Abstract
Tree peonies (Paeonia Sect Moutan DC) are an emerging oil crop because of their high oil and α-linolenic acid (ALA) content. This research was to investigate the potential use of near infrared reflectance spectroscopy (NIRS) for estimating the major fatty acids contents, such as palmitic acid (C16:0), oleic acid (C18:1), linoleic acid (C18:2) and linolenic acid (C18:3) in tree peonies. A total of 115 small seed samples and 447 single seeds were selected to calibrate the predictive models. NIRS absorbance spectra were collected using a Fourier transform near infrared (FT-NIR) spectrometer for the small seed samples, and acousto-optic tunable filter-near infrared (AOTF-NIR) for the single seed samples. Statistical analysis was performed with partial least squares (PLS). For the husked samples, C18:3, C18:1 and C18:2 showed the highest correlation coefficient of validation (R v; = 0.9756, 0.9467 and 0.8485, respectively) and the ratio of performance to deviation (RPD; = 3.58, 1.91 and 2.17, respectively); however, C16:0 did not reach expectations (R v = 0.7783, RPD = 1.32). For intact samples, C18:3 showed the best prediction (R v = 0.9096, RPD = 3.14), followed by C18:2 (R v = 0.8479, RPD = 1.96). The results for C18:1 were poor (R v = 0.7237, RPD = 1.70). For single seeds, only the results for C18:3 (R = 0.9150, RPD = 1.73) were good in the husked seed samples. It was concluded that NIRS can be used to rapidly assess the content of the major fatty acids in small samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tree peony (Paeonia sect Moutan DC) is a well-known ornamental plant native to China that has recently emerged as an oilseed plant [1]. Tree peonies have shown enormous potential to be developed as a new oil crop because of their high content of oil (27–33 %) and α-linolenic acid (ALA > 40 %) [2, 3]. Due to the extreme consumption of oil in China, the potential use of tree peony oil has gained attention from breeders, growers and traders. However, there are no specific cultivars of the tree peony for oil use because the breeding work was only aimed at ornamental [4, 5] and herbal [6] use in the past. China has the richest resources for tree peonies including all 8 of the wild species and more than 1500 widely cultivated varieties [7]. However, presently there are only two tree peony seedling plants that are cultivated for oil in China: the P. ostii (distributed throughout eastern and southern China) and the P. rockii (distributed throughout northern and northwestern China). It is worth mentioning that there is significant natural genetic variability within these species. In addition, seeds from the species of P. qiui also show significant potential for propagation. To discover target genotypes from the uncharacterized tree peony varieties, bulk seeds and plants need to be analyzed to meet the urgent breeding and commercial needs. Although conventional methods such as gas chromatography (GC) offer a high level of precision, they seem to be less suitable since they are time consuming, costly, and a large amount of sample destruction. Therefore, effective and rapid methods are required to screen seeds in breeding programs.
Near infrared reflectance spectroscopy (NIRS) is a fast, low cost, analytical technique that can be utilized without using hazardous chemicals. It is widely used for the analysis of oil content and fatty acid composition of seeds in many crops, such as rapeseed [8], sunflower [9], and maize [10]. NIRS can also be accurately applied in single seed selection using a special adapter, which has been used to measure oleic and linoleic acid in rapeseeds [11] and stearic acid in sunflower seeds [12]. Single seed analysis is popular for plant breeders, who often focus on single seed analyses across seed generations for plant selection.
It has been reported that sample conditions can influence NIRS results. Rudolphi et al. [13] developed NIRS calibrations for oil seed content and fatty acid composition from seed hull fractions and empty seed samples in safflower. Quampah et al. [14] demonstrated the prediction of total oil content (RPDv = 11.495) and linoleic acid (RPDv = 5.026) in cottonseed powder. Xi et al. [15] determined the oil content in Camellia oleifera seeds and the correlation coefficient reached 0.88 for intact seeds and 0.98 for seed powder. These results suggest NIRS has great feasibility for detecting oil content and fatty acid composition in bulk samples (in the form of whole grain or powder) and can also be applied, with modifications, for detection in single-seed; however, the potential to use NIRS to analyze intact seeds with a large shape, size or hard hull is unclear.
The tree peony seed has an irregular oval shape, a black hard hull and a relatively large size, making it difficult to analyze with NIRS. The ability to conduct analysis with a small sample size is of paramount importance in the breeding of oil tree peonies, and the potential of NIRS to predict the necessary parameters has not been reported. Therefore, we employed NIRS using an adapter that could hold approximately 20 g of seeds (as small sample size for individual plants) to determine if we could assess the target genotypes, which are of interest to breeders and researchers. Due to the limited availability of seeds in cross breeding programs, it is also important to develop single seed analysis tool to screen seeds at an early stage in the breeding process.
The objective of this study was to systematically study the feasibility of using NIRS to predict the fatty acid composition in tree peony seeds using 20-g seed samples and single seeds as well as both intact and husked seed samples.
Materials and Methods
Materials
The tree peony seed samples used in this study were harvested from tree peony plants cultivated on the forestry experiment station of Beijing Forestry University (located in Beijing, China) in 2013 and 2014. The seed samples were collected from plants with known oil use potential including from P. ostii, P. rockii and P. qiui, and 25 hybrid plants. In addition, germplasm from the Chinese herbaceous peony were also collected to broaden the calibration coverage. In total, 115 seed samples (approx. 20 g of seeds per plant) were used for small seed sample analysis. Additionally, a total of 447 single seeds were selected and used for single seed calculation. All samples were oven-dried for 8 h at 60 °C before experimental use.
NIR Scanning
The 20-g seed samples were analyzed by diffuse reflectance using a Fourier transform near infrared spectrometer (FT-NIR) (ANTARIS II, Thermo Nicolet Co., USA). The seeds were placed in the auto-rotating cup (32 mm diameter, 15 mm thickness) and each spectrum was the average of 64 successive scans at a spectral range from 4000 to 10000 cm−1 with a 2-cm−1 data interval. Each sample was run in triplicate to minimize the particular effects of sample size.
The acousto-optic tunable filter-near infrared (AOTF-NIR) was used for the single seed scanning. To limit the effects of seed size and shape, we chose the adapter with a 5 mm diameter hole that could only hold a single seed. The absorbance spectrum (log1/R) was the average of 100 successive scans in the spectral range from 1100 to 2300 nm at a wavelength interval of 2 nm.
Fatty Acid Analysis
The tree peony seed oil from each 20-g seed sample was extracted by Soxhlet extraction after NIRS scanning. The oil from single seeds was extracted according to described methods [8, 16]. Briefly, a single seed was crushed using a stainless steel rod and placed in a centrifuge tube (1.5 ml). Hexane (1 ml) was added and the tubes were shaken for 1 min and then incubated at room temperature for 5 h. Afterwards, the supernatant was transferred to a fresh centrifuge tube and shaken for 2 min. K-Methylate (0.5 ml in methanol) was added, and the tube was incubated for 1 h at room temperature. The tube was centrifuged for 5 min at 6000 rpm, and the supernatant was removed for analysis.
The extracted oils were analyzed by (GC; Agilent, 7890A, USA) under the following conditions: capillary column, BPX-70 (60 m × 0.25 mm × 0.25 μm); 250 °C injection port temperature and 280 °C detector port; column temperature programmed from 80 to 230 °C at 5 °C/min, holding for 15 min at 230 °C; H2, 40 ml/min; air, 300 ml/min; injection sample amount, 1 μl; split ratio, 30:1. Each sample was analyzed in triplicate.
Data Analysis
Spectra corresponding to the samples were sorted on the basis of the GC values for each component. From the lowest to the highest, one out of every three samples was selected into the validation set. As a result, the calibration and validation sets were divided at a 2:1 ratio.
Statistical analysis on the spectral information was performed using TQ Analyst V7.2 (Thermo Nicolet Co., USA) software with a partial least squares (PLS) regression to develop calibrations. To enhance the accuracy of the PLS models, several pre-treatments were carried out, such as the multiple scatter correction (MSC), the standard normal variate (SNV), and the first and second derivatives. Calculated calibration statistics included the root mean square error of calibration (RMSEC) and correlation coefficient of calibration (R c). For predictions, the calibrations were tested on the validation group analysis. The correlation coefficient of validation (R v), the root mean square error of validation (RMSEP), and the ratio of performance to deviation (RPD) were the main methods to appraise the predicting effects. According to previous studies [17, 18], a high R v value and a low RMSEP value always predict a good calibration model. Furthermore, RPD values of 2.5 can be used for screening purposes. We selected ten samples from the 20 g seed samples and 20 single seed samples (originating from F1 plants collected in 2015) to evaluate the reliability of the NIR models.
Result and Discussion
Variability for Fatty Acid Composition
There were approximately 10 components of fatty acids in the tree peony seed oil (Fig. 1). The dominant components included palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2) and linolenic acid (C18:3). NIRS cannot be used to predict components of less than 4 %; therefore, we focused on the four dominant components, C16:0, C18:1, C18:2 and C18:3.
The gas chromatography of extracted oil from tree peony seeds of experimental samples. Upper left: the lowest content of small seed sample; upper right: the lowest content of small seed sample: low left: the lowest content of single seed; low right: the highest content of single seed. Peaks: 21.6 min: myristic acid (C14:0); 24.9 min: palmetic acid (C16:0); 25.3 min: hexadecenoic acid (C16:1); 26.1 min: heptadecanoic acid (C17:0); 26.7 min: heptadecenoic acid (C17:1); 27.9 min: stearic acid (C18:0); 28.3 min: oleic acid (C18:1); 29.2 min: linoleic acid (C18:2); 30.1 min: linolenic acid (C18:3); 30.6 min: eicosenoic acid (C20:1)
All the samples exhibited large variation in fatty acid composition ranging from 3.3891 to 49.8066 % in the calibration set and from 4.1801 to 49.4510 % in the validation set (Table 1). The widest range was observed for C18:3 (SD = 3.93 %), followed by C18:2 (SD = 3.25 %), C18:1 (SD = 2.74 %) and C16:0 (SD = 0.49 %). Furthermore, we observed minimal differences in the means and SD between the calibration set and the validation set, with the exception of C18:3, which had a relatively wide range between the mean and SD. This indicated that both the calibration and validation sets were appropriately selected and were able to represent the total variation in fatty acid composition between the samples; furthermore, this indicated that the population was suitable for NIRS analysis.
Similarly, the GC analyses of the 447 single seeds (Table 2) showed the widest variation for C18:3 (SD = 5.82 %), followed by C18:2 (SD = 5.15 %), C18:1 (SD = 4.10 %) and C16:0 (SD = 0.83 %). The variation in SD values between the single seeds compared to the small (20 g) seed samples, indicated that there was larger variability in fatty acid contents between single seeds compared to between individual plants.
Reflectance Spectra of Tree Peony Seeds
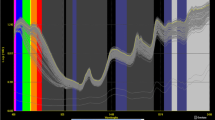
The original spectra of the intact and husked samples showed a similar trend and broad peaks at 8286.6, 6882.8, and 5176.3 cm−1 (Fig. 2). In reports [19, 20], the peak at 8286.6 cm−1 is assigned to the second overtone of C-H, the peak at 6882.8 cm−1 is associated with the first overtone of C–H, and the peak at 5176.3 cm−1 is assigned to the second overtone of C=O. This indicates that it is necessary to choose an optimal spectrum region in subsequent calculations. The diversity of the absorbing strengths between the intact and husked samples also showed that the hull impeded the production of absorption bands for inner part of seed. Thus, it was necessary to calculate the samples with different pre-treats via separate methods in following research.
Calibration for Small Seed Samples
Each component required repeated calculations to determine the optimum calibration. For example, in the C18:3 model calculation (Fig. 3), PLS analysis was performed on the transformed spectral data of the calibration group, and the optimal number of PLS factors was determined by the validation group. We used R v and RMSEP to determine the optimal mathematical pretreatment. The increasing value of R v, and the minimizing value of RMSEP indicated better results for the model. In the first step, the R v of the husked samples reached 0.9349 in full wavelength (from 4000 to 10000 cm−1) after MSC to reduce the spectral noise. We then selected the optimum wavelength situated at 8018.5–8630.8 and 5954.1–5603.1 cm−1, and the R v rose to 0.9756. At the same time, the RMSEP declined from 1.35 to 1.08. These values did not improve after FD, SD or SNV pre-treatment. It confirmed that the best pre-treatment was MSC, and the optimum wavelength was 8018.5–8630.8 and 5954.1–5603.1 cm−1 with 10 factors used. Similarly, we confirmed that for the intact sample model the best pre-treatment was MSC combined with FD, and the optimum wavelength was 7233.0–5417.0 cm−1 with 6 factors used.
The fatty acid composition models for the 20-g seed samples are shown in Table 3. For the husked samples, the C18:3, C18:1 and C18:2 showed high R v (0.9756, 0.9467 and 0.8485, respectively) and RPD (3.58, 1.91 and 2.17, respectively) values, but C16:0 did not reach expectations (R v = 0.7783, RPD = 1.32). For the intact samples, the calibrations were less accurate. C18:3 showed the best correlation (R v = 0.9096, RPD = 3.14), followed by C16:1 (R v = 0.8951, RPD = 2.42) and C18:2 (R v = 0.8479, RPD = 1.96). The results for C18:1 were poor (R v = 0.7237, RPD = 1.70). All the validation reference illustrated the predictions for the husked samples were more accurate compared to the intact samples with the exception of C16:0. In the tree peony seeds, the relative content of C18:3 was the highest compared to other fatty acid components while the lowest levels were observed for C16:0. Due to its high relative content, the C18:3 model showed the greatest accuracy and could be used for value determination in nondestructive way. Because of its low level, C16:0 was the least accurate and coincided with the limited of detection for NIRS [16]. Similar to published results, we confirmed that the spectrum range between 6250–5555 cm−1 coincided with C18:2 and C18:1 [21].
Calibration for Single Seeds
The single seed models are shown in Table 4. Overall, the single seed models were less accurate compared to the small sample models. The R v of C16:0 was 0.5395 for intact and 0.5928 for husked, and the RPD values were less than 1, suggesting this was not a good model of prediction. The best result for the single seed NIRS was the C18:3 model, with a R v of 0.7382 for intact samples and 0.9150 for husked samples. In addition, the intact single seed model of C18:1 was better compared to the C18:2 model (R v = 0.6675 and 0.6000, respectively), and was worse for the husked single seed model (R v = 0.6775 and 0.8992, respectively). For the RPD values, the best model was C18:2, with the RPD values greater than 2.5 for both the intact (2.88) and husked (2.60) samples. The next best model was C18:1 (RPD of 2.56 for intact samples and 2.28 for husked samples). Although the C18:3 had the best R v values than other fatty acids, the RPD value was 1.98 for intact samples and 1.73 for husked samples, indicating inconsistency in the results.
Evaluation of NIRS Models
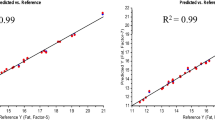
We selected 10 small seed samples and 20 single seeds harvested in 2015 to evaluate the reliability of the NIRS models. Those seeds originating from F1 plants were not used in the calibration or validation sets. After NIRS scanning, the samples were checked by GC. The NIRS values are shown in Tables 5 and 6. We observed that the fatty acid components acquired by NIRS were similar to the GC values. No significant difference (P > 0.05; ANOVA) was observed in the composition of the four fatty acids between the GC and NIRS results in the small seed samples; furthermore, no significant difference (P > 0.05; ANOVA) was also observed for the single seeds with the exception of C16:0 (P = 0.033). The ANOVA results indicated that the NIRS had a high level to predict the content of fatty acids in tree peony seeds. The GC vs NIRS concentration plots were performed using the R statistical software (R 3.2.1) and are shown in Fig. 4 and Fig. 5. The correlation coefficient between the NIRS and GC showed the highest consistency for C16:0, C18:2 and C18:3 (Fig. 4). For C16:0, one sample showed a high bias in intact prediction. For C18:2, two samples exhibited high prediction error (one intact and one husked sample). Specially, the reliability of the NIRS prediction for C18:3 suggested this model could be directly used to select for target traits during oil cultivar breeding and seed testing. In general, the single seed models were not predictive of fatty acid content. However, the calibration of C18:3 could roughly predict the value in husked single seed samples.
In this study, the models were constructed using seeds harvested in 2013 and 2014, and we used seeds harvested in 2015 to test the effects. This showed the stability of the results across several seasons. The prediction results suggest that the fatty acid composition can be tested using a small amount of sample, such as a single seed. The results in our study could speed up breeding progress and processing through the commercialization chain.
Conclusion
This is the first study to examine NIRS testing in tree peony seeds. We concluded that NIRS is a very useful, rapid screening tool for oil plant selection by tree peony breeding programs using small sample sizes. In this study, we accurately describe the analysis of dominant fatty acid composition, including C16:0, C18:1, C18:2 and C18:3 in both 20-g seed samples and single seeds from tree peonies. We show, in particular, the content of C18:3 can be precisely predicted nondestructively, rapidly, and cost effectively, which will contribute to oil tree peony breeding and commercial use.
References
Li SS, Yuan RY, Chen LG, Wang LS, Hao XH, Wang LJ, Zheng XC, Du H (2015) Systematic qualitative and quantitative assessment of fatty acids in the seeds of 60 tree peony (Paeonia section Moutan DC.) cultivars by GC–MS. Food Chem 173:133–140
Han JG, Li XQ, Liu Z, Hu YH (2014) Potential applications of tree peony as an oil plant. Cereals Oils 5:21–25 (in Chinese)
Shi GA, Jiao FX, Jiao YP, Yang HA, Han MW, Wu YQ, Shi BR (2014) Development prospects and strategies of oil tree peony industry in China. J Chin Cereals Oils Assoc 29:124–128 (in Chinese)
Li JJ, Zhang XF, Zhao XQ (2011) Tree peony of China. Encyclopaedia of China Publishing House, Beijing, pp 17–44 (in Chinese)
Cheng FY, Li JJ, Chen DZ, Zhang ZS (2005) Chinese Flare Mudan. China Forestry Publishing House, Beijing, pp 77–83 in Chinese)
Guo BL, Hong DY, Xiao PG (2008) Further research on chemotaxonomy of paeonol and analogs in Paeonias (Ranunculaceae). J Systmat Evolut 46:724–729
Yuan JH, Cornille A, Giraud T, Cheng FY, Hu YH (2014) Independent domestications of cultivated tree peonies from different wild peony species. Mol Ecol 23:82–95
Hom NH, Becker HC, Möllers C (2006) Non-destructive analysis of rapeseed quality by NIRS of small seed samples and single seeds. Euphytica 153:27–34
Sato T, Takahata Y, Noda T, Yanagisawa T, Morishita T, Sakai S (1995) Nondestructive determination of fatty acid composition of husked sunflower (Helianthus annua L.) seeds by near-infrared spectroscopy. J Am Oil Chem Soc 72:1177–1183
Huang YY, Zhu LW, Li JH, Wang JH, Sun BQ, Sun Q (2011) Rapid and nondestructive discrimination of hybrid maize seed purity using infrared spectroscopy. Spectrosc Spectra Anal 3:661–664 (in Chinese)
Niewitetzki O, Tillmann P, Becker HC, Möllers C (2010) A new near-infrared reflectance spectroscopy method for high-throughput analysis of oleic acid and linolenic acid content of single seeds in oilseed rape (Brassica napus L.). J Agric Food Chem 58:94–100
Leonardo V, Begona PR, Nez JMFN (2004) Use of near-infrared reflectance spectroscopy for selecting for high stearic acid concentration in single husked achenes of sunflower. Crop Sci 44(1):93–97
Rudolphi S, Becker HC, Schierholt A, von Witzke-Ehbrecht S (2012) Improved estimation of oil linoleic and oleic acid and seed hull fractions in safflower by NIRS. J Am Oil Chem Soc 89:363–369
Quampah A, Huang ZR, Wu JG, Liu HY, Li JR, Zhu SJ, Shi CH (2012) Estimation of oil content and fatty acid composition in cottonseed kernel powder using near infrared reflectance spectroscopy. J Am Oil Chem Soc 89:567–575
Xi RC, Zhong YM, Deng XM, Chen XY, Mo BY, Yi LF (2013) Models for determining oil contents in Camellia oleifera seeds by using near infrared spectroscopy. Sci Silvae Sin 4:1–6 (in Chinese)
Velasco L, Llers CM, Becker HC (1999) Estimation of seed weight, oil content and fatty acid composition in intact single seeds of rapeseed (Brassica napus L.) by near-infrared reflectance spectroscopy. Euphytica 106:79–85
Kovalenko IV, Rippke GR, Hurburgh CR (2006) Measurement of soybean fatty acids by near-infrared spectroscopy: linear and nonlinear calibration methods. J Am Oil Chem Soc 83:421–427
Kovalenko IV (2005) Determination of amino and fatty acid composition of soybeans using near-infrared spectroscopy. Iowa State University, Iowa
Yang H, Irudayaraj J, Paradkar M (2005) Discriminant analysis of edible oils and fats by FTIR, FT-NIR FT-Raman spectroscopy. Food Chem 93:25–32
Kasemsumran S, Thanapase W, Punsuvon V, Ozaki Y (2012) A feasibility study on non-destructive determination of oil content in palm fruits by visible-near infrared spectroscopy. J Near Infrared Spectrosc 20:687–694
Sato T, Uezono I, Morishita T, Tetsuka T (1998) Nondestructive estimation of fatty acid composition in seeds of Brassica napus L. by near-infrared spectroscopy. J Am Oil Chem Soc 75:1877–1881
Acknowledgments
The authors are grateful to Prof. Han Donghai, College of Food Science & Nutritional Engineering, China Agricultural University, for NIRS instruments support and Mr Cheng Xinyun, Beijing National Beauty Tree peony Technology Co. Ltd., for experimental materials support. This work was financially supported by the National Science and Technology Support Program of China (2012BAD01B07), and the Key Technology Research of Oil Peony Industry in Shaanxi Province (2014HXFWYL016).
Author information
Authors and Affiliations
Corresponding author
Additional information
This study investigates the potential use of near infrared reflectance spectroscopy (NIRS) as a rapid screening method to estimate the major fatty acids contents, such as palmitic acid (C16:0), oleic acid (C18:1), linoleic acid (C18:2) and linolenic acid (C18:3) in tree peonies.
About this article
Cite this article
Huliang, C., Fangyun, C. & Liping, P. Determination of the Fatty Acid Composition in Tree Peony Seeds Using Near-Infrared Spectroscopy. J Am Oil Chem Soc 93, 943–952 (2016). https://doi.org/10.1007/s11746-016-2839-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11746-016-2839-0