Abstract
To evaluate the effectiveness of a germin-like protein (GLP) in legumes against the serious soil-borne pathogen Fusarium oxysporum f. sp. lentis, an Oryza sativa root-expressed GLP (OsRGLP1) was expressed in the model legume Medicago truncatula using the recombinant vector pCOsRGLP1. The transgene was highly expressed in M. truncatula transformed lines as assessed by RT-qPCR. Consistent with the active status of the transgene there was an elevated accumulation of H2O2 in transformed progeny. Enzymatic characterization of T1 transgenic progeny showed increased superoxide dismutase (SOD) activity. The additional SOD activity in transgenic lines was insensitive to potassium cyanide and sensitive to H2O2 indicating its resemblance to FeSOD. The effectiveness of the OsRGLP1 gene was tested by monitoring the root disease after infection of wild-type and transgenic lines. Wild-type plants were greatly affected by the pathogen infection showing a percent disease index value of 50 compared to 10–18 for the transgenic lines. The tolerance of the transgenic lines leads to recovery in fresh weight and pod production to an almost normal level. Analysis of defense-related genes downstream of hydrogen peroxide (H2O2) in transgenic plants showed induction of salicylic acid and jasmonate signaling pathways and increased expression of some pathogenesis-related-1 (PR-1) genes and a plant defensin gene. Overall, the findings suggest that OsRGLP1 provides protection against the fungal pathogen F. oxysporum that may involve the direct influence of H2O2 on signaling pathways leading to the activation of defense-related genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Survival of plants is dependent on their ability to tolerate harsh environmental conditions, resulting from biotic or/and abiotic stresses. To reduce the damage, plants have developed sophisticated stress response mechanisms. The molecular mechanism involved in these stress responses needs to be better understood and is the subject of intense research. Genes which show changes in expression under stress conditions include germins and germin-like proteins (GLPs) (Dunwell et al. 2008). Despite sequence similarity with germins and their apoplastic localization, GLPs do not usually possess oxalate oxidase activity; instead most GLPs are reported to have superoxide dismutase (SOD) activity (Christensen et al. 2004; Zimmerman et al. 2006; Chen et al. 2011). SODs are responsible for dismutation of superoxide to H2O2 and O2 and protection of cells from the oxidative burst. Production of H2O2 is also part of the defense mechanism of the cell and is well known as a signaling molecule for induction of systemic acquired resistance (SAR) in un-inoculated tissues (Tran et al. 2014) as well as promoting lignification of the cell wall which restricts invasion of pathogens by cell wall reinforcement through the cross-linkage of proteins and carbohydrates (Christensen et al. 2000). Hydrogen peroxide can initiate salicylic acid (SA) and jasmonate signaling pathways which ultimately result in synthesis of pathogenesis-related proteins (PR proteins) and plant defensins, respectively (Leon et al. 1995). It was, therefore, no surprise when GLPs were reported to be upregulated and/or implicated in defense against plant pathogens including fungi (Rietz et al. 2012) and viruses (Park et al. 2004). Heterologous expression of GLPS or gene silencing of endogenous GLPs had also provided evidence for GLP defense against fungal pathogens in wheat, barley and rice (Christensen et al. 2004; Banerjee and Maiti 2010). Yasmin et al. (2008) found several regulatory elements putatively associated with biotic stress response through in silico analysis of the promoter region of OsRGLP1.
Grain legumes are important since people depend on these crops to meet their needs for protein, micronutrients and oil. However, fungal diseases have emerged as the most important factor influencing their production, among them the widely distributed soil-borne root-infecting pathogens Fusarium oxysporum, Fusarium solani, and Rhizoctonia solani are of major concern. The problem is being addressed through management practices and exploration of available variability in breeding programs; however, expansion of the genetic base through the introduction and overexpression of stress responsive genes from heterologous sources has emerged as a powerful tool for crop improvement.
Legume root biology and legume–pathogen interactions are quite complex as compared to other plant families. The studies conducted in non-legume model plants such as Arabidopsis or tobacco are less likely to explain these interactions comprehensively. Leguminous plants, including lentil, soya bean, green pea and chickpea, are relatively recalcitrant towards genetic transformation techniques. To overcome these limitations M. truncatula was adopted as a model plant for the study of legume biology (Barker et al. 1990). The M. truncatula genome has now been largely sequenced and a range of resources for molecular analysis are available (Young et al. 2011). While M. truncatula has served as a major model for the rhizobium–legume symbiosis it has this capacity to distinguish pathogen from symbiont which gives an interesting perspective to pathogenesis (Rose 2008). M. truncatula is a permissive host of F. oxysporum (Ramírez-Suero et al. 2010) and can serve as a pathogenesis model for legumes, as well as for the disease being of direct economic importance. While GLPs are thought to be part of a basal defense mechanism, they can be particularly important in resistance against specific pathogens (Rietz et al. 2012). The present study was, therefore, designed to investigate the possible role for a rice root-expressed GLP gene, OsRGLP1 in providing defense against the root-infecting fungal pathogen F. oxysporum in M. truncatula.
Materials and methods
Expression vector and bacterial strain
Plant expression vector pCOsRGLP1 was used in the present study. A 958-bp fragment of the OsRGLP1 cDNA isolated from rice Japonica cultivar root tissue (Gene bank accession no. AF141878.1) was cloned in pCAMBIA1301 by replacing the GUS gene to construct pCOsRGLP1 (Yasmin et al. 2015). The vector expressed OsRGLP1 under control of the CaMV35S promoter and carried the hygromycin phosphotransferase resistance gene as a selectable marker for the selection of transformed plants (Online Resource Fig. 1a).
Agrobacterium tumefaciens AGL-1 (Lazo et al. 1991) was used for the harboring and subsequent transformation of the recombinant vector pCOsRGLP1. The vector was electroporated into AGL-1 by applying a voltage of 2.5 kV with a pulse resistance of 400 Ω. Selection for transformed AGL-1 was achieved on 50 mg/L rifampicin and 100 mg/L ampicillin, with additional 50 mg/L kanamycin. The transformation of the pCOsRGLP1 vector in AGL-1 cells was verified by amplifying the 958 bp OsRGLP1 gene fragment using the primers CGLP1-F and CGLP1-R. All primers used in the study are shown in Online Resource Table 1. For PCR, an initial pre-amplification denaturation at 94 °C for 3 min was followed by 35 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 40 s. A final extension was performed at 72 °C for 7 min.
Plant transformation and confirmation
A highly embryogenic M. truncatula line 2HA was used for transformation (Rose et al. 1999). Healthy leaves from 2-month-old glasshouse grown plants were collected and surface sterilized. Plant transformation was carried out by the leaf explant method, according to Song et al. (2013), using the more recent phytohormone combination suggested by Nolan et al. (2014). Using a sharp blade, rectangular-shaped leaf pieces, each containing the midrib, were obtained. The explants were infected with AGL-1 harboring the pCOsRGLP1 construct and co-cultured at 27 °C. After 3 days in the dark, the explants were washed and transferred on two concentrations of hygromycin, i.e., 15 and 20 mg/L. After 8 weeks of the selection on antibiotics, putatively transformed whitish somatic embryos were transferred to P4 10:4:1:5 medium (P4 is the basal medium (Song et al. 2013) plus 10 μM 1-naphthalene acetic acid, 4 μM benzylaminopurine, 1 μM abscisic acid and 5 μM gibberellic acid) with 100 mg/L timentin. The plantlets regenerated from these embryos were shifted to Magenta pots with liquid P40 medium (Song et al. 2013) after 14–16 weeks. These plantlets reached a five- to six-branch stage after another 4 weeks, when they were removed from the medium and transferred to pots after washing the roots under tap water to remove the medium. The potting mix was composed of coarse sand, perlite, coir-peat (1:1:1) plus 5 g of Osmocote Exact Standard slow release fertilizer. The plants were covered by a polyethylene tent with wooden sticks to maintain the humidity. Small holes were punched in the polyethylene at intervals followed by complete removal after 3–4 weeks. The well-grown and healthy plants were shifted to the glass house. The whole process of transformation of leaf explants to shifting of well-grown plants to glass house took 6 months. Seeds were collected from the pods and stored at 4 °C.
The DNA was extracted from leaf tissues using the CTAB method (Richards et al. 1997). PCR analysis with gene-specific primers was performed to confirm the presence of the OsRGLP1 gene in the transformed plants. Primers were also designed outside of the left and right border on the backbone of the expression vector, i.e., Kan-F and Kan-R in the kanamycin resistance gene (Online Resource Table 1). The PCR analysis with these primers was used to establish the integration of the transgene into the genome of the transgenic plant and to rule out the presence of contamination of A. tumefaciens in transgenic plants. Transformation efficiency was calculated as percentage of PCR-positive plants produced per number of explants cultured.
Expression analysis of the OsRGLP1 gene using RT-qPCR
Total RNA was extracted at the 4–5-branch stage of plant development using the SV Total RNA Isolation System (Cat.# K0801, Thermo Scientific, Lithuania). The cDNA was synthesized using the Superscript III First-Strand synthesis system (Cat.# 18080-0510, Invitrogen, USA). One microgram of RNA was used for cDNA synthesis. cDNAs were 1:10 diluted and used as template for PCRs with 28 cycles of amplification using the thermal profile given above. Housekeeping gene GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) was amplified as the endogenous control as used and described by Nolan et al. (2003). Primers used for targeting the 679-bp fragment (GGLP1-F and GGLP1-R) and 125-bp GAPDH region are mentioned in Online Resource Table 1.
Quantitative expression analysis of the OsRGLP1 gene utilized the Maxima SYBR Green qPCR Master Mix (Thermo Scientific, Lithuania). The gene-specific primers RTGLP1-F and RTGLP1-R were used to amplify a 267-bp gene fragment. The thermal profile included one cycle of pre-amplification denaturation at 95 °C for 10 min, 30 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 30 s and extension at 72 °C for 40 s followed by a final extension of 10 min at 72 °C. Line-Gene K Fluorescence Quantitative PCR Detection System (BIOER) was used for relative quantification. The data were analyzed using the 2−∆∆Ct relative quantification method (Livak and Schmittgen 2001), with GAPDH as a reference gene for data normalization. The transgenic line with the lowest expression (predicted from its Ct value) was taken as a calibrator. The experiment was done with three repeats.
Functional analysis of T0 progeny for enzymatic activity
To verify that the introduced gene was biologically active in transformed plants, the level of H2O2 was measured by the luminescence assay as described by Murphy and Huerta (1990). Leaf discs of equal size were prepared with a sharp blade and placed for one min in 0.5 ml of assay buffer containing 10 mM MES, pH 7.0, 0.6 M mannitol, 0.1 mM KCl and 1 mM CaCl2. The reaction was initiated by the addition of 5 μl of 25 mM luminol and 5 μl of horseradish peroxidase. The tubes were immediately loaded into a luminometer (AutoLumat Plus LB953, Berthold Technologies, Germany). Each assay took a total of 2 min with 9 (5 s) measurements. The samples were run in triplicates, each with two blank controls run in parallel. Calculation of each measurement was taken from the average of the final seven readings after subtracting the average of two blank controls.
Functional evaluation of T1 transformants
For functional analysis, T1 progeny of selected transgenic lines was used. Seeds of transformed plants were selected on the basis of hygromycin resistance by culturing the seeds on MS medium containing 20 mg/L of hygromycin and verified by PCR amplification with gene-specific primers.
Superoxide dismutase activity
SOD activity was measured according to Dhindsa et al. (1981). Leaf samples were homogenized in 0.2 M phosphate buffer (pH 7.0) at 4 °C. Centrifugation was performed at 20,000g at 4 °C for 10 min and the supernatant was taken for SOD activity. The SOD reaction mixture contained 75 µM nitro-blue tetrazolium (NBT), 13 mM methionine, 0.1 mM EDTA, 50 mM potassium phosphate buffer (pH 7.8) 2 µM riboflavin and the enzyme sample. Cuvettes without enzyme served as controls. The cuvettes were placed under a light source for 15 min with a similar set of cuvettes placed in dark to serve as blanks. All the tubes were then covered with a black cloth to stop the reaction. The absorbance was taken at 560 nm at 25 °C. One unit of enzyme activity was defined as the quantity of enzyme responsible for reduction of the absorbance to 50 % in comparison with control samples. Reduction in absorbance due to enzyme activity was compared with wild-type plants and in different transgenic lines.
To establish the type of transgene-associated SOD activity in transgenic plants, the leaf samples were treated with either 3 mM potassium cyanide or 10 mM H2O2 prior to SOD assay. Heat sensitivity of transgene-associated SOD activity was determined by heating leaf extract at 90 °C for 15 min prior to activity testing. SOD activity was measured and change in SOD activity due to treatment was compared with samples without treatment.
Oxalate oxidase activity
The procedure developed by Liang et al. (2001) was used to detect oxalate oxidase activity. Leaf cuttings were taken and incubated in a solution containing 2.5 mM oxalic acid, 25 mM succinate buffer (pH 4.0) and staining reagent 4-chloro-1-naphthol (6 mg/ml) at room temperature in the dark for 24 h. Wheat seeds were used as the positive control. The data were recorded as photographs of leaf or seed subjected to the procedure.
Determination of antifungal activity conferred by the OsRGLP1 gene
Antifungal activity induced by OsRGLP1 in M. truncatula plants after F. oxysporum infection was assessed by the method described by Stoilova and Chavdarov (2006) with slight modification. Highly virulent isolates of F. oxysporum f. sp. lentis stored in gel beads were obtained from the Plant Pathology Department of the University. The fungus was revived on Potato Dextrose Agar (PDA) plates and allowed to grow at 25 °C in the dark for 15–20 days. Spore formation in fungus culture was observed under the microscope. Fungus grown on plates was washed in sterilized water and fungal suspension at a concentration 1 × 106 conidia/ml was prepared. The suspension was vortexed for about 1 min before inoculating into pots filled with sterilized potting mixture. The susceptibility of M. truncatula genotype 2HA to the F. oxysporum isolate was determined by dipping the roots of 9-week-old transformed and wild-type plantlets in fungal culture for 5 min after wounding four to five roots on each plant by trimming before planting, and by adding culture to the soil near the rhizosphere of these plantlets. The same number of plants injured and dipped in sterilized water served as non-infected controls.
The symptoms of infection on leaves in the form of yellowing, wilting and drying were recorded weekly for 5 weeks. Severity of infection on the plants from mild to severe was recorded on a scale of 1–5 which means no symptoms, greenish yellow leaves, yellow leaves, yellow and mildly wilted leaves, and wilted and completely dried leaves, respectively. The numbers of leaves in each grade were recorded. Percent disease index (PDI) was calculated by the following formula:
where S1–S5 stand for the number of leaves in each scale. Morphological and growth data on infected transgenic plants were compared with infected wild-type plants.
Expression level of defense-related genes
Primers were designed using bioinformatics tools Primer3 and IDT’s Oligo analyzer version 3.1 for transcriptional analysis of defense-related genes PR-1, PR-1(580), PR-1(590), PR-1A, PR-1a and PDF-1.1 (Online Resource Table 1). The transcript level of transformed and wild-type plants was measured by real-time PCR using total RNA as template isolated from leaf tissues according to the procedure already mentioned.
Statistical analysis
Analysis of variance (ANOVA) was carried out using statistical software MSTAT-C (Michigan State University, USA). Means were compared by LSD/DMRT wherever applicable.
Results
Plant transformation and confirmation
The transformation efficiency of the OsRGLP1 gene introduced into the M. truncatula 2HA line through Agrobacterium-mediated transformation was determined at two concentrations of hygromycin (Hyg). Fifty-five percent of explants produced Hyg-resistant embryogenic calli while 35.5 % of explants regenerated Hyg-resistant plantlets as shown in Table 1. Morphologically no visual phenotypic difference was observed between transgenic and positive control wild-type plants. All transgenic plant lines were fertile and developed flowers and pods with normal shape and size (Fig. 1a). Hyg-resistant plantlets were verified for successful transformation by PCR amplification of the OsRGLP1 gene using gene-specific primers amplifying 958-bp gene fragment (Online Resource Fig. 1b). No amplification was seen in the wild-type plants. To rule out the possibility of false positives and the presence of A. tumefaciens in transgenic plants developed from somatic embryos, the amplification was performed with primers designed on the plasmid backbone outside the T-DNA borders. In case of contamination with Agrobacterium, these primers were to generate a 650-bp amplicon from the pCOsRGLP1 backbone (Online Resource Fig. 1b). The transgenic plants with no or a very light band were considered putative positives, and were used for RNA extraction and expression analysis.
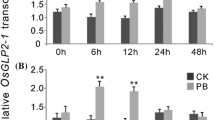
a Different stages of M. truncatula transformation using AGL-1 harboring the OsRGLP1 gene. A Positive control plate; somatic embryos observed on induced callus on P4 10:4:1:5 medium, B negative control plate; callus formation on selection medium, C Transformed calli; somatic embryos observed on induced callus on selection medium, D transformed plant grown in magenta pots on liquid P40 medium, E non-transformed control plant grown in magenta pots on liquid P40 medium, F transgenic plants growing in glass house, G flower formation on transgenic plants, H pod formation on transgenic plants (bars in A–E represent 1 cm). b Quantification of OsRGLP1 gene expression in transgenic lines by RT-qPCR. WT wild-type control; T1-10 transgenic lines. Error bars represent standard error of the mean. Means followed by the same letter indicate no significant difference (P < 0.05)
Transformation efficiency was then calculated in terms of number of PCR-positive plants regenerated per number of explants. Out of total explants cultured after transformation, 21.6 % of explants produced PCR-positive plants (Table 1). Two concentrations of hygromycin were used for selection of transformants, higher transformation efficiency was at 20 mg/L such as 24.4 %. Transgenic plants had very well developed roots and showed a high survival rate of 90 % upon transfer to soil.
Expression analysis of the OsRGLP1
The expression of the OsRGLP1 transgene was analyzed by gel RT-PCR and RT-qPCR analysis in ten selected lines. Reverse transcriptase PCR and gel analysis revealed that the OsRGLP1 gene was actively expressed in transgenic lines. Light bands observed in some lines were considered low-expression lines, such as T4 and T5 (Online Resource Fig. 2). Transgene transcripts were not detectable in the WT plants. Expressions of the transgene in the same lines were further verified by RT-qPCR, Variation in expression level was observed among the transgenic lines (Fig. 1b). Six transgenic lines, with the observed range of expression levels, were selected for further analysis.
Functional analysis of T0 progeny for enzymatic activity through H2O2 accumulation
Given that GLPs can have oxalate oxidase (OXO) and/or superoxide dismutase (SOD) activity leading to H2O2 production; assays were conducted for the estimation of H2O2 production in six transgenic lines with relatively high GLP expression. Substantially increased H2O2 accumulation was observed in these lines as compared to wild-type control (Fig. 2a).
H2O2 accumulation and SOD activity assay of leaf tissues of wild-type and transgenic plants. a H2O2 accumulation as measured by luminescence assay. b SOD activity assay with and without 10 mM H2O2 treatment. c SOD activity assay with and without 3 mM potassium cyanide treatment. d SOD activity assay with and without heat treatment. WT wild-type control, T3-10 transgenic lines. Bars represent the standard error of the mean of three experimental repeats with six plants in each experiment. Means followed by the same symbol indicate no significant difference (P < 0.05)
Functional evaluation of T1 transformants
T1 progeny of six selected transgenic lines showing elevated expression of the transformed gene was used for analysis. Transformed plants among T1 progeny were selected on the basis of hygromycin resistance and verified through PCR amplification with gene-specific primers and the active expression of the transgene was demonstrated through RT-PCR analysis (Figs. not shown).
Superoxide dismutase activity
Transgenic plants with overexpression of the OsRGLP1 gene showed significantly enhanced SOD activity compared to wild-type plants. However, depending on the metal cofactor ion, SOD exists in one of the three isoforms that can be discriminated by different inhibitors. Cu/ZnSOD is sensitive to both the cyanide and H2O2 (Scandalios 1993), whereas FeSOD and MnSOD are sensitive to only H2O2 and KCN, respectively (Cheng and Song 2006).
In the H2O2 sensitivity assay, SOD activity of treated and untreated wild-type samples did not vary significantly, indicating that the native SOD activity is H2O2 insensitive. However, in transgenic samples a significant decrease in activity was observed following H2O2 treatment. The residual activity after treatment became nearly equal to wild type (Fig. 2b), highlighting the sensitivity of transgene-induced SOD activity to H2O2 and pointing to its nature that it may either be FeSOD or Cu/ZnSOD type. In transgenic KCN-treated plant samples, the decrease in SOD activity was about the same amount as in wild-type plants, while the residual activity was still significantly higher than the wild type. Therefore, this residual activity which was sensitive to H2O2 and resistant to KCN was not native, and not due to either Cu/Zn or Mn type SOD (Fig. 2c), rather it is suggested to be of FeSOD type attributed to the transferred gene.
GLPs are reported to be heat-resistant proteins (Dunwell et al. 2008). After heat treatment the SOD activity of wild-type samples decreased significantly as compared to their untreated controls, which showed that M. truncatula native SOD activity is heat sensitive. On the contrary, the SOD activity in transgenic plants differed between control and heat treated, but with the SOD activity of transgenic samples remaining significantly higher than wild type (Fig. 2d). Thus, it may be concluded that the native SOD activity is both KCN and heat sensitive while additional SOD activity conferred by OsRGLP1 gene in the transgenic plants is both KCN and heat resistant. These findings strongly suggested that the SOD activity acquired due to the transgene is likely due to an FeSOD-like GLP.
Oxalate oxidase activity
All the germins are reported to possess oxalate OXO activity while very few GLPS show OXO activity. To determine if there was any OXO activity associated with the OsRGLP1 gene, the OXO activity assay was performed on different tissues of M. truncatula as well as 2 days post-imbibition stage of wheat seeds. Blue staining of tissues in the presence of oxalate and the chromogenic substrate 4-chloro-1-naphthol demonstrates the presence of oxalate oxidase activity (Liang et al. 2001). Two and five days post-imbibition, wheat seeds served as a positive control. Blue staining was observed on the surface of wheat seeds as well as on root and shoot in the presence of oxalic acid, whereas no color was seen in its absence (Online Resource Fig. 3a). The activity assay with M. truncatula wild-type seed after two days post-imbibition with oxalic acid showed no blue color, demonstrating lack of any native oxalate oxidase activity. Similarly assay with seeds of M. truncatula transgenic lines under study did not develop the blue color both in the absence or the presence of oxalic acid, indicating that the transformed OsRGLP1 gene has no association with OXO activity. Likewise, no detectable OXO activity was observed in leaf discs, and stem cuttings of mature wild-type and transgenic plants with and without oxalic acid (Online Resource Fig. 3b).
Antifungal activity of the OsRGLP1 gene in M. truncatula T1 progeny
Before evaluating the resistance against fungal infestation, the susceptibility of wild-type M. truncatula genotype 2HA was determined against pathogenic fungus F. oxysporum f. sp. lentis by root inoculation. Infection symptoms started appearing 3–4 days after inoculation in the form of yellowing of leaves which gradually wilted and then dried. Plants mock inoculated with water showed no symptoms of infection.
Nine-week-old transgenic and wild-type plants were then inoculated with the F. oxysporum. The symptoms of infection appeared 3–4 days post-inoculation in the form of yellowing, wilting and drying of leaves in wild type, but in six transgenic lines these symptoms were milder and appeared 5–7 days post-inoculation. The number of affected leaves gradually increased in infected plants while the non-infected controls remained healthy. The Fusarium wilt disease incidence and severity was estimated using the Percent Disease Index (PDI). Data for the damaging effects of the disease were collected weekly. In wild-type plants, the PDI value which was 24 % over the first week, thereafter gradually increased. On the other hand, the PDI in different transgenic lines was significantly different from wild type (except T10 line) ranging from 12 to 20 % after 1 week post-infection (Fig. 3). After the second week, the number of affected leaves was significantly higher in infected wild-type plants and their growth was also restricted. In contrast, the disease symptoms in transgenic plants developed on only a few leaves (Fig. 4). Afterwards, the symptoms in transgenic plants gradually subsided and eventually the resistance was developed, while the PDA went on increasing in wild-type plants and reached 50 % after 5 weeks (Fig. 3). The infection not only damaged the developed leaves but also caused the death of existing branches, while restricting the development of new leaves and branches. On the contrary, very few disease symptoms were detectable on the older leaves and branches of transgenic plants (Fig. 4) which kept growing well with the development of new branches. PDI in different transgenic lines ranged from 10 to 18 %, which was significantly lower than wild type as shown in Fig. 3.
Percent disease index of M. truncatula plants infected with F. oxysporum in soil. Six selected transgenic lines and a wild type were infected with F. oxysporum. WT wild-type plant, T3-10 transgenic lines. Data for disease incidence were collected at weekly intervals. Bars represent standard error of the mean based on two independent experiments with six plants in each group. Means followed by the same symbol indicate no significant difference (P < 0.05)
Post-infection growth and development
The effect of fungal infection on the growth and development of transgenic and wild-type plants was also recorded post-infection. Higher fresh weight and more pods were recorded for infected transgenic plants than wild-type infected plants (Fig. 5a, b). Flower shape, size and physical appearance were similar in infected and non-infected plants. Pod development was normal in infected transgenic plants while in wild-type infected plants, pods either remained absent or if formed were shorter in length and produced smaller and fewer seeds per pod. The number of basal branches and leaves developed on these branches was high in transgenic plants as compared to wild type after infection (Fig. 5c, d). These results clearly suggest that the expression of OsRGLP1 in M. truncatula increased its resistance against F. oxysporum.
Comparison of transgenic and wild-type plants with or without infection a for fresh weight, b for number of pods developed, c for number of leaves developed, d for number of basal branches developed. Bars represent standard error of the mean based on two independent experiments with six plants in each group. Means followed by the same symbol indicate no significant difference (P < 0.05)
Expression level of defense-related genes
To investigate, whether H2O2 produced in transgenic plants correlates with the expression of PR protein genes and defensins, the transcription of some selected PR protein genes and a defensin gene was determined by RT-PCR. The transcript levels in two independent transgenic lines T7 and T9 were compared to non-infected wild type. Both transgenic plants showed significantly upregulated expression of PR-1, PR-1(580), PR-1A and PDF-1.1 (Fig. 6).
Discussion
The expression of several GLP genes has been reported to be induced by fungal pathogens (Zimmermann et al. 2006). The antifungal role of GLPs has also been demonstrated by the transgenic approach through overexpression or gene silencing, predominantly in cereals, against powdery mildew in wheat and barley (Christensen et al. 2004), and against blast in rice (Banerjee and Maiti 2010), as well as against Verticillium longisporum and Rhizoctonia solani in A. thaliana (Knecht et al. 2010). In the present study, this approach was extended to explore whether a rice root-expressed OsRGLP1 gene can confer resistance or tolerance in a leguminous plant against the soil-borne fungal pathogen F. oxysporum.
Researchers exploring plant functional genomics generally use Arabidopsis thaliana as a model for determining the molecular basis of plant gene function. However, the plant kingdom is remarkably diverse, and members of the legume family (Fabaceae) are especially interesting in having numerous specialized traits such as symbiotic nitrogen fixation and mycorrhizal symbiosis. To evaluate OsRGLP1 gene function in members of this family upon heterologous expression; the legume model M. truncatula was exploited. A number of transgenic lines were generated using A. tumefaciens, regeneration from leaf explants, and verified through PCR amplification of the OsRGLP1 gene. Transformation efficiencies were similar to those reported by Chabaud et al. (2003) who also used A. tumefaciens AGL1 and the 2HA line of M. truncatula. Reverse transcriptase PCR and RT-qPCR studies demonstrated active expression of the transgene in transgenic T0 plants. The variation observed among transgenic lines in terms of expression and biochemical functionality can be attributed to a number of factors including somaclonal variations, different copy numbers of the transgene, and/or site of integration in the genome. Changes in the organization of the introduced gene (Bundock and Hooykaas 1996) and methylation of the transgene may also cause variation (Vaucheret and Fagard 2001). Importantly, the constitutive expression of the transgene resulted in additional accumulation of H2O2 compared to wild-type control plants.
For functional analysis, the T1 progeny was used. Primary transgenic are not considered suitable for functional analysis, as plants regenerated from tissue culture may show altered physiological response compared to their counterparts (Lee et al. 1999). Seeds collected from high- and moderate-expression lines were selected for analysis. The OsRGLP1 possessed SOD, but no OXO activity and the transgenic lines exhibited significantly higher levels of SOD activity compared to wild type. GLPs, in contrast to germins, are mostly associated with SOD activity (Christensen et al. 2004; Zimmermann et al. 2006; Godfrey et al. 2007). The additional SOD activity observed in transgenic plants was H2O2 sensitive, so it was considered an FeSOD-like enzyme. This has also been very recently reported by Yasmin et al. (2015).
The present study clearly revealed that the expression of OsRGLP1 in M. truncatula interferes with F. oxysporum root infection and provides substantive resistance/tolerance against the fungal infection. There are fewer disease symptoms and OsRGLP1 expression completely overcomes the yield reduction, as expressed by both fresh weight and pod production. While the use of the CaMV35S promoter, a strong constitutive promoter, with a rice gene provides a successful approach for F. oxysporum resistance/tolerance, we note that it may not unequivocally interpret the in vivo situation. An endogenous promoter may simply be a weak promoter or only target specific organs. Similarly, a rice promoter with OsRLGP1 may have had minimal effect. It is, therefore, at least possible that the rice gene coding sequence itself may have a superior capacity against F. oxysporum, as opposed to a legume GLP.
The increased presence of H2O2 in the apoplast has been proposed to play multiple roles. First, the cell wall reinforcement by abundant callose (a polymer of β (1,3)-glucan) appositions during papilla formation at the infection site serves as a physical barrier against pathogen invasion (Thordal‐Christensen et al. 1997). Accumulation of H2O2 has been detected in the mesophyll cells directly underlying the infected epidermal cells in barley with mlo (mildew resistance locus O)-mediated resistance against the fungal pathogen Blumeria graminis (Piffanelli, et al. 2002). Second, the H2O2 may also cause inactivation of haustorium/appressorium localized in the intracellular space by oxidation. Cheng et al. (2012) reported that accumulation of H2O2 and callose materials is responsible for encasement of the haustorium in broad bean interaction with the wheat stripe rust pathogen. Third, the H2O2 may influence resistance against pathogens by acting as a signaling molecule for inducing expression of defense-related genes. H2O2 acts as a diffusible signal for the induction of defense-related genes in surrounding cells (Levine et al. 1994), and mediates production of salicylic acid (Leon et al. 1995) and Jasmonate (Hu et al. 2003). SA then induces transcriptional activation of pathogenesis-related proteins-1 (PR-1) genes, while jasmonate is responsible for the induction of defensins (Turner et al. 2002). The antifungal activity of these proteins as a part of plant defense pathways is well documented (Van Loon et al. 2006). Some PR proteins have chitinase activity which attacks fungal cell walls (Legrand et al. 1987). PR proteins are also necessary for the onset of SAR (Le Henanff et al. 2009). Upregulation of transcript levels for members of the PR1 gene family (PR-1, PR-1(580), PR-1A) as well as that of plant defensin (PDF-1.1) in transgenic plants as compared to wild type was correlated with expression of the OsRGLP1 gene in the present study, demonstrating activation of members of both PR1 and defensin pathways operative in plants.
Conclusion
Conclusively, the transfer of OsRGLP1 into M. truncatula seems to have pleiotropic effects on plant defense mechanisms through expression of apoplastic FeSOD-like heat-resistant catalytic activity responsible for the generation of H2O2. The H2O2 so generated is most likely responsible for protection/tolerance against the root fungal pathogen F. oxysporum f. sp. Lentis in the investigation being reported. Nevertheless, it points to a generalized mechanism of defense capable of initiating different signaling cascades and thus less prone to the development of resistance in pathogens. The GLP genes may, therefore, serve as a reliable target for incorporating transgenic resistance in legumes for generating fungal tolerance in species greatly damaged by at least F. oxysporum.
Author contribution statement
Conceived and designed the experiments: TS SMSN. Performed the experiments: TS FD. Supervised the study: SMSN RJR FN. Analyzed the data: TS FN SMSN. Wrote the paper: TS SMSN. Critically revised the manuscript: RJR.
References
Banerjee J, Maiti MK (2010) Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem Biophys Res Commun 394(1):178–183
Barker DG, Bianchi S, Blondon F, Dattée Y, Duc G, Essad S et al (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Report 8(1):40–49
Bundock P, Hooykaas PJ (1996) Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc Natl Acad Sci 93(26):15272–15275
Chabaud M, de Carvalho-Niebel F, Barker D (2003) Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell Rep 22(1):46–51
Chen X, Wang ML, Holbrook C, Culbreath A, Liang X, Brenneman T et al (2011) Identification and characterization of a multigene family encoding germin-like proteins in cultivated peanut (Arachis hypogaea L.). Plant Mol Biol Report 29(2):389–403
Cheng HY, Song SQ (2006) Species and organ diversity in the effects of hydrogen peroxide on superoxide dismutase activity in vitro. J Integ Plant Biol 48(6):672–678
Cheng Y, Zhang H, Yao J, Wang X, Xu J, Han Q et al (2012) Characterization of non-host resistance in broad bean to the wheat stripe rust pathogen. BMC Plant Biol 12(1):96
Christensen JH, Baucher M, O’Connell A, Van Montagu M, Boerjan W (2000) Control of lignin biosynthesis. In: Jain SM, Minocha S (eds) Molecular biology of woody plants, vol 1. Kluwer Academic Publishers, Dordrecht, pp 227–267
Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjær MF, Dudler R, Schweizer P (2004) The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact 17(1):109–117
Dhindsa RS, Plumb-Dhindsa P, Thorpe TA (1981) Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot 32(1):93–101
Dunwell JM, Gibbings JG, Mahmood T, Naqvi SMS (2008) Germin and germin-like proteins: evolution, structure, and function. Crit Rev Plant Sci 27(5):342–375
Godfrey D, Able AJ, Dry IB (2007) Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: a possible role in defense? Mol Plant Microbe Interact 20(9):1112–1125
Hu X, Neill S, Cai W, Tang Z (2003) Hydrogen peroxide and jasmonic acid mediate oligogalacturonic acid-induced saponin accumulation in suspension-cultured cells of Panax ginseng. Physiol Plant 118(3):414–421
Knecht K, Seyffarth M, Desel C, Thurau T, Sherameti I, Lou B et al (2010) Expression of BvGLP-1 encoding a germin-like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol Plant Microbe Interact 23(4):446–457
Lazo GR, Stein P, Ludwig RA (1991) A DNA transformation competent Arabidopsis genomic library in Agrobacterium. Nat Biotechnol 9(10):963–967
Le Henanff G, Heitz T, Mestre P, Mutterer J, Walter B, Chong J (2009) Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biol 9:54
Lee SH, Shon YG, Kim CY, Chun HJ, Cheong YH, Kim ZH et al (1999) Variations in the morphology of rice plants regenerated from protoplasts using different culture procedures. Plant Cell Tiss Org 57(3):179–187
Legrand M, Kauffmann S, Geoffroy P, Fritig B (1987) Biological function of pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Nat Acad Sci USA 84(19):6750–6754
Leon J, Lawton MA, Raskin I (1995) Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol 108(4):1673–1678
Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79(4):583–593
Liang H, Maynard CA, Allen RD, Powell WA (2001) Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol Biol 45(6):619–629
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Murphy TM, Huerta AJ (1990) Hydrogen peroxide formation in cultured rose cells in response to UV-C radiation. Physiol Plant 78(2):247–253
Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133(1):218–230
Nolan KE, SongY Liao S, Saeed NA, Zhang X, Rose RJ (2014) An unusual abscisic acid and gibberellic acid synergism increases somatic embryogenesis, facilitates its genetic analysis and improves transformation in Medicago truncatula. PLoS One 9:e99908
Park CJ, An JM, Shin YC, Kim KJ, Lee BJ, Paek KH (2004) Molecular characterization of pepper germin-like protein as the novel PR-16 family of pathogenesis-related proteins isolated during the resistance response to viral and bacterial infection. Planta 219(5):797–806
Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U et al (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129(3):1076–1085
Ramírez-Suero M, Khanshour A, Martinez Y, Rickauer M (2010) A study on the susceptibility of the model legume plant Medicago truncatula to the soil-borne pathogen Fusarium oxysporum. Eur J Plant Pathol 126(4):517–530
Richards E, Reichardt M, Rogers S (1997) Preparation of plant DNA using CTAB Short protocols in molecular biology 3:2.10–2.11
Rietz S, Bernsdorff FE, Cai D (2012) Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J Exp Bot 63(15):5507–5519
Rose RJ (2008) Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Funct Plant Biol 35(4):253–264
Rose RJ, Nolan KE, Bicego L (1999) The development of the highly regenerable seed line Jemalong 2HA for transformation of Medicago truncatula implications for regenerability via somatic embryogenesis. J Plant Physiol 155(6):788–791
Scandalios JG (1993) Oxygen stress and superoxide dismutases. Plant Physiol 101(1):7
Song Y, Nolan KE, Rose RJ (2013) Stable transformation of Medicago truncatula cv. Jemalong for gene analysis using Agrobacterium tumefaciens. In: Rose RJ (ed) Legume genomics, methods and protocols, methods in molecular biology series 1069. Humana Press, Springer, New York, pp 203–214
Stoilova T, Chavdarov P (2006) Evaluation of lentil germplasm for disease resistance to Fusarium wilt (Fusarium oxysporum f. sp. lentis). J Central Eur Agric 7(1):121–126
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley powdery mildew interaction. Plant J 11(6):1187–1194
Tran PT, Choi H, Kim SB, Lee HA, Choi D, Kim KH (2014) A simple method for screening of plant NBS-LRR genes that confer a hypersensitive response to plant viruses and its application for screening candidate pepper genes against Pepper mottle virus. J Virol Methods 201:57–64
Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell 14(suppl 1):S153–S164
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Vaucheret H, Fagard M (2001) Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet 17(1):29–35
Yasmin T, Mahmood T, Hyder MZ, Akbar S, Naqvi SMS (2008) Cloning, sequencing and in silico analysis of germin-like protein gene 1 promoter from Oryza sativa L. ssp. indica. Pak J Bot 40:1627–1634
Yasmin T, Mumtaz A, Mahmood T, Hyder MZ, Naqvi SMS (2015) A germin-like protein gene of rice increased superoxide dismutase activity in transformed tobacco. Biol Plant 59(3):456–462
Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, Van de Peer Y et al (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480(7378):520–524
Zimmermann G, Baumlein H, Mock HP, Himmelbach A, Schweizer P (2006) The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance. Plant Physiol 142(1):181–192
Acknowledgments
This work was financially supported by Higher Education Commission under Indigenous PhD 5000 Fellowship Program (No. 17-5-4(Ps4-279) and International Research Scholarship Initiative (No. IRSIP-21-BMS-47). The authors are grateful to Center for Application of Molecular Biology to International Agriculture, Australia for the generous gift of pCAMBIA vectors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Kuzniak-Gebarowska.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sultana, T., Deeba, F., Naz, F. et al. Expression of a rice GLP in Medicago truncatula exerting pleiotropic effects on resistance against Fusarium oxysporum through enhancing FeSOD-like activity. Acta Physiol Plant 38, 255 (2016). https://doi.org/10.1007/s11738-016-2273-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2273-9