media plates were further studied for morphological and cultural characterizations by inoculating in sterile NA media plates. After 48 h of incubation at 37 °C, their morphological characteristics such as Grams staining, shape and arrangements were observed in a compound microscope under 100X magnification. Colony characteristics like colony size, form, margin and elevation were noted. The pure cultures were preserved on a nutrient agar slant at 4 °C temperature.
DNA extraction, PCR amplification and identification
DNA was extracted and amplified using universal primers (forward primer 27F, 5’ AGAGTTTGATCCTGGCTCAG 3’, and reverse primer 1492R, 5’ TACGGTTACCTTGTTACGACTT 3’) for 16 S rRNA gene amplification at 94 °C for 5 min initial denaturation, 94 °C for 20s denaturation, 58 °C for 20s annealing, 72 °C for 10 min extension. The size of the fragments was confirmed using agarose gel electrophoresis. The Sanger sequencing was performed for 16 S rRNA gene sequencing. The sequence was submitted to NCBI (National Center for Biotechnology Information). The phylogenetic tree was constructed using MEGA X software.
Qualitative screening of isolates for cellulase production
Purified strains were inoculated as a regular spot into media plates containing, Bushnell Haas agar supplemented with 1% carboxymethylcellulose Na salt (medium viscosity) and incubated for 48 h at 37 °C temperature. Clearance zone surrounding colonies were observed by flooding plates with Gram’s iodine. The relative enzyme activity of isolates was calculated by following the formula:
REA = Diameter of clearance zone (mm)/ Diameter of bacterial colony (mm).
Qualitative screening of isolates for amylase production
The starch agar medium was used to screen extracellular amylase production. The bacterial suspension was inoculated for 24 h at 37 °C temperature. Gram’s iodine was used as a differentiating stain to observe the zone of clearance. Above mentioned formula was used to calculate REA.
Qualitative screening of isolates for protease production
Gelatine agar medium was used to screen extracellular protease production. The bacterial suspension was inoculated in gelatin agar and incubated for 24 h at 37 °C temperature. Frazier’s reagent [containing (g L− 1) concentrated HCl (200 mL); mercuric chloride (150 gm)] was used to flood the plate. The clearance zone indicated protease secretion. Above mentioned formula was used to calculate REA.
Qualitative screening of isolates for pectinase production
Qualitative screening of extracellular pectinase was carried out on the pectin agar medium. Bacterial strains were spot inoculated and incubated for 24 h at 37 °C temperature. Detection of extracellular pectinase was carried out by flooding the plates with Grams iodine. The clearance zone surrounding colonies was measured and REA was calculated by the above-mentioned formula.
Qualitative screening of isolates for ligninases production
Lignin degrading ability was measured using the ABTS agar method. Bacterial strains were grown on Lignin Basal Medium (LBM) containing (g L− 1) (KH2PO4; 1, C4H12N2O6; 0.5, MgSO4. 7H2O; 0.5, CaCl2.2H2O; 0.01, yeast extract; 0.01, CuSO4.H2O; 0.001, Fe2 (SO4)3; 0.001, MnSO4.H2O; 0.001, agar; 16) supplemented with 0.1% w/v ABTS. 1 mL of 20% glucose (separately sterilized) was aseptically added to 100 mL of LBM. Cells were inoculated on LBM and plates were incubated at 28 °C temperature for 24 h. ABTS (2, 2- Azino-bis 3-ethylbenz-thiazoline-6-sulfonic acid) was converted to ABTS- azine a green colour substance by laccase-producing isolates. Thus, colonies with a green colour indicate ligninase-secreting isolates (Neethu et al. 2017).
Biochemical characterization
Strains producing the mentioned enzymes were further studied for biochemical characterization. Positive and negative results were noted for biochemical tests viz. indole test, MR test, VP test, citrate utilization test, catalase test, oxidase test, urea hydrolysis test, gelatine hydrolysis test, ammonia production test, nitrate reduction test, H2S production test, dehydrogenase test and haemolysis production test.
Salt profile
The impact of NaCl on extracellular cellulase secretion was examined at NaCl concentrations ranging from 0 to 15% w/v. The study used an agar medium containing 1% carboxy-methylcellulose Na salt (medium viscosity) as the sole carbon source, supplemented with Bushnell Haas agar. After 48 h of incubation at 37 °C, the plates were flooded with Gram’s iodine, and the ratio of the zone of clearance to the colony diameter was calculated.
Temperature growth profile
The optimal growth temperature of bacterial isolates was determined by inoculating uniform inoculums (1 absorbance) in nutrient broth and incubating at temperatures ranging from 30 to 50 °C for 48 h. The medium pH was adjusted to 7 using 1 N HCl and 1 N NaOH. Bacterial growth was periodically estimated spectrophotometrically at 600 nm wavelength after 0, 24, and 48 h of incubation using 5 mL harvested suspensions.
pH growth profile
The effects of pH on the growth behaviour of bacterial isolates were determined in nutrient broth. The pH of the medium was adjusted in the range of acidic to alkaline (4, 5, 6, 7, 8, 9, 10, and 11) by 1 N HCl and 1 N NaOH. The inoculum (1 absorbance) was added to media broth of various pH values and incubated for 48 h at 35 °C temperature. Aliquots of 5 mL inoculated suspension were periodically harvested at 0, 24 and 48 h of incubation and turbidity of bacterial growth was estimated spectrophotometrically at 600 nm wavelength. Growth assessment involved a comparison of the turbidity in the growth medium with that of the controls.
Statistical analysis
The physiological characteristics along with the enzymatic profile were examined in triplicate and both standard deviation and standard error were calculated to estimate variability within a sample and across the samples respectively. The statistical analysis was performed using SPSS software. Moreover, the ANOVA (Analysis of Variance) was performed for physiological characteristics and enzymatic profile.
Results
The synergistic action of endoglucanases, exoglucanases and β-glucosidase is required to break cellulose polysaccharides. However, some fungal cellulase systems lack β-glucosidase, causing feedback inhibition and repression due to cellobiose accumulation. Bacteria have rapid growth rates and multienzyme expression potential; therefore, emphasis has been given to the isolation of bacterial genera having highly active and specific secretion systems (Petre et al. 2000).
Enrichment and isolation of bacteria
Cellulolytic and lignolytic bacteria were enriched in CMC-MSM and L-MSM enrichment broth. CMC was used as the sole carbon and energy source in the CMC-MSM enrichment broth. However, in addition to lignin, glucose and peptone were used as additional carbon and nitrogen sources in L-MSM enrichment broth, which worked as a co-substrate to enhance lignolytic bacteria (Zainithet al. 2019). Variations in the total culturable heterotrophic bacterial populations (CFU g− 1) were observed in these samples. The highest population density of total culturable heterotrophic bacteria was observed as 1.3 × 108 and 1.2 × 108 CFU g− 1on L-MSM and NA plates respectively, while the culturable cellulolytic bacterial population displayed 4 × 106 CFU g− 1 population densities in the compost sample. However, culturable heterotrophic bacterial population density of sugarcane rhizospheric samples on L-MSM and NA plates were observed as 1.2 × 107 CFU g− 1and 1.7 × 107 CFU g− 1respectively, while culturable cellulolytic bacterial population density was 2.5 × 105 CFU g− 1. The proportion of the population of cellulose-degrading bacteria to the total culturable heterotrophic bacteria in compost and rhizospheric soil was 3.33% and 1.47% respectively.
In the present investigation, cultivation strategies resulted in 96 bacterial isolates from compost and sugarcane rhizospheric soil. Enrichment techniques yielded 36.45% isolates (L-MSM; n = 21, 21.87% and CMC-MSM; n = 14, 14.58%) whereas, the rest of the n = 61, 63.54% isolates by direct plating on NA medium. Results are shown in Fig. 1. A total of 21 isolates were recovered on an L-MSM isolation medium containing supporting carbon and nitrogen sources. These isolates were further streak plated on a minimal medium supplemented with lignin (300 mg L− 1) as a sole carbon and energy source to study lignin utilisation. Remarkable growth was observed in four isolates CP2, CP4, CP9 and CP11 showing the indication of lignin hydrolysis. Additionally, 14 isolates in this study, recovered from CMC-MSM plates were considered cellulolytic bacterial isolates.
Morphological and cultural characterization of isolates
With the aim of morphological and cultural characterization in the present investigation, 35 isolates recovered on L-MSM and CMC-MSM mediums were selected and cultivated on nutrient agar for 24 h at 37 °C temperature. The morphological and cultural characterization of these isolates recovered on CMC-MSM and L-MSM media plates are shown in Table 1. Microscopic observations revealed that 75% of isolates were Gram-positive. Furthermore, rod shape was observed in 82.85% of isolates while 22% of isolates were arranged in chains and 48% of isolates displayed isolated cell arrangements. Colony characteristics revealed that 45% of colonies have intermediate size whereas, 31% large and 15% small colonies were observed. The irregular and round shape was observed in 34% and 65% of colonies respectively.
Identification of isolates
In order to identify the potent isolates SRS11, SRS8, CP11, CP9, CP5, and CP2, the 16S rRNA gene was first amplified using the universal primers 1492R (5’ TACGGTTACCTTGTTACGACTT 3’) and 27F (5’ AGAGTTTGATCCTGGCTCAG 3’). Subsequently, sequencing of the 16 S rRNA gene amplified product of the isolates SRS11, SRS8, CP11, CP9, CP5, and CP2 resulted in 882 bp, 1516 bp, 1510 bp, 1510 bp, 1507 bp, 1504 bp sequences respectively, which was closely related to the sequences of Bacillus licheniformis, Priestia megaterium, Bacillus subtilis, Priestiaflexa, Priestia megaterium, and Priestia flexa respectively with the sequence similarity of 98.61%, 99.97%, 99.60%, 99.54%, 99.53%, and 99.73% respectively. Further, the 16 S rRNA gene sequences of potent isolates were submitted to NCBI and the assigned Genbank accession numbers to the isolated SRS11, SRS8, CP11, CP9, CP5, and CP2 strains were OR856070, OR856075, OR856071, OR856072, OR856073, and OR856074 respectively. Moreover, the 16 S rRNA gene sequences were used to construct a phylogenetic tree by a neighbour-joining approach in Mega X software which represents three clusters of closely related microbial isolates (Fig. 2). The isolates’ sharing ancestry and evolutionary divergence were distinguished in the clusters. The main three clusters observed in the phylogeny tree contained Priestia sp. and Bacillus sp. In the first cluster, Priesta sp. and in the third cluster Bacillus sp. were predominant, while both genera were observed in the second cluster which exhibited a higher diversity of microbial species. The genetic diversity seen in the microbial communities was revealed by this analysis, which also provided insights into the taxonomy and evolutionary background of the microbial strains isolated from the sugarcane rhizospheric soil and compost.
Qualitative screening of isolates for hydrolytic enzyme secretion
Organic solid wastes are varied in composition and are made up of heterogeneous organic polymers. Multiple enzyme systems are required for the saccharification of these wastes. Morphologically distinct isolates were further studied for in-vitro qualitative screening of enzymes to study waste saccharification. 1% CMC, starch and pectin were used as a substrate; differentiating stain iodine has a binding ability with these sugars and produces a bluish-black complex but not with hydrolyzed sugars. The details of isolates having multiple enzyme secretion potentials are shown in Table 2. Out of the total isolates, 16.66% of isolates were found positive for at least one lytic enzyme. Cellulase, ligninase, amylase, protease and pectinase production were observed in 14.53%, 4.16%, 9.37%, 7.29% and 5.20% isolates respectively. The results revealed that the compost isolates CP2 secreted all five lytic enzymes, whereas isolates CP4 and CP11 secreted all lytic enzymes except pectinase. Moreover, isolate CP9 secreted all enzymes except protease. The least relative enzyme secretion was observed in the isolates of sugarcane rhizosphere SRS3, which displayed positive results for cellulase and protease, and SRS11, which showed positive results for cellulase and pectinase, while, SRS8 was observed positive for cellulase, amylase and protease. Multiple enzyme secretion was recorded optimum in isolates of compost samples. The analysis of cellulase, amylase, protease, and pectinase secretion as variables in a Completely Randomized Design (CRD) revealed significant results with a p-value < 0.01 (Table 3). The highest levels of cellulase, amylase, and pectinase secretion were observed in isolates CP2, SRS8, and CP9, respectively, and none of the other isolates were at par with it. However, the highest levels of protease were secreted by isolates CP11 and CP2 and were found statistically at par with it. The mean values along with standard error and Tukey test results are shown in Table 3.
Biochemical characterization of isolates
Biochemical characterization of isolates having lytic activity was carried out on various mediums and results are reflected in Fig. 3. Biochemical tests following the protocol outlined by Cowan and Steel (1974), were conducted to identify the unknown genus. Microscopic observation and cultural characteristics were also considered for the identification of isolates. Isolate CP4 was included in Gram-negative rods, catalase and oxidase-positive category. It was found catalase, oxidase, MR, VP, and citrate were positive, while, indole, urease, nitrate reduction, ammonia production, H2S production, dehydrogenase and hemolysis production were negative. According to Cowan and Steel’s manual, isolate CP4 was tentatively identified as Pseudomonas spp. Isolate CP5 was included in Gram-positive rods, catalase-negative; oxidase-positive category. Bacilli was observed in chain arrangement and found indole, VP, citrate, urease, nitrate reduction, ammonia production, H2S production, dehydrogenase, and hemolysis production negative while, MR test and starch hydrolysis positive. All other isolates are categorised in Gram-positive rods, catalase and oxidase-positive category. They were found oxidase and catalase positive, whereas, indole, MR, ammonia production, H2S production, and dehydrogenase negative. A haemolysis pattern was observed in the CP11 isolate. Nitrate reduction was observed in isolates SRS3, SRS8, and CP9. Citrate utilization was displayed by SRS3, SRS8, CP2 and CP11, whereas, VP negative was displayed by CP2 and CP9 isolates. As per the Cowan and Steel manual, isolates SRS3, SRS8, SRS11, CP2, CP9 and CP11 were tentatively identified as Bacillus spp.
Salt profile
The effect of NaCl concentration on enzyme activity was carried out at the range of 0 to 15% by using a CMC-BH medium and relative enzyme activity was calculated after 48 h in terms of zone ratio displayed in Table 4. The results revealed that the sugarcane rhizospheric isolate SRS8 displayed the lowest tolerance limit of up to 3% NaCl concentration, with its optimum activity (zone ratio 1.76 ± 0.028) observed at 0% NaCl concentration. Additionally, colony growth (4 mm) was observed at 5% NaCl, but a hydrolytic zone was not observed. Similarly, isolates SRS3 and SRS11 exhibited relative activity up to 5% NaCl concentration. However, SRS11 showed optimum activity (zone ratio 2.00 ± 0.000) at 0% and 3% NaCl (w/v), while SRS3 displayed optimum activity (zone ratio 1.56 ± 0.057) at 0% NaCl. Colony growth (3 mm) was observed at 7% NaCl, but no hydrolysis zone was formed by SRS3. A similar pattern to SRS3 was also observed in compost sample isolate CP4, with a 5% NaCl tolerance limit. The optimum activity (zone ratio 1.80 ± 0.200) was observed at 0% NaCl, while growth without activity was observed at 7% NaCl. Isolate CP5 displayed optimum activity at 0% NaCl (zone ratio 3.40 ± 0.200), which was decreased up to 5% NaCl. Compost isolates CP2, CP9, and CP11 showed activity across a wide range of NaCl concentrations (0 to 7%), with optimum activity at 0%, 0%, and 3% NaCl with zone ratios of 3.56 ± 0.055, 2.60 ± 0.000, and 2.62 ± 0.203 respectively. In the presence of 9%, 11%, and 15% NaCl, none of the isolates exhibited any growth, indicating their limited tolerance to high salinity conditions. Among all the isolates, CP2 displayed the highest relative activity, followed closely by CP5 and CP9, suggesting their superior enzymatic capabilities in response to the tested conditions. Moreover, the compost isolates demonstrated a greater relative activity compared to the isolates from sugarcane rhizospheric soil, indicating their better adaptability and efficiency in enzyme secretion under the given salinity levels. The analysis of the isolate factor component, NaCl factor component and interaction component variables in a Completely Randomized Design (CRD) revealed significant results with a p-value < 0.01 (Table 5). For the isolate variable, the highest zone ratio was observed for CP2 and none of the treatment combinations were at par with it. Additionally, for the NaCl variable, the highest zone ratio was observed at 0% NaCl (w/v) and none of the treatment combinations were at par with it. However, for interaction component variables (Isolate x NaCl) highest zone ratio was observed for CP2 × 0% NaCl and CP5 × 0% NaCl, CP2 × 3% NaCl, were found statistically at par with it. The mean values along with standard error and Tukey test results are shown in Table 5.
Temperature growth profile
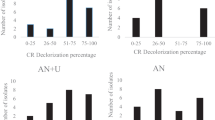
The isolates displaying multiple enzyme secretion potentials were further examined to determine their temperature growth optima. The results revealed that sugarcane rhizospheric isolates SRS3 exhibited rapid growth with an absorbance of 1.15 ± 0.028 at 35 °C within 48 h, followed by SRS8 with an absorbance of 0.936 ± 0.003 at 40 °C. However, the growth of SRS11 was comparatively slower, indicated by an absorbance of 0.36 ± 0.010 at 35 °C within 48 h. Additionally, compost sample isolates CP5 exhibited rapid growth with an absorbance of 1.34 ± 0.032 at 40 °C within 24 h, followed by CP9 and CP2, which showed absorbance of 1.29 ± 0.023 and 0.87 ± 0.003 respectively at 45 °C within 48 h. On the other hand, CP11 and CP4 exhibited slower growth, indicated by absorbance of 0.71 ± 0.019 and 0.86 ± 0.003 at 40 °C and 35 °C respectively within 48 h. The analysis of the isolate factor component, time factor component, temperature factor component and interaction component variables of rhizospheric and compost isolates in a Completely Randomized Design (CRD) revealed significant results with a p-value < 0.01. The highest absorbance values for the isolate variable, time variable, and temperature variable were observed for SRS3, 48 h, and 40 °C, respectively, for rhizospheric isolates. For compost isolates, the highest absorbance values were observed for CP5 at 48 h and 40 °C temperature, and none of the treatment combinations were at par with it. Additionally, for isolates x time, isolate x temperature and time x temperature interaction component variables, the highest absorbance values were observed for SRS3 × 48 h, SRS3 × 35 °C and 48 h x 40 °C respectively for rhizospheric isolates and none of the treatment combinations were at par with it. For compost isolates the highest absorbance values were observed for CP5 × 48 h, CP 5 × 40 °C and 48 × 40 °C respectively, and none of the treatment combinations were at par with it. However, for the isolates x time x temperature interaction component variables, the highest absorbance value was observed for SRS3 × 48hx35°C, and none of the treatment combinations in rhizospheric isolates were found to be at par with it. Regarding compost isolates, the highest absorbance values were observed for CP5 × 48 × 40 °C and CP5 × 24 × 40 °C, CP9 × 48 × 45 °C, CP5 × 48 × 30 °C, CP5 × 48 × 35 °C, CP5 × 24 × 35 °C were statistically at par with the highest values. The absorbance values along with standard error and Tukey test results of rhizospheric and compost isolates are shown in Figs. 4a and b and 5.
The comparative discussion on temperature growth optima of bacterial isolates from compost and sugarcane rhizospheric soil revealed interesting findings. The results indicated that the optimal growth temperature for most bacterial isolates from compost was higher, with 45 °C being the optimum for CP2 and CP9. In contrast, some isolates showed optimal growth at 40 °C, such as CP5 and CP11, while CP4 exhibited optimal growth at 35 °C as indicated in Fig. 4a,b.
On the other hand, the bacterial isolates from sugarcane rhizospheric soil displayed their highest growth rate at lower temperatures. SRS3 and SRS11 exhibited optimal growth at 35 °C, and SRS8 showed the highest growth at 40 °C as shown in Fig. 5.
pH growth profile
The pH growth optima of multiple enzyme-secreting isolates were determined. Potent isolates from sugarcane rhizospheric soil (SRS3, SRS8, and SRS11) exhibited their optimum growth at pH 7. Isolate SRS3 displayed rapid growth with an absorbance of 1.14 ± 0.015 within 48 h, while SRS8 and SRS11 showed delayed growth with an absorbance of 1.00 ± 0.057 and 0.78 ± 0.002, respectively, within the same time frame as shown in Fig. 6. In contrast, potent isolates from compost samples CP2, CP4, and CP5 showed optimum growth at pH 7, while isolates CP9 and CP11 exhibited their best growth at pH 8. The isolate CP5 demonstrated rapid growth with an absorbance of 1.25 ± 0.037 within 24 h at pH 7, while CP2 and CP4 showed delayed growth with absorbance of 1.018 ± 0.045 and 1.01 ± 0.026, respectively, within 48 h at pH 7. Similarly, CP11 and CP9 displayed delayed growth, with absorbance of 1.27 ± 0.031 and 0.85 ± 0.020, respectively, within 48 h at pH 8 as indicated in Fig. 7a,b. The analysis of the isolate factor component, time factor component, pH factor component and interaction component variables of rhizospheric and compost isolates in a Completely Randomized Design (CRD) revealed significant results with a p-value < 0.01. The highest absorbance values for the isolate variable, time variable, and pH variable were observed for SRS3, 48 h, and 7 pH, respectively, for rhizospheric isolates. For compost isolates, the highest absorbance values were observed for CP5, 48 h, and none of the treatment combinations were at par with it. For the pH factor, the highest absorbance value was observed for pH 8 and pH 7, which were found statistically at par with it. Additionally, for isolates x time, isolate x pH and time x pH interaction component variables, the highest absorbance values were observed for SRS3 × 48 h, SRS3 × 7 pH and 48 h x 7 pH, respectively, for rhizospheric isolates. The highest absorbance values were observed for CP5 × 48 h, CP5 × 7pH and 48 × 8pH respectively for compost isolates and none of the treatment combinations were at par with it. However, for the isolates x time x pH interaction component variables, the highest absorbance value was observed for SRS3 × 48 × 7pH and CP5 × 48 × 7pH respectively, for rhizospheric and compost isolates and none of the treatment combinations were found to be at par with it. The mean values along with standard error and Tukey test results of rhizospheric and compost isolates are shown in Figs. 5 and 6a and b. Optimum physiological conditions for potent isolates are summarized in Table 6.
Discussion
The present investigation showed that densities of cellulolytic bacteria were lower in rhizospheric soil and higher in compost than those reported by Saini et al. (2012) who detected 6.6 × 104 and 2.8 × 107 CFU g− 1 in rhizospheric soil and compost respectively. Additionally, in support to present investigation, they have reported low cellulolytic as well as total bacterial population density in rhizospheric soil compared to the compost. Previously, Kim et al. (2013) stated that the high organic matter composition of compost was responsible for the elevated population of lignocellulolytic bacteria in compost than other samples. More recently, Rinady et al. (2023) reported a lower population of 1.71 × 105 and 4.24 × 104CFU g− 1 of heterotrophic and cellulolytic bacteria, respectively at the pine-coffee agroforestry system of the plantation. In another study, Cabrera et al. (2013) reported a higher abundance of cellulolytic bacteria (106 CFU g− 1) and total culturable bacteria (107 CFU g− 1) in the alfalfa rhizosphere. They also conclude that the cellulolytic bacterial density in the rhizosphere enhanced with the growth of plants, and different plant spp. have different rhizospheric effects at various stages of growth according to the composition of root exudates. Recently, Mohammadipour et al. (2021) reported 3 × 109 CFU mL− 1 heterotrophic and 94 × 104 CFU mL− 1 cellulolytic population densities in compost leachate samples. Temperature, methods of composting, raw material and decomposition steps consisting of mesophilic and thermophilic microorganisms affect the population density and diversity of microbes in compost (Roy et al. 2018). More recently, Breza-Boruta et al. (2023) reported that soil treated with manure exhibited the highest abundance of heterotrophic bacteria (165.1 × 106 CFU g− 1) and cellulolytic bacteria (17.2 × 106 CFU g− 1). Additionally, using the CMC-MSM enrichment medium, Udume et al. (2023) obtained an average value of 2.92 ± 0.10 × 106 CFU g− 1 of cellulolytic organisms from water hyacinth compost enriched samples. In a microbial population study of various waste organic materials, Dąbrowska et al. (2022) reported that spent mushroom compost contained 1.47 × 1011 CFU g− 1 heterotrophic and 4.46 × 109 CFU g− 1 cellulolytic bacteria, while composted sawdust contained 2.38 × 107 CFU g− 1 heterotrophic and 2.17 × 106 CFU g− 1cellulolytic bacteria. Moreover, raw sawdust and wood chips contained 8.20 × 103 CFU g− 1 and 7.02 × 104 CFU g− 1 heterotrophic bacteria, as well as 1.17 × 102 CFU g− 1 and 1.76 × 103 CFU g− 1 cellulolytic bacteria.
In the present investigation, enrichment techniques yielded 36.45% isolates on an L-MSM isolation medium containing supporting carbon and nitrogen sources. In a similar finding Verma et al. (2020) conducted a study on bacterial decolourization of Kraft lignin using glucose and peptone as supplementary carbon and nitrogen sources. The initial stage exhibited limited decolourization of KL, attributed to the utilization of peptone and glucose as nitrogen and carbon sources present in L-MSM. However, as these nutritional sources were gradually depleted in L-MSM, the bacterial culture shifted towards utilizing lignin as a co-substrate for further decolourization. In the current finding, these isolates were further screened for lignin utilization on a minimal medium supplemented with lignin (300 mg L− 1) as a sole carbon and energy source and remarkable growth was observed in 4 isolates. A similar study reported very fast growth of Bacillus spp. Ochrobactrum sp. and Leucobacter sp. on agar plates containing lignin (500 mg L− 1) without the addition of glucose and peptone (Rahman et al. 2013). In support to present investigation, Chen et al. (2012) used a maximum lignin concentration (300 mg L− 1) for Comomonas sp. due to inhibition of bacterial growth at high lignin concentration. Moreover, bacterial isolates have the competency to efficiently assimilate and degrade sole carbon source lignin as confirmed by the findings of Dube et al. (2023) who reported ten bacterial species isolated from compost with lignin utilization ability, whereas optimum growth was observed in Pseudomonas aeruginosa (CP031449.2) and E. coli (LR025096.1). Additionally, Radhika et al. (2023) conducted a study involving the selective enrichment of lignin-degrading bacterial consortia through a forced laboratory adaptation method. This method utilized Kraft lignin as the sole carbon source and involved samples from compost and soil. They identified lignin utilization potential in various bacterial genera, including Bacillus, Pseudomonas, Brevibacillus, Paenibacillus, and Aneurini bacillus. Moreover, in accordance with the current findings, Azman et al. (2019) studied the depolymerization of alkali lignin to demonstrate the capacity of thermophilic bacterial strains for utilizing and breaking down alkali lignin beyond a monomeric form within a 7 days incubation period. Findings revealed that Stenotrophomonas sp. S2 and Bacillus subtilis S11Y achieved around 50% and 20% reduction in alkali lignin over 7 days of incubation without the need for supplementary carbon sources. Additionally, 14 isolates in this study, recovered from CMC-MSM plates were considered cellulolytic bacterial isolates. Similar to the present finding, Mohammadipour et al. (2021) purified 15 isolates from compost leachates sample on CMC media plates according to differences in morphological characteristics. Nevita et al. (2018) also reported the isolation and identification of Stenotrophomonas maltophilia RSD6 from the rhizospheric region of O. sativa L by CMC and Lignin–MSM nutrient enrichment techniques. Moreover, Sekhar et al. (2022) screened 24 cellulase-positive isolates from sugarcane rhizosphere on a CMC-MSM medium and identified two potential isolates Bacillus amyloliquefaciens and Pseudomonas putida. Moreover, Nur et al. (2020) reported positive screening of 125 cellulolytic bacteria on CMC agar medium using decomposing rice straw with varying C/N ratios. They found that the total number of isolates from rice straw with a higher degree of decomposition exceeded those from rice straw with a lower level of decomposition.
Findings of microscopic, cultural and biochemical characteristics revealed that 75% of isolates were Gram-positive and the majority of them were tentatively identified as a Bacillus spp. Similar to the present study, Ojo-Omoniyi et al. (2016) reported the optimum occurrence of Bacillus (42.5%) and related Gram-positive genera (22.5%) in dump site waste and compost compared to other isolates because they can thrive in harsh environments and they are indigenous to soil environments. In addition, Dube et al. (2023) reported the abundance of the Bacillus genus commonly found in plant litter, soil as well as compost samples and they were known for lignocellulolytic activity and responsible for the bioconversion of macromolecules. Henry et al. (2020) conducted a metagenomics analysis of compost samples and revealed the optimum occurrence of firmicutes, chloroflexi, bacteroidetes, actinobacteria, and acidobacteria. The highest abundance (59%) of firmicutes in the heating phase of tomato stalk composting was also reported by Zhang et al. (2021). In another study, Grenier et al. (2023) reported seventy-two lignocellulolytic species from compost samples, out of them 34 spp. belonging to the phylum Firmicutes were dominated by the order Bacillales with 22 spp. Lignocellulolytic bacteria, including Arthrobacter, Brevibacterium, Bacillus, Chryseobacterium, Pseudomonas, Xanthomonas, Paenibacillus, Stenotrophomonas, and Streptomyces, were reported in the rhizosphere of Phleum pretense (Saleh et al. 2019), Streptomyces in the rhizosphere of Zea mays (Adegboye et al. 2018), Arthrobacter and Pseudomonas in the rhizosphere of Quercus sp. (Lasa et al. 2019), and Bacillus, Pseudomonas, and Kocuriain the rhizosphere of Salsola stocksii and Atriplex amnicola (Mukhtar et al. 2019). It has long been known that Bacillus species produce large amounts of different enzymes with a wide range of catalytic activity. The Bacillus sp. produces enzymes that are used extensively in industry and aid in the development of economical and environmentally friendly procedures. The versatility of Bacillus enzymes, encompassing their stability and activity under diverse situations, renders them indispensable instruments in a multitude of sectors, encompassing bioremediation and the manufacturing of bio-based commodities. The Bacillus species demonstrate a noteworthy ability to decompose solid waste, underscoring its potential for use in waste management techniques. These bacteria are important decomposers of a variety of organic materials in both natural and artificial contexts because of their adaptability, enzymatic activity, and versatility.
Phylogenetic analysis is essential for the taxonomy classification of microorganisms (Gohel et al. 2023). Organisms can be arranged and classified according to their evolutionary relatedness with the assistance of a phylogenetic tree. It is essential to preserve a uniform and widely recognized system of naming and categorizing living things. The phylogenetic tree derived from genomic data provides a more accurate depiction of the branching patterns, making it easier to distinguish between closely related species and strains (Pearson et al. 2009; Patwardhan et al. 2014). In this study, the species belonging to the genera Bacillus were predominantly observed in the phylogenetic tree, constructed using isolates obtained from the sugarcane rhizosphere and compost. Numerous bioactive chemicals are known to be produced by Bacillus species (Kaspar et al. 2019). The variety of secondary metabolite biosynthesis gene clusters found in Bacillus genomes are shown by genomic investigations (Belbahri et al. 2017). The diversity of Bacillus species enhances their capacity to generate new bioactive chemicals that find use in industry, agriculture, and medicine (Mondol et al. 2013; Kaspar et al. 2019 ). The adaptation of Bacillus to a variety of ecological habitats, such as compost and rhizospheric soil, was reflected by variations of genomic content, such as the presence of certain transporters or stress response genes (Zhang et al. 2016; Iqbal et al. 2021). The taxonomic categorization of Bacillus species is improved and species boundaries are drawn using genomic data. Numerous details, including functional features and evolutionary links, can be obtained from genomic and molecular insights into the phylogeny of Bacillus species (Alcaraz et al. 2010; Khurana et al. 2020). These discoveries are significant due to their implications in environmental science, agriculture, and medicine as well as for understanding microbial ecology and the biotechnology potential of microorganisms.
The present investigation unveiled that cellulase, ligninase, amylase, protease, and pectinase production of isolates were observed in varying percentages. In a more or less similar outcome, Mukhtar et al. (2019) conducted enzymatic screening of bacterial isolates from the rhizosphere of Salsola stocksii. They observed protease, lipase, cellulase, amylase, urease, gelatinase, and catalase activity in 53%, 68%, 40%, 33%, 29%, 26%, and 34% of the isolates, respectively. Similarly, in a study published by Semira et al. (2021), 31% amylase-positive isolates were observed to give a zone of clearance from various rhizospheric soil samples. The rhizospheric microbes exhibited a preferential utilization of D-galacturonic acid and its reduced form, D-galactonic acid (Lopes et al. 2016). D-galacturonic acid, the most abundant component of pectin and a major constituent of plant cell walls is consequently released in the rhizosphere (Zhang et al. 2011). Furthermore, D-galacturonic acid has been identified as a component of root exudates by Tawaraya et al. (2015). Additionally, in sugarcane field soils, the abundance of hemicellulose is notably higher due to the continuous deposition of leaves on the surface (Chandel et al. 2012). Hemicellulose is a significant component of lignocellulose, which constitutes the most recalcitrant part of the plant cell wall and requires the participation of complex lignocellulase enzymes for degradation (Mansour et al. 2016). In support of the present investigation, Neethu et al. (2017) reported positive qualitative screening in 15 cellulolytic (14.56%) and 10 lignolytic (9.70%) bacterial isolates from various compost samples on CMC and ABTS plates. Similarly, Bambharolia et al. (2021) reported that among a total of 56 bacterial isolates, 73% exhibited distinct clear zones around their colonies on CMC agar, while only 8% isolates displayed coloured zones around their colonies on ABTS agar. Contradictory to the described trends, 38% protease and 28% cellulase-positive screening from various enriched samples was published by Admassie et al. (2022). Additionally, Jiang et al. (2020) reported that a commensal relationship between lignocellulolytic bacteria occurs through the enzymatic removal of lignin from lignocellulosic biomass. This process facilitates the availability of cellulose for degradation by cellulase-producing organisms. More recently, Chukwuma et al. (2023) reported that landfill bacteria, such as Bacillus proteolyticus, Bacillus Sanguinis, Bacillus spizizenii, Bacillus paramycoides, Bacillus paranthracis, and Neobacillus fumarioli, exhibited multi-enzymatic activity, including amylase, cellulase, xylanase, and protease. Earlier, Priyanka et al. (2021) also reported positive screening of 22.47% cellulolytic and 13% lignolytic bacterial isolates from rhizospheric soil and compost-enriched samples. In a metagenomic analysis conducted by Martins et al. (2013) on São Paulo Zoo compost samples, 56% of genomes were found to contain genes associated with β-glucosides, and 24% of the total genomes contained cellulases and β-glucosides. This finding suggests that the chemical nature and availability of carbohydrates in the habitat influence microbial adaptation and their carbohydrate-active enzyme composition. Soil and compost ecosystems have been extensively studied in this regard (López-Mondéjar et al. 2020). In a study by Mironov et al. (2021), abundant proteolytic microorganisms were reported throughout composting. During the initial 14 days, active growth of amylolytic microorganisms was observed, while cellulolytic microorganisms thrived during the thermophilic period, reaching their peak at the beginning of maturation. Similarly, in a study conducted by Chroni et al. (2009), a significant increase in amylolytic, cellulolytic, and proteolytic microorganisms was observed during the thermophilic stage. Similarly, López-González et al. (2014) reported distinct variations in activities related to the degradation of lignocellulosic and starchy polymers. Approximately 33% of the total isolates exhibited both amylolytic and hemicellulolytic activities. However, the percentage of microorganisms capable of hydrolyzing cellulose, lignin, and pectin did not exceed 4% in the composting process. Identification of cellulolytic Pseudomonas sp. was also performed by Poly et al. (2022) using the Cowan and Steel manual. Also, Pan et al. (2021) characterized Thermohalobaculum Xanthum Gen. Nov., Sp. Nov. using the method described by Cowan and Steel.
NaCl significantly affected CMCase activity, except SRS8 all the isolates exhibited activity between 0 and 7% NaCl (w/v), and no activity at higher NaCl concentrations. A similar study reported supporting evidence as they identified 98 cellulolytic bacterial isolates from the soil (Cabrera et al. 2013). CMCase hydrolytic zone was observed in 100%, 82.6%, 50%, 24%, and 1% of the isolates at 1%, 3%, 7%, and 9% NaCl concentration, respectively, while growth was not observed at 12%. Balla et al. (2022) examined the influence of NaCl on 12 cellulolytic bacterial isolates from the rhizosphere. The results revealed that 58%, 34%, and 8% of the isolates displayed optimum growth at 0 mM, 400 mM, and 800 mM NaCl concentrations, respectively. Furthermore, Mukhtar et al. (2019) also reported that rhizospheric bacterial strains HL1RS13, AT2RP4, NRS4HaP9, and LK3HaP7 exhibited stable cellulase activity in the presence of NaCl concentrations ranging from 0.5 to 2.5 M. Moreover, Vijayalaxmi et al. (2013) demonstrated the ability of lignocellulolytic compost-isolated Exiguobacterium sp. strain VSG-1 to grow across a wide range of NaCl concentrations (1–16%), with the best growth observed at 1% NaCl (w/v).
Compost isolates such as CP5 and CP11 showed optimum growth at 40 °C, CP9 and CP2 showed optimum growth at 45 °C, while CP4 showed optimum growth at 35 °C. Rhizospheric isolates on the other hand such as SRS3 and SRS11 thrived at 35 °C. Previously, Buraimoh et al. (2015) reported the identification of three strains from a composting site such as Bacillus sp. (KF977555), Bacillus megaterium (KF977554) and Streptomyces aureus (KF977549). These strains were found to grow within a temperature range of 28 to 60 °C, with optimum growth occurring at 37 °C, 60 °C, and 50 °C respectively. López et al. (2021) reported the lignocellulolytic bacterial strains Bacillus thermoamylovorans 1141 and Geobacillusthermodenitrificans 3781 from compost samples, which displayed a temperature growth range of 30 to 60 °C and 20 °C to 60 °C, respectively, with optimum growth temperatures of 40 and 50 °C. Additionally, during the heating phase, Bacillus sp. has been investigated as the predominant genus in the compost, which can form endospores and has growth optima in the thermophilic range. In contrast, the rhizospheric region was moisturized due to root exudates, and mesophiles were the prominent flora (Mohammadipour et al. 2021; Villar et al. 2016). Furthermore, Mukhtar et al. (2019) reported that more than 90% of bacterial isolates from the rhizosphere of Salsola stocksii and Atriplex amnicola grew well at temperatures ranging from 25 to 40 °C, with some also capable of growing at 4 and 42 °C. Moreover, Balla et al. (2022) also revealed that 58%, 17%, and 25% of rhizospheric isolates exhibited optimum growth at temperatures 30 °C, 45 °C, and 50 °C, respectively.
According to the present study, sugarcane rhizospheric isolates preferred pH 7. Most compost isolates favoured pH 7, except CP9 and CP11, which showed best growth at pH 8. In support of the present finding, Zhang et al. (2021) reported an increase in pH from 4.5 to 9 during the composting process due to microbial activities, production of lactic acid, and release of ammonia. Studies have indicated that lignocellulose mineralization is more favourable in neutral or slightly acidic environments. However, the solubilization of lignin and the release of lignin by-products are most pronounced in alkaline pH conditions (Verma et al. 2017). Furthermore, several carbohydrate-active enzymes exhibited alkaliphilic behaviour and are most active in higher pH ranges (Mello et al.2017). Mironov et al. (2021) in their study reported the optimum occurrence of waste-degrading Sphingobacterium, Bacillus, Planifilum, and Novibacillus at pH 8 and Leuconostoc and Lactococcus at pH7 in different stages of composting. Kulkarni et al. (2019) reported growth of lignocellulolytic Sphingobacterium sp. ksn-11 in the range of pH 4 to pH 9 while optimum growth was observed within 12 h at pH7 and 40 °C temperature. Moreover, Vijayalaxmi et al. (2013) identified a lignocellulolytic Exiguobacterium sp. strain VSG-1, which displayed optimal growth at alkaline pH 9.0. The strain exhibited maximum growth between 30 and 50 °C, with the optimum temperature at 37 °C. Balla et al. (2022) unveiled that a higher proportion of rhizospheric isolates, specifically 66%, exhibited optimal growth at pH 7, whereas 44% demonstrated their optimal growth at pH 9. Some of the major lignocellulolytic bacteria with their substrates and enzymes are presented in Table 7.
Conclusion
In conclusion, this study demonstrated successful cultivation strategies for lignocellulolytic bacterial strains from compost and sugarcane rhizospheric soil using selective enrichment media. The use of CMC-MSM and L-MSM facilitated the growth of bacteria capable of lignin and cellulose degradation. Compost samples showed higher lignocellulolytic and heterotrophic bacterial density compared to rhizospheric soil. The majority of isolates from compost were identified as Bacillus species due to their potential to survive in diverse and dynamic habitats by producing various hydrolytic enzymes. The isolates displayed significant variations in enzyme secretion potential, NaCl tolerance, temperature preference, and pH growth optima. Cellulase production was prevalent among the isolates. The findings suggest that the potential of these isolated bacteria can be screened for the identification of valuable bioactive compounds, including enzymes and antibiotics, with potential commercial applications. This research contributes to the understanding of lignocellulolytic microbial communities and their biotechnological prospects in diverse environmental habitats.
Abbreviations
- ABTS:
-
2, 2- Azino-bis 3-ethylbenz-thiazoline-6-sulfonic acid
- CMC-MSM:
-
Carboxymethylcellulose - Minimal salt medium
- CMC:
-
Carboxymethylcellulose sodium
- CP:
-
Compost pits
- CPCB:
-
Central pollution control board
- EC:
-
Enzyme commission number
- KL:
-
Kraft lignin
- L-MSM:
-
Lignin - Minimal salt medium
- LBM:
-
Lignin basal medium
- MSW:
-
Municipal solid wastes
- NCBI:
-
National Center for Biotechnology Information
- OSW:
-
Organic solid waste
- REA:
-
Relative enzyme activity
- SRS:
-
Sugarcane rhizospheric soil
- TCI:
-
Tokyo chemical industry
References
Abdel-Rahman MA, Nour El-Din M, Refaat BM, Abdel-Shakour EH, Ewais EED, Alrefaey HMA (2016) Biotechnological application of thermotolerant cellulosedecomposingbacteria in composting of rice straw. Ann Agric Sci 61(1):135–143. https://doi.org/10.1016/j.aoas.2015.11.006
Abu-Tahon MA, Arafat HH, Isaac GS (2020) Laundry detergent compatibility and dehairing efficiency of alkaline thermostable protease produced from aspergillus terreus under solid-state fermentation. J Oleo Sci 69(3):241–254. https://doi.org/10.5650/jos.ess19315
Adegboye MF, Lobb B, Babalola OO, Doxey AC, Ma K (2018) Draft genome sequences of two novel cellulolytic streptomyces strains isolated from South African rhizosphere soil. Genome Announc 6(26):e00632–e00618. https://doi.org/10.1128/genomea.00632-18
Admassie M, Woldehawariat Y, Alemu T (2022) In vitro evaluation of extracellular enzyme activity and its biocontrol efficacy of bacterial isolates from pepper plants for the management of Phytophthora capsici. Biomed Res IntArticle ID 6778352. https://doi.org/10.1155/2022/6778352
Ahmad M, Taylor CR, Pink D, Burton KS, Eastwood DC, Bending GD (2010) Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders. Mol Biosyst 6(5):815–821. https://doi.org/10.1039/b908966g
Ahmad W, Tayyab M, Aftab MN, Hashmi AS, Ahmad MD, Firyal S, Wasim M, Awan AR (2020) Optimization of conditions for the higher level production of protease: characterization of protease from Geobacillus SBS-4S. Waste Biomass Valori 11:6613–6623. https://doi.org/10.1016/j.bcab.2020.10163
Alcaraz LD, Moreno-Hagelsieb G, Eguiarte LE, Souza V, Herrera-Estrella L, Olmedo G (2010) Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics 11:1–17. https://doi.org/10.1186/1471-2164-11-332
Azman NF, Noor MJMM, Akhir FNM, Ang MY, Hashim H, Zakaria Z, Hara H (2019) Depolymerization of lignocellulose of oil palm empty fruit bunch by thermophilic microorganisms from tropical climate. Bioresour Technol 279:174–180. https://doi.org/10.1016/j.biortech.2019.01.122
Balla A, Silini A, Cherif-Silini H, Bouket AC, Boudechicha A, Luptakova L, Alenezi FN, Belbahri L (2022) Screening of cellulolytic bacteria from various ecosystems and their cellulases production under multi-stress conditions. Catalysts 12:769. https://doi.org/10.3390/catal12070769
Bambharolia RP, Vyas TK, Deshmukh AJ (2021) Isolation and screening of lignocellulolytic microorganisms from different locations of Dang (Gujarat) India. Int J Econ Plants 8(4):217–221. https://doi.org/10.23910/2/2021.0434
Belbahri L, Chenari Bouket A, Rekik I, Alenezi FN, Vallat A, Luptakova L, Rateb ME (2017) Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front Microbiol 8:1438. https://doi.org/10.3389/fmicb.2017.01438
Book AJ, Lewin GR, McDonald BR, Takasuka TE, Wendt-Pienkowski E, Doering DT (2016) Evolution of high cellulolytic activity in symbiotic Streptomycesthrough selection of expanded gene content and coordinated geneexpression. PLoSBiol 14(6):e1002475. https://doi.org/10.1371/journal.pbio.1002475
Breza-Boruta B, Bauza-Kaszewska J (2023) Effect of microbial preparation and biomass incorporation on soil biological and chemical properties. Agriculture 13(5):969. https://doi.org/10.3390/agriculture13050969
Buraimoh OM, Ilori MO, Amund OO, Michel FC, Grewal SK (2015) Assessment of bacterial degradation of lignocellulosic residues (sawdust) in a tropical estuarine microcosm using improvised floating raft equipment. Int Biodeterior Biodegradation 104:186–193. https://doi.org/10.1016/j.ibiod.2015.06.010
Cabrera YT, Mendoza AP, Vásquez-Murrieta MS, Rivera-Orduña FN, Wang ET (2013) Diverse cellulolytic bacteria isolated from the high humus, alkaline-saline chinampa soils. Ann Microbiol 63:779–792. https://doi.org/10.1007/s13213-012-0533-5
Chandel AK, Silva SS, Carvalho W, Singh OM (2012) Sugarcane bagasse and leaves: foreseeable biomass of biofuel and bio-products. J Chem Technol Biotechnol 87:11–20. https://doi.org/10.1002/jctb.2742
Chandra R, Singh R, Yadav S (2012) Effect of bacterial inoculum ratio in mixed culture for decolourization and detoxification of pulp paper mill effluent. J Chem Technol Biotechnol 87(3):436–444. https://doi.org/10.1002/jctb.2758
Chauhan JV, Gohel SD (2020) Molecular diversity and pharmaceutical applications of free-living and rhizospheric marine actinobacteria. Mar Niche: Appl Pharm Sciences: Translational Res 111–131. https://doi.org/10.1007/978-981-15-5017-1_6
Chauhan JV, Mathukiya RP, Singh SP, Gohel SD (2021) Two stepspurification, biochemical characterization, thermodynamics and structureelucidation of thermostable alkaline serine protease from Nocardiopsis alba strain OM-5. Int J Biol Macromol 169:39–50. https://doi.org/10.1016/j.ijbiomac.2020.12.061
Chen YH, Chai LY, Zhu YH, Yang ZH, Zheng Y, Zhang H (2012) Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. J Appl Microbiol 112(5):900–906. https://doi.org/10.1111/j.1365-2672.2012.05275.x
Chroni C, Kyriacou A, Georgaki I, Manios T, Kotsou M, Lasaridi K (2009) Microbial characterization during composting of biowaste. Waste Manag 29(5):1520–1525. https://doi.org/10.1016/j.wasman.2008.12.012
Chuanjiao D, Chenxu L, Peng C (2021) Massiliacellulosiltytica sp. a novel cellulose-degrading bacterium isolated from rhizosphere soil of rice (Oryza sativa L.) and its whole genome analysis. Antonie Van Leeuwenhoek 114:1529–1540. https://doi.org/10.1007/s10482-021-01618-3
Chukwuma OB, Rafatullah M, Kapoor RT, Tajarudin HA, Ismail N, Siddiqui MR, Alam M (2023) Isolation and characterization of lignocellulolytic bacteria from municipal solid waste landfill for identification of potential hydrolytic enzyme. Fermentation 9(3):298. https://doi.org/10.3390/fermentation9030298
Contesini FJ, Melo RR, Sato HH (2018) An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol 38(3):321–334. https://doi.org/10.1080/07388551.2017.1354354
Cowan I, Steel ST (1974) Cowan and Steel’s manual for the identification of medical bacteria. 3rd edn. Cambridge University Press, London. https://doi.org/10.1017/CBO9780511527104
Dąbrowska M, Jacek R, Uhrynowski W, Drewniak L (2022) Use of lignocellulosic waste materials in the passive treatment of highly alkaline wastewater contaminated with sulphates and metals – from a laboratory study to pilot scale. J Environ Manage 321:115967. https://doi.org/10.2139/ssrn.4106715
Dahal B, NandaKafle G, Perkins L, Brözel VS (2017) Diversity of free-living nitrogen fixing Streptomyces in soils of the badlands of South Dakota. Microbiol Res 195:31–39. https://doi.org/10.1016/j.micres.2016.11.004
Datta S, Saha D, Chattopadhyay L, Majumdar B (2020) Genome comparison identifies different Bacillus species in a best fibre-retting bacterial consortium and provides insights into pectin degrading genes. Sci Rep 10:8169. https://doi.org/10.1038/s41598-020-65228-1
Dube SL, Osunsanmi FO, Ngcobo BP, Mkhwanazi LB, Jobe ZH, Aruleba RT, Mosa RA, Opoku AR (2023) Isolation and characterization of potential lignin peroxidase producing bacteriafrom compost samples at Richards Bay (South Africa). Pol J Microbiol 72(2):117–124. https://doi.org/10.33073/pjm-2023-003
Gohel SD, Singh SP (2018) Molecular phylogeny and diversity of the salt-tolerant alkaliphilic actinobacteria inhabiting coastal Gujarat, India. Geomicrobiol J 35(9):775–789. https://doi.org/10.1080/01490451.2018.1471107
Gohel SD, Majithiya VR, Singh SP (2023) Genetic and physiological diversity of marine actinobacteria from the Okha Coast, Gujarat, India. Geomicrobiol J 1–15. https://doi.org/10.1080/01490451.2023.2218383
Grenier V, Gonzalez E, Brereton NJB, Pitre FE (2023) Dynamics of bacterial and archaeal communities during horse bedding and green waste composting. PeerJ 11:e15239. https://peerj.com/articles/15239/
Henry BA, Maung CEH, Kim KY (2020) Metagenomic analysis reveals enhanced biodiversity and composting efficiency of lignocellulosic waste by thermoacidophilic effective microorganism (tEM). J Environ Manage 276:111252. https://doi.org/10.1016/j.jenvman.2020.111252
Iqbal S, Vollmers J, Janjua HA (2021) Genome mining and comparative genome analysis revealed niche-specific genome expansion in antibacterial bacillus pumilus strain SF-4. Genes 12(7):1060. https://doi.org/10.3390/genes12071060
Jiang C, Cheng Y, Zang H, Chen X, Wang Y, Zhang Y, Wang J, Shen X, Li C (2020) Biodegradation of lignin and the associated degradation pathway by psychrotrophic Arthrobacter sp. C2 from the cold region of China. Cellulose 27:1423–1440. https://doi.org/10.1007/s10570-019-02858-3
Kaspar F, Neubauer P, Gimpel M (2019) Bioactive secondary metabolites from Bacillus subtilis: a comprehensive review. J Nat Prod 82(7):2038–2053. https://doi.org/10.1021/acs.jnatprod.9b00110
Khurana H, Sharma M, Verma H, Lopes BS, Lal R, Negi RK (2020) Genomic insights into the phylogeny of Bacillus strains and elucidation of their secondary metabolic potential. Genomics 112(5):3191–3200. https://doi.org/10.1016/j.ygeno.2020.06.005
Kim TI, Jeong KH, Ham JS, Yang CB, Chung IB, Kim MK, Kim KN (2013) Isolation and characterization of cellulase secreting bacterium from cattle manure: application to composting. Compost Sci Utilization 12(3):242–248. https://doi.org/10.1080/1065657X.2004.10702189
Kulkarni SN, Kumar S, Jayalakshmi SK, Kuruba S (2019) Optimization of conditions for the production of lignocellulolytic enzymes by Sphingobacterium sp. ksn-11 utilizing agro-wastes under submerged condition. Prep Biochem Biotechnol 49(9):927–934. https://doi.org/10.1080/10826068.2019.1643735
Lasa AV, Mašínová T, Baldrian P, Fernández-López M (2019) Bacteria from the endosphere and rhizosphere of Quercus spp use mainly cell wall-associated enzymes to decompose organic matter. PLoS ONE 14(3):e0214422. https://doi.org/10.1371/journal.pone.0214422
Latt ZK, Yu SS, Kyaw EP, Lynn TM, Nwe MT, Mon WW, Kyaw NA (2018) Using cellulolytic nitrogen fixing bacterium, Azomonasagilis is for effective degradation of agricultural residues. Open Microbiol 12(1):154–162. https://doi.org/10.2174/1874285801812010154
Li J, Zhang F, Li J, Zhang Z, Bai F, Chen J, Zhao X (2019) Rapid production of lignocellulolytic enzymes by Trichoderma Harzianum LZ117 isolated from Tibet for biomass degradation. Bioresour Technol 292:122063. https://doi.org/10.1016/j.biortech.2019.122063
Lopes LD, Pereira MC, Andreote FD (2016) Bacterial abilities and adaptation toward the rhizosphere colonization. Front Microbiol 7:1341. https://doi.org/10.3389/fmicb.2016.01341
López MJ, Jurado MM, López-González JA, Estrella-González MJ, Martínez-Gallardo MR, Toribio A, Suárez-Estrella F (2021) Characterization of thermophilic lignocellulolytic microorganisms in composting. Front Microbiol 12:697480. https://doi.org/10.3389/fmicb.2021.697480
López-González JA, Vargas-García MC, López MJ, Suárez-Estrella F, Jurado M, Moreno J (2014) Enzymatic characterization of microbial isolates from lignocellulose waste composting. Chronological evolution. J Environ Manage 145:137–146. https://doi.org/10.1016/j.jenvman.2014.06.019
López-Mondéjar R, Tláskal V, Vìtrovský T, Štursová M, Toscan R, Nunes T (2020) Metagenomics and stable isotope probing reveal thecomplementary contribution of fungal and bacterial communities in therecycling of dead biomass in forest soil. Soil Biol Biochem 148:1078775. https://doi.org/10.1016/j.soilbio.2020.107875
Machado AS, Valadares F, Silva TF, Milagres AMF, Segato F, Ferraz A (2020) The secretome of Phanerochaete chrysosporium and Trametes Versicolor grown in microcrystalline cellulose and use of the enzymes for hydrolysis of lignocellulosic materials. Front Bioeng Biotechnol 8:826. https://doi.org/10.3389/fbioe.2020.00826
Majithiya V, Gohel S (2020) Isolation and screening of extracellular enzymes producing actinobacteria associated with sea weed. In Proceedings of the National Conference on Innovations in Biological Sciences (NCIBS). https://doi.org/10.2139/ssrn.3560095
Majithiya VR, Gohel SD (2022a) Isolation and characterization of marine actinobacteria associated with the seaweeds, Codium dwarkense and Sargassum Cinereum, collected from the Veraval coastline, Gujarat, India. J Mar Biol Assoc India 64(1):33–37. https://doi.org/10.6024/jmbai.2022.64.1.2188-05
Majithiya VR, Gohel SD (2022b) Actinobacteria associated with marine invertebrates: diversity and biological significance in actinobacteria diversity, applications and medical aspects. IntechOpen. https://doi.org/10.5772/intechopen.106642
Mansour AA, Da Costa A, Arnaud T, Lu-Chau TA, Fdz-Polanco M, Moreira MT (2016) Review of lignocellulolytic enzyme activity analyses and scale down to microplate based assays. Talanta 150:629–637. https://doi.org/10.1016/j.talanta.2015.12.073
Martins LF, Antunes LP, Pascon RC, de Oliveira JCF, Digiampietri LA, Barbosa D (2013) Metagenomic analysis of a tropical composting operation at the São Paulo Zoo Park reveals diversity of biomass degradation functions and organisms. PLoS ONE 8(6):0061928. https://doi.org/10.1371/journal.pone.0061928
Mironov V, Vanteeva A, Merkel A (2021) Microbiological activity during co-composting of food and agricultural waste for soil amendment. Agronomy 11(5):928. https://doi.org/10.3390/agronomy11050928
Mohammadipour Z, Enayatizamir N, Ghezelbash G, Moezzi A (2021) Bacterial diversity and chemical properties of wheat straw-based compost leachate and screening of cellulase producing bacteria. Waste Biomass Valoriz 12(3):1293–1302. https://doi.org/10.1007/s12649-020-01119-w
Mondol MAM, Shin HJ, Islam MT (2013) Diversity of secondary metabolites from marine Bacillus species: chemistry and biological activity. Mar Drugs 11(8):2846–2872. https://doi.org/10.3390/md11082846
Mukhtar S, Mehnaz S, Mirza MS, Malik KA (2019) Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola Stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Braz J Microbiol 50:85–97. https://doi.org/10.1007/s42770-019-00044-y
Murad HA, Azzaz HH (2011) Microbial pectinases and ruminant nutrition. Res J Microbiol 6(3):246–269. https://scialert.net/abstract/?doi=jm.2011.246.269
Naveed M, Nadeem F, Mehmood T, Bilal M, Anwar Z, Amjad F (2021) Protease-a versatile and ecofriendly biocatalyst with multi-industrial applications: an updated review. Catal Lett 151:307–323. https://doi.org/10.1007/s10562-020-03316-7
Neethu TM, Patel KG, Vyas TK, Sree G (2017) Exploring microbes for their cellulolytic and lignolytic enzyme activity for manure preparation. Int J Curr Microbiol App Sci 6(12):3808–3816. https://doi.org/10.20546/ijcmas.2017.612.438
Nevita T, Sharma GD, Pandey P (2018) Composting of rice-residues using lignocellulolytic plant-probiotic Stenotrophomonas maltophilia, and its evaluation for growth enhancement of Oryza sativa L. Environ Sustain 1(2):185–196. https://doi.org/10.1007/s42398-018-0017-z
Nur SK, Syaiful A, Dwi AS (2020) Isolation and selection of cellulolytic bacteria from rice straw for consortium of microbial fuel cell. Biodiversitas 21(4):1686–1696. https://doi.org/10.13057/biodiv/d210450
Ojo-Omoniyi, Olusola A, Okwa OO, Junaid IO, Ikuoye AO (2016) Sustainable solid waste management: isolation of cellulolytic microorganisms from dumpsites in Lagos, Southwest Nigeria. Int J Curr Microbiol Appl Sci 11(5):842–853. https://doi.org/10.1136/bmjopen-2015-forum2015abstracts.105
Pan X, Li F, Li Z, Huang Y, Wang Q, Huang S, Hu W (2021) Thermohalobaculum Xanthum gen. Nov, sp. Nov, a novel moderately thermophilic bacterium isolated from mangrove sediment. Antonie Van Leeuwenhoek. https://doi.org/10.21203/rs.3.rs-651509/v1
Patwardhan A, Ray S, Roy A (2014) Molecular markers in phylogenetic studies-a review. J Phylogenetics Evol Biol 2(2):131. https://doi.org/10.4172/2329-9002.1000131
Paul JS, Gupta N, Beliya E, Tiwari S, Jadhav SK (2021) Aspects and recent trends in microbial α-amylase: a review. Appl Biochem Biotechnol 193:2649–2698. https://doi.org/10.1007/s12010-021-03546-4
Pearson T, Okinaka RT, Foster JT, Keim P (2009) Phylogenetic understanding of clonal populations in an era of whole genome sequencing. Infect Genet Evol 9(5):1010–1019. https://doi.org/10.1016/j.meegid.2009.05.014
Petre M, Zarnea G, Adrian P, Gheorghiu E (2000) Biodegradation and bioconversion of cellulosewastes using bacterial and fungal cellsimmobilized in radiopolymerized hydrogels. Resour Conserv Recy 27:309–332. https://doi.org/10.1016/S0921-3449(99)00028-2
Pfenning N, Lippert KD (1966) Uber das vitamin B-12-bedrurbins phototrophers chwefelbakterien. Arch Microbiol 55:245–256. https://doi.org/10.1007/BF00410246
Poehlein A, Funkner K, Schüler MA, Daniel R (2017) First insights into the genome sequenceof the cellulolytic bacterium Clostridium hungatei DSM 14427. Genome Announc 5(20):e00363–e00317. https://doi.org/10.1128/genomea.00363-17
Pokhrel BP, Wanjare S, Singh B, Purushotham, Kumara SM (2013) Isolation, screening and characterization of promising α-amylase producing bacteria from sewage enriched soil. Int J Adv Biotechnol Res 4(2):286–290
Poly NY, Sabrina M, Khan MMH, Hoque MN, Azad AK, Hasan M (2022) Isolation, documentation, and biochemical characterization of cellulolytic bacteria from rumen fluid of cattle. J Adv Biotechnol Exp Ther 5(2):433–444. https://doi.org/10.5455/jabet.2022.d126
Priyanka AS, Subhashini R, Jayashree R, Vendan T (2021) Bioprospecting of microorganisms with lignocellulolytic enzyme activity for leaf litter degradation. J Pharm Innov 10(11):465–470. https://doi.org/10.22271/tpi.2021.v10.i11g.8702
Radhika NL, Sachdeva S, Kumar M (2023) Development of potential consortia for biotransformation of lignin and polyaromatic hydrocarbons (PAHs). https://doi.org/10.1007/s13399-023-04486-1. Biomass Conv Bioref
Rahman NHA, Rahman NAA, Aziz SA, Hassan MA (2013) Production of lignolytic enzymes by newly isolated bacteria from oil plantation soil. BioResource 8(4):6136–6150
Rinady MVP, Nuraini Y, Prayogo C, Arfarita N (2023) The effect of landmanagement and organic matter inputs on bacterial population and soil nutrientsacross different types of agroforestry system. Biodivesitas 24(3):1333–1345. https://doi.org/10.13057/biodiv/d240302
Roy D, Azaïs A, Benkaraache S, Drogui P, Tyagi RD (2018) Composting leachate: characterization, treatment, and future perspectives. Rev Environ Sci Biotechnol 17(2):323–349. https://doi.org/10.1007/s11157-018-9462-5
Saini JK, Arti, Lakshmi T (2012) Simultaneous isolation and screening of cellulolytic bacteria: selection of efficient medium. J Pure Appl Microbiol 6(3):1339–1344
Saleh D, Jarry J, Rani M, Aliferis K, Seguin P, Jabaji SH (2019) Diversity, distribution and multi-functional attributes of bacterial communities associated with the rhizosphere and endosphere of timothy (Phleum pratense L). J Appl Microbiol 127:794–811. https://doi.org/10.1111/jam.14334
Sekhar CV, Varma KP, Swapna G, Vamsi K, Lakshmi BM (2022) Cellulolytic activity of Bacillus and Pseudomonas species isolated from sugarcane rhizoplane and its correlation with carbohydrate utilization. Biol Forum Int J 14(2):240–246
Sellstedt A, Richau KH (2013) Aspects of nitrogen-fixing actinobacteria, in particularfree-living and symbiotic frankia. FEMS Microbiol 342(2):179–186. https://doi.org/10.1111/1574-6968.12116
Semira NY, Tamene MJ, Meera I (2021) Screening and characterization of thermostable amylase producing bacteria isolated from soil samples of Afdera, Afar region, and molecular detection of amylase coding gene. Int J Microbiol. https://doi.org/10.1155/2021/5592885
Shishtawy RM, AsiriS AM, Gomaa AM, Ibrahim IH, Al-Talhi HA (2014) Solid fermentation of wheat bran for hydrolytic enzymes production and saccharification content by a local isolate Bacillus megatherium. BMC Biotechnol 14:29. https://doi.org/10.1186/1472-6750-14-29
Tao J, Chen Q, Chen S, Lu P, Chen Y, Jin J, Li j, Xu Y, He W, Long T, Deng X, Yin H, Li Z, Fan J, Cao P (2022) Metagenomic insight into the microbial degradation of organic compounds in fermented plant leaves. Environ Res 214:113902. https://doi.org/10.1016/j.envres.2022.113902
Tawaraya K, Horie R, Saito A, Shinano T, Wagatsuma T, Saito K (2015) Metabolite profiling of shoot extracts, root extracts, and root exudates of rice plant under phosphorus deficiency. J Plant Nutr 36:1138–1159. https://doi.org/10.1080/01904167.2013.780613
Udume OA, Abu GO, Stanley HO, Vincent-Akpu IF, Momoh Y, Eze MO (2023) Biostimulation of petroleum contaminated soil using organic and inorganic amendments. Plants 12:431. https://doi.org/10.3390/plants12030431
Vaghela N, Gohel S (2023) Medicinal plant-associated rhizobacteria enhance the production of pharmaceutically important bioactive compounds under abiotic stress conditions. J Basic Microbiol. https://doi.org/10.1002/jobm.202200361
Varma VS, Das S, Sastri CV, Kalamdhad AS (2017) Microbial degradation of lignocellulosic fractions during drum composting of mixed organic waste. Sustain Environ Res 27:265–272. https://doi.org/10.1016/j.serj.2017.05.004
Verma M, Ekka A, Mohapatra T, Ghosh P (2020) Optimization of kraft lignin decolorization and degradation by bacterial strain Bacillus velezensis using response surface methodology. J Environ Chem Eng 8(5):104270. https://doi.org/10.1016/j.jece.2020.104270
Vijayalaxmi S, Anu Appaiah KA, Jayalakshmi SK, Mulimani VH, Sreeramulu K (2013) Production of bioethanol from fermented sugars of sugarcane bagasse produced by lignocellulolytic enzymes of Exiguobacterium sp. VSG-1. Appl Biochem Biotechnol 171:246–260. https://doi.org/10.1007/s12010-013-0366-0
Villar I, Alves D, Garrido J, Mato S (2016) Evolution of microbialdynamics during the maturation phase of the composting of different types of waste. Waste Manag 54:83–92. https://doi.org/10.1016/j.wasman.2016.05.011
Weselowski B, Nathoo N, Eastman AW, MacDonald J, Yuan ZC (2016) Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide,biofertilization, biomass degradation and biofuel production. BMC Microbiol 16:244. https://doi.org/10.1186/s12866-016-0860-y
Wu X, Wang Y, TongB, Chen X, Chen J (2018) Purification and biochemical characterization of a thermostable and acid-stable alpha-amylase from Bacillus licheniformis B4-423. Int J Biol Macromol 109:329–337. https://doi.org/10.1016/j.ijbiomac.2017.12.004
Wu L, Che S, Qin X, Xu Y, Tian S, Zhu Y, Song J, Guan Y, Wang D, Wu M, Yang X, Wu Z, Yang M (2023) Identification, characteristics and rice growth promotion of a highly efficient cellulolytic bacterial strain, Cellulomonas iranensis ZJW-6, isolated from paddy soil in Central China. Front Microbiol 14:1152966. https://doi.org/10.3389/fmicb.2023.1152966
Xia C, Li Z, Xu Y, Yang P, Gao L, Yan Q, Song X (2019) Introduction of heterologous transcription factors and their target genes into Penicillium oxalicum leads to increased lignocellulolytic enzyme production. Appl Microbiol Biotechnol 103:2675–2687. https://doi.org/10.1007/s00253-018-09612-y
Zainith S, Purchase D, Saratale GD, Ferreire FLR, Bilal M, Bhargava RN (2019) Isolation and characterization of lignin-degrading bacterium Bacillus aryabhattai from pulp and paper mill wastewater and evaluation of its lignin-degrading potential. 3 Biotech 9:92. https://doi.org/10.1007/s13205-019-1631-x
Zhang L, Thiewes H, Kan JAL (2011) The D-galacturonic acid catabolic pathway in Botrytis Cinerea. Fungal GenetBiol 48(10):990–997. https://doi.org/10.1016/j.fgb.2011.06.002
Zhang N, Yang D, Kendall JR, Borriss R, Druzhinina IS, Kubicek CP, Shen Q, Zhang R (2016) Comparative genomic analysis of Bacillus amyloliquefaciens and Bacillus subtilis reveals evolutional traits for adaptation to plant-associated habitats. Front Microbiol 7:2039. https://doi.org/10.3389/fmicb.2016.02039
Zhang X, Zhu Y, Li J, Zhu P, Liang B (2021) Exploring dynamics and associations of dominant lignocellulose degraders in tomato stalk composting. J Environ Manage 294:113162. https://doi.org/10.1016/j.jenvman.2021.113162
Acknowledgements
The authors acknowledge the Department of Biosciences, Saurashtra University for financial and infrastructural facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhimani, A.A., Bhimani, H.D., Vaghela, N.R. et al. Cultivation methods, characterization, and biocatalytic potential of organic solid waste degrading bacteria isolated from sugarcane rhizospheric soil and compost. Biologia 79, 953–974 (2024). https://doi.org/10.1007/s11756-023-01592-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-023-01592-3