Abstract
In this work, a fermentation starter involving autochthonous strains obtained from an artisanal dry sausage from Colonia Caroya (Córdoba, Argentina) was formulated, with the aim of preserving the typicity that characterizes this artisanal meat product. Isolates of lactic acid bacteria (LAB) and coagulase-negative-cocci (CNC) were obtained, identified and typified using 16S rRNA gene sequences and RAPD-PCR reactions. Some technological and safety-related properties were also studied: 1-resistance to low pH and high salt concentration, 2-proteolytic and lipolytic activities, 3-antibiotic resistance against tetracycline, kanamycin, vancomycin and ampicillin, and 4-absence of histidine decarboxylase (hdc) gene. A bacterial consortium of one LAB (Latilactobacillus sakei UNQLs16) and five CNC selected strains with the best features was used to formulate a meat fermentation starter (UNQ-MFS), which was compared with a commercial one (C-MFS), in a pilot-scale sausage production. Certain technological parameters (pH and weight reduction, microbial counts, and final texture profiles) were assessed in both batches to ensure the appropriate development of the process. The strains implantation was followed using RAPD-PCR: L. sakei UNQLs16 showed dominance throughout the process and its implantation was confirmed in the batch inoculated with UNQ-MFS, whereas isolates obtained from the C-MFS were not detected in the batch inoculated with this commercial starter. The fermentative starter design with native strains is not only relevant for the regional producers, but also to expand the knowledge and increase the number and diversity of available strains capable of leading the fermentative processes, contributing to safety and quality of foodstuffs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fermented dry sausages are meat products with stable microbial characteristics, a singular flavor, and a long shelf life. They are prepared by mixing minced pork or beef with fat, sugar, salt, spices, and other ingredients, which are then introduced into a casing, where the fermentation and the ripening process will take place [1, 2]. Due to the variety of combinations of types, amounts, and proportions of meats used, and raw materials, as well as different drying conditions, the sensory characteristics of the final products are diverse [3].
Lactic acid bacteria (LAB) and coagulase-negative cocci (CNC) are the most microbiologically important groups involved in the fermentation of dry sausages. The main function of LAB is to ferment sugars, leading to the production of acid products (mainly lactic acid), which is the main cause of the decrease in pH during the fermentation process [4,5,6]. This acidification is highly relevant since it is necessary for the fibrillar proteins to coagulate, and this improves the cohesiveness and hardness of the final product, facilitating slicing [7]. The CNC, especially Staphylococcus spp., contribute to the stabilization of the red coloration, and the devolvement of the characteristic flavor and aroma through lipolytic and proteolytic activities [8,9,10,11,12]. However, it is worth mentioning that the proteolytic activity of LAB through endopeptidases, proteinases, and exopeptidases has also been reported in different food matrices, which may contribute to the organoleptic and texture profile of the final product [13, 14]. Lipolysis causes the release of free fatty acids by the action of lipases, which, when oxidized, give rise to volatile compounds such as alcohols, aldehydes, and aromatic hydrocarbons, responsible for the distinctive flavor of these fermented products [15,16,17]. Proteolysis is directly related to the characteristic flavor of dry-fermented sausages due to the formation of free amino acids, and it also contributes indirectly to aroma formation, since these free amino acids are precursors of numerous aromatic compounds [15, 18, 19]. Proteolysis is also extremely important to determine the texture characteristics of the product, since the several low molecular weight components formed are influenced by both muscle and microbial enzymes [20,21,22,23]. Additionally, the reduction in pH as a consequence of the production of lactic acid during the LAB fermentation process, the decrease in the water activity of the meat through the addition of salt, the antioxidant and antimicrobial role of nitrates and nitrites, and the elimination of oxygen during chopping, constitute the main factors related to the safety and lifespan of fermented meat products, avoiding the proliferation of pathogenic and deteriorating bacteria [4,5,6, 24].
Latilactobacillus sakei strains seem to be the prevailing LAB in fermented meat products [25], and the most competitive LAB representing half to two-thirds of all LAB isolates during an artisanal fermented sausage [12]. Latilactobacillus curvatus and Lactiplantibacillus plantarum are also LAB frequently found in meat fermentation processes [26,27,28]. On the other hand, there is a greater diversity of CNC species, the most relevant being Staphylococcus equorum, Staphylococcus saprophyticus and Staphylococcus xylosus [28,29,30].
The spontaneous meat fermentation process in artisanal sausages is led by a microbiota that depends on the raw materials used, the manufacturing environment and the process conditions and, therefore, the final product obtained is widely variable [26, 31, 32]. Alternatively, the use of a meat fermentation starter (MFS), which are commonly employed by the sausage industry, can improve the quality and safety of its products when combined with a strict control of temperature and relative humidity conditions [33]. The MFS are composed of selected strains of LAB and CNC that can lead the fermentative process [34, 35], and rapidly and persistently colonize the meat batches [36, 37]. Studying the viability of LAB and knowing their concentration at the end of the fermentation process is interesting since strains with beneficial properties (for example probiotic strains) could be included in the MFS, given that the consumer’s preference for healthier products has increased in recent years [38, 39]. In addition, an MFS could also be constituted by autochthonous strains isolated from a spontaneously dry-fermented sausage, which could help to exacerbate the artisan-like flavors [26].
Among the safety-related criteria used to select strains for MFSs, the absence of biogenic amines production as well as the absence of transferable antibiotics resistance is desirable. Histamine is one of the most important biogenic amines and one of the most studied due to its toxicological effects (vasoactive and psychoactive properties) [40, 41]. Histamine production depends on the histidine decarboxylase activity in the microorganisms present in fermented foods. Since phenotypic detection could lead to false positives, the detection by PCR of the histidine decarboxylase (hdc) gene is preferable. On the other hand, resistance to antibacterial drugs has become a global problem for both human and animal health, being the food chain one of the key routes of emergence and spread of antimicrobial resistant bacterial populations [42, 43]. In this context, screening tests to identify resistance to antimicrobials should be included as a criterion for strain selection.
Artisanal dry-fermented sausages from Colonia Caroya are culturally and economically relevant food products, which have obtained the quality certificate of Geographical Indication in Argentina [44,45,46]. Several producers follow a series of ancient European recipes that have been handed down from generation to generation. This important food is a coarsely-chopped sausage, with a length of 25–40 cm, and 4–6 cm in diameter, with a little salt and slightly spicy, usually stuffed in natural casings, matured in a cellar for at least 21 days, with a dark red final color. Molds often grow on the surface of these fermented meat products; this fungal microflora is usually dominated by Penicillium nalgiovense, which confers it a desirable white or whitish grey surface color, contributes to the formation of the flavor and prevents the development of other mycotoxigenic fungi. A greenish color can be attributed to the presence of other, less desirable, though not deteriorating, molds [46,47,48]. Since the fermentation process occurs spontaneously, sometimes the quality of the final products cannot be ensured, leading to significant economic losses. Although a solution would be to use commercial fermentation starters that could promote the fermentation process in the desired way, and contribute to the safety of the final product, these starters have the disadvantage of reducing the typicity that characterizes a regional sausage [49]. In this work, we set out to formulate a fermentation starter that would preserve the advantages of using a starter but avoiding the disadvantages of the commercial ones, by developing a multi-strain starter of autochthonous LAB and CNC, tested in a pilot-scale production under controlled conditions, with the goal of obtaining a final dry-fermented sausage with characteristics similar to those from Colonia Caroya. We aim to achieve in the future a technology transference that can increase the value of the product while widening the options of starter strains capable of implantation.
Materials and methods
Strains isolation and identification
LAB and CNC isolation and growth conditions
Mechanical breaks (stomacher AES CHEMUNEX, Easy MIX, Quebec, Canada) were performed on 25 g of samples of a Colonia Caroya sausage throughout the fermentation process. Serial dilutions were done and sown in Mann–Rogosa–Sharpe (MRS, Biokar Diagnostics, Allonne, France) and Mannitol Salt (MS, Biokar Diagnostics, Allonne, France) agar to isolate LAB and CNC respectively. Gram positive, catalase negative and non-sporulating bacilli were selected as possible LAB, while Gram-positive, catalase positive, coagulase negative cocci were selected as CNC candidates (according to Argentinian Farmacopea [50]). LAB were grown at 28 °C under microaerophilic conditions for 2 days whereas CNC were grown at 37 °C under aerobic conditions.
Identification and typing of isolates
Genomic DNA from LAB and CNC was obtained according to Bravo Ferrada et al. [51]. Polymerase chain reaction (PCR) amplification of a 16S rRNA gene fragment was carried out using the primers pA16SF and pH16SR [52]. The amplicons were purified using the QIAquick PCR purification kit (QIAGEN Corp. California, Redwood City, USA) and sent to be sequenced (Macrogen Corp., Korea). Sequences were identified by comparing them with those deposited in GenBank, using BLAST software.
The isolates typing was performed by Random Amplified Polymorphic DNA-PCR (RAPD-PCR) analysis using primers Coc [53], M13 [54], and 1254 [55] according to Delfederico et al. [56] and the PCR products were resolved by electrophoresis in 1.5% (w/v) agarose gel. Primers were purchased from Genbiotech SRL (Buenos Aires, Argentina). All the PCR amplifications were performed in an Eppendorf Mastercycler equipment (Hamburg, Germany).
Technological properties
CNC and LAB growth
In order to emulate the environmental conditions during the fermentation process, LAB and CNC strains were cultured at different pH (4.2 and 4.8) and salt concentrations (4% and 8% NaCl, Reagents S.A., San Lorenzo, Argentina), in MRS and in Brain–Heart Infusion (Biokar Diagnostics, Allonne, France), respectively. The growth was followed using OD600 values (spectrophotometer Bio-Rad, SmartSpec 3000, Hercules, CA, USA) at the beginning and after 22 h. An arbitrary criterion regarding the control without salts or adverse pH was established, wherein high resistance (value 3): decrease < 2 log units; medium resistance (value 2): decrease < 3 log units; and low resistance (value 1): decrease > 4 log units.
Proteolytic activity
Agar plates were prepared for CNC and LAB, supplemented with skimmed milk powder (Oxoid—Thermo Fisher Scientific, Basingstoke, UK) 20% (previously pasteurized for 5 min at 90 °C), and bacteriological gelatin (Oxoid—Thermo Fisher Scientific, Basingstoke, UK) 10 g/L. The plates were inoculated with 10 µL of a bacterial suspension (previously grown to OD = 1.0 in their corresponding culture medium) and incubated under the adequate conditions. The proteolytic activity was determined by measuring the radius of the halo generated according to: “3” for a halo > 4 mm, “2” for a halo between 2 and 4 mm, “1” for a halo between 1 and 2 mm, “0” when no halo was detected [57].
Lipolytic activity
Lipolytic activity was measured by the agar diffusion technique in test tubes. The corresponding culture medium for LAB and CNC was supplemented with 0.1% tributyrin (Sigma-Aldrich Company Ltd, Dorset, UK). The agar surface was inoculated with 10 µL of a bacterial suspension of OD600 = 0.1, and then incubated for 5 days at 37 °C for CNC and 28 °C for LAB. Lipolytic activity was arbitrarily assigned “3” when was present, and “0” when absent.
Strains safety-related properties
Determination of the presence of hdc (histidine decarboxylase) gene
The amplification of a 375 bp fragment of the histidine decarboxylase (hdc) gene was performed in the selected colonies. For this, the JV16Hc and JV17Hc primers and the amplification conditions described by Marcobal et al. [40, 41] were used. In addition, the ST2A strain of L. buchneri was used as a positive control. Primers were purchase from Genbiotech SRL (Buenos Aires, Argentina).
Determination of antibiotic susceptibility
The LAB and CNC strains with the best technological features were selected to be part of the UNQ-MFS. For these strains, resistance to antibiotics was determined by means of the minimum inhibitory concentration (MIC) test; we assayed two inhibitors of protein synthesis: tetracycline (Stanton, CABA, Argentina) and kanamycin (Sigma-Aldrich Company Ltd, Dorset, UK); and two cell wall inhibitors: vancomycin (Klonal Laboratories, Quilmes, Argentina) and ampicillin (Sigma-Aldrich Company Ltd, Dorset, UK). First, the strains were grown until the exponential phase. The cultures were washed twice with a sterile physiological solution, then the pellet was resuspended in its corresponding culture medium. Subsequently, the DO600 (Spectrophotometer, Ultrospec Amersham Biosciences SE-751-84, Amersham, UK) was measured to calculate the CFUs in each culture. Dilutions were made until reaching 0.5 on the McFarland scale for each inoculum [58] (108 CFU/mL). On the other hand, twofold serial dilution for each antibiotic were realized ranging from 0.25 up to 256 µg/mL. Once the 96-well plates were prepared, DO600 was measured in a plate reader (RAYTO, RT-6000, Nanshan, China) at time 0. Then the plates were incubated at 30 °C and after 48 h, DO600 was measured again. Cut-off values were taken from the Clinical and Laboratory Standard Institute (CLSI) [59] for Staphylococci spp., and from the EFSA Guidance [60] for the L. sakei. Since M. caseolyticus and Rothia sp. are of no clinical relevance, their cut-off values for different antibiotics have not been reliably established, to the best of our knowledge.
Meat-batches preparation
Starters
The UNQ starter of meat fermentation (UNQ-MFS) was constituted by a LAB, Latilactobacillus sakei UNQLs16, and five CNC: Macrococcus caseolyticus UNQMca2, Staphylococcus saprophyticus UNQSscb2, Rothia sp. UNQRcb1o, Staphylococcus equorum UNQSeco12, Staphylococcus xylosus UNQSxco16, were all selected for their technological characteristics. A commercial starter of meat fermentation (C-MFS) was used as a control, from which two catalase negative cocci were isolated. In both cases, colony forming unit (CFU) and optical density (OD) relation were determined according to Miles and Misra [61] in order to standardize the inoculum. The microorganisms selected for UNQ-MFS were grown until the exponential phase, and manufacturer’s instructions were followed for the C-MFS.
UNQ and commercial meat batches
In the pilot plant of the Center of Research and Industrial Technology of Meat, National Institute of Industrial Technology (INTI), two meat batches of 10 kg each were prepared with the composition described in Appendix 1. One part of the processed meat was kept refrigerated in a chamber at 0–2 °C, while the other part was frozen at − 18 °C. Bacon (from a local supplier), which was cut with a knife, was used as pork fat and kept in a chamber at − 18 °C until use. The components were added in the following order: frozen meat, additives, ferments, bacon and refrigerated meat. The two batches were differentially inoculated, one batch with UNQ-MFS (107 CFU UNQLs16, 108 CFU mix-CNC) and the other with C-MFS (107 CFU). The obtained meat batter was introduced into casings and the pieces were closed with clips, obtaining sausages of approximately 15 cm long, and weighing between 250 and 300 g each. The dry-out was carried out at the conditions detailed in Appendix 2. During the fermentative process, samples were taken after 0, 1, 2, 3, 4, 7 and 15 days.
Physicochemical characteristics
pH
The pH was monitored during the fermentation process (pH meter Testo, model 205, CABA, Argentina). The measurements were made in triplicate and were carried out in the following time interval: 0, 1, 2, 3, 4, 8, 11, 15, 21 and 27 days.
Weight loss
The drying process was followed by monitoring weight loss by measuring three sausages of each batch (Ohaus Balance model Explorer Pro EP 6102, CABA, Argentina) for each drying time (TS) (same time interval as pH). The percentage of weight loss was calculated using the following equation [62]:
Texture analysis
Final products from each batch (UNQ-MFS and C-MFS) were analyzed by 8 instrumental texture measurements (texturometer TMS-Pro, FTC Food Technology Corp., Virginia, USA). Those were taken in a temperature range between 15 and 20 °C, on 1-mm thick slices, using an aluminium plate of 10 mm in diameter. A texture profile analysis was carried out (TPA: performance of two successive compression cycles on the sample, imitating the action of the jaws), measuring the parameters: hardness, adhesiveness, elasticity, and chewiness.
Microbial monitoring
Counts and microscopy
The survival of microorganisms in both batches of meat (UNQ-MFS and C-MFS), was followed on samples taken at times 0, 1, 2, 3, 4, 7 and 15 days. Ten g were aseptically taken from the matrix and placed in a sterile stomacher bag with 10 mL of physiological solution. After mechanical disintegration, serial dilutions were seeded on MRS agar plates for LAB, MS for CNC, and eosin and methylene blue (EMB, Laboratorios Britania S.A., Buenos Aires, Argentina) for Gram-negative bacteria. The MRS plates were incubated at 28 °C, and MS and EMB at 37 °C for 48 h. The CFUs for each culture media were counted, after which Gram tests and catalase activity assessments of colonies were performed.
Implantation of the strains
The implantation analysis was carried out from twenty colonies randomly taken from MRS and MS plates at each sampling time. The colonies selected from the UNQ batch were: Gram positive, catalase negative bacilli growing on MRS, and Gram positive, catalase positive, mannitol fermenting cocci, growing in MS. From the commercial batch, the selection was random to ensure a representative fraction of bacteria present.
Implantation ability was evaluated by RAPD-PCR method, with M13 primer. The electrophoretic profiles obtained were compared between the strains comprising each fermentation starter with isolates recovered from batches inoculated with UNQ-MFS and C-MFS, for each sampling time (0, 1, 2, 3, 4, 7 and 15 days) and each culture medium analyzed (MRS and MS). From the twenty LAB and CNC previously selected, at least 5 were randomly chosen for this analysis. The initial number of colonies for screening was increased only for those cases where it was not possible to find a RAPD-PCR electrophoretic profile that matched the inoculated strain(s). To consider that a strain was implanted, we established a cutoff at a minimum coincidence of 60% between the electrophoretic profiles of the isolates analyzed and the strains constituent of a fermentation starter [63].
Statistical analysis
Variations in pH, weight loss and microbial counts through time were analyzed by means of Repeated Measures ANOVA using the statistical software Infostat v2017. Texture analysis data was analyzed by means of an analysis of variance (ANOVA) using the statistical software Infostat v2017 [64]. We set the significance level at 0.05 for all analyses.
Results
Strains isolation and identification
Samples obtained through the fermentation process of an artisanal dry-fermented sausage from Colonia Caroya (Córdoba, Argentina) were processed. We obtained and preliminarily identified 53 microorganisms according to the criteria established in 2.1.1 (Fig. 1). For further assays, 12 autochthonous strains (5 LAB and 7 CNC) were chosen, based on genetic diversity analysis by RAPD-PCR, and identified by sequencing of a 16S rRNA gene fragment.
Technological and safety-related properties
From these twelve autochthonous strains, 1 LAB and 5 CCN exhibiting the best performance in growth assays under stressful conditions of pH and salt concentrations were selected: L. sakei UNQLs16 (Accession Number MK478379), M. caseolyticus UNQMca2 (MK478377), S. saprophyticus UNQSscb2 (MK478378), Rothia sp. UNQRcb1o (MK478385), S. equorum UN-QSeco12 (MK478384) and S. xylosus UNQSxco16 (MK478387). Further characterization assays (proteolytic and lipolytic) were performed on these six selected strains, and the results are shown in Fig. 2. Safety-related properties were also assayed, hdc gene on genomic DNA, and antibiotic susceptibility tests, which are summarized in Table 1.
Radar chart of the technological properties assayed. Proteolytic activity was assayed using either casein (Proteolysis 1) or bacteriological gelatin (Proteolysis 2) as substrates. Lipolysis activity with 0.1% tributyrin. Resistance to pH and NaCl were performed upon 22 h of growth, and then compared to controls without salts or adverse pH. For more details see “Materials and methods” (section “Technological properties”)
Pilot scale evaluation of autochthonous strains as starter culture
To study the effectivity of the UNQ-MFS in a fermentative process, two meat batches were prepared at pilot-scale at the National Institute of Industrial Technology (INTI), Argentina. One of them was inoculated with a starter involving autochthonous strains (UNQ-MFS), and the other one with a commercial starter (C-MFS, see section “Meat-batches preparation”), for comparison purposes. Both batches were monitored over time, evaluating the modifications of physicochemical parameters, changes of macroscopic and textural characteristics, and bacterial counts of LAB and CNC, as well as potentially harmful Gram-negative bacteria. The results obtained are described below.
Physicochemical characteristics
The pH and weight loss evolution for both batches did not show significant differences (Fig. 3A and B, respectively). In both, a minimum pH value lower than 5 was reached 72 h after inoculation, and then took place a subsequent restoration to values close to neutrality (between 6.6 and 6.8). The weight loss reached values close to 30% in the final product for each batch studied. Among the variables measured in texture profiles analysis, the chewiness and hardness parameters of the sausages obtained with UNQ-MFS were statistically lower than those obtained with C-MFS at the end of the process (Fig. 3C), whereas there were no significant differences for adhesiveness and elasticity. Regarding the qualitative characteristics of the final sausages, an intense and pleasant aroma stood out. The surface of sausages obtained with the UNQ-MFS had a predominance of white mycelium, while the one obtained with the C-MFS was mostly greenish (Online Resource 1).
Physicochemical characteristics during (A, B) and at the end of the fermentation process (C). A pH evolution of UNQ-MFS and C-MFS, while the vertical line shows the lowest pH point obtained for both batches. B Weight loss during the time for both batches (UNQ and Commercial). C Texture indicators for the final products obtained with UNQ-MFS and with C-MFS. **p = 0.0095; ***p < 0.0001
Microbial counts throughout the fermentation process
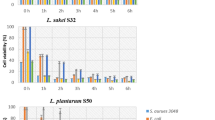
Figure 3 shows the counts of LAB, CNC and Gram-negative bacteria throughout the fermentation process of two batches studied (UNQ and commercial). The BAL count for the batch inoculated with UNQ-MFS was slightly but significantly higher during the first 72 h than the obtained with the C-MFS (Fig. 4A). Then, LAB counts reached values close to 10 log CFU/g in both cases, which remained until the end of the ripening (differences between UNQ and C-MFS treatments: F (1, 12) = 21.13, p = 0.0006; time: F (5, 12) = 309.4, p < 0.0001; interaction: F (5, 12) = 16.57, p < 0.0001). Counts obtained for CNC indicate that growth of this group was similar in both batches until the 4th day (Fig. 4B). Then, counts remained stable for the C-MFS batch, whereas it decreased significantly in the UNQ-MFS batch (differences between UNQ and C-MFS treatments: F (1, 12) = 20.71, p = 0.0007; time: F (5, 12) = 124.0, p < 0.0001; interaction: F (5, 12) = 30.89, p < 0.0001). Finally, as a criterion of quality and safety of the final products, the Gram-negative flora was monitored, and its counts became negative by the 7th day (Fig. 4C).
Implantation of the strains
To evaluate the implantation ability of the strains inoculated in each batch, colonies isolated throughout the fermentation process were analyzed by RAPD-PCR (see section “Microbial monitoring”). Most RAPD patterns obtained from LAB colonies (UNQ-MFS batch, for example electrophoretic profile 1, 2 and 3) matched the pattern (P) of L. sakei UNQLs16 (Fig. 5A). At the end of the ripening, other electrophoretic profiles non-matching to L. sakei UNQLs16 were also detected; however, these profiles were sporadic and not repeated over time. In the implantation analysis of the CNC for the same batch, none of the electrophoretic profiles of the isolates matched those obtained for the strains included in UNQ-MFS formulation (data not shown). In the batch inoculated with C-MFS, none of the electrophoretic profiles matched those obtained from isolates of this commercial starter (b and c patterns, Fig. 5D). However, we detected one electrophoretic pattern consistently repeated over time (electrophoretic profile 4) (Fig. 4C).
Implantation analysis followed by RAPD-PCR throughout time for sausages obtained with UNQ-MFS (A, B) and C-MFS (C, D). A and C show the growth of LAB over time and pictures of gels with representative RAPD-PCR profiles. B and D correspond to an analysis of repeated profiles found over time. References of the electrophoretic profiles: M, marker; P, L. sakei UNQLs16; b and c, strains from C-MFS; 1–3, native strains from batch inoculated with UNQ-MFS; 4, native strain from batch inoculated with C-MFS
Discussion
Since both LAB and CNC are relevant microorganisms for the fermentation process, genetically diverse autochthonous LAB and CNC strains, obtained from sausages of Colonia Caroya, were subjected to screening to select those that exhibited the best technological characteristics. It has been widely reported that CNC, particularly Staphylococcus spp., are involved in stabilizing the red color and producing compounds that contribute to flavor and aroma through their lipolytic and proteolytic activity [8,9,10,11,12]. These findings are consistent with ours, which show proteolytic and lipolytic activities on different substrates for S. saprophyticus UNQSscb2, S. equorum UNQSeco12 and S. xylosus UNQSxco16. Furthermore, M. caseolyticus UNQMca2 and Rothia spp. UNQRcb1o were positive for these metabolic activities using the same substrates. S. xylosus and S. equorum are the most commonly CNC species found in dry-sausages, followed by S. saprophyticus, also frequently isolated from this type of meat product [28,29,30]. Three strains belonging to these species were selected as the most significant representatives of the CNC group in the starter formulation. On the other hand, M. caseolyticus and Rothia spp. are less abundant in meat fermented products, or they are not frequently included in meat fermentation starters. However, given that the strains belonging to these species showed proteolytic and lipolytic activities similar to the staphylococci, and with the aim of trying to reproduce the complex microbial consortia of the food matrix from which they were obtained, these CNC strains were also selected to formulate a multi-strain fermentation starter (UNQ-MFS). Other authors have demonstrated that M. caseolyticus could contribute to accelerate the degradation and oxidation of lipids and proteins, and improve the flavor characteristics of Cantonese sausage [65]. Additionally, the potential of M. caseolyticus to generate diverse volatile flavor compounds has also been demonstrated [66].
Despite other authors reported L. sakei strains with proteolytic and lipolytic activities [3, 67, 68], UNQLs16 did not show this behavior under the in vitro conditions tested.
In addition to the technological characteristics that make the strains successful in leading a fermentation process, it is also important to evaluate the safety aspects of the strains to be used in a food product. Although autochthonous starters have been claimed to lead sausage production with desirable sanitary and sensory characteristics [32, 42], there are some concerns about their safety, since some LAB have been recognized as reservoirs of antibiotic resistant genes that could be horizontally transmissible to pathogens through the food chain [69, 70]. There are several reports describing the antibiotic resistance of LABs [71,72,73,74,75]. Intrinsic resistance is estimated to pose a minimal potential risk of horizontal transfer between different bacterial species, and could be shown mainly for aminoglycosides, quinolones, and glycopeptides [42, 72, 76]. Most Lactobacillus species are intrinsically resistant to vancomycin [60, 77]. Additionally, the transfer of antibiotic resistance within LAB from food has been studied [78, 79]. In previous reports about antibiotic resistances profiles and related genetic determinant of LAB from dry-sausages, the most assessed antibiotics were tetracycline and erythromycin, followed by chloramphenicol, streptomycin, ampicillin, and vancomycin [42]. Our results show that L. sakei UNQLs16 is inhibited by all the antibiotics tested at a concentration lower than the established cut-off value. For screening purpose, we chose some of the antimicrobials relevant to human and veterinary health, according to EFSA [60].
The MIC values observed, for all the antibiotics assayed, were low, except for S. saprophyticus UNQSscb2, resistant to vancomycin, and S. xylosus UNQSxco16, resistant to both vancomycin and kanamycin, according to cut-off values stablished by CLSI [59]. Since dry-sausages provide an environment where close contact among bacteria could facilitate horizontal genetic transfer [42], further assays at molecular level would be required to assess the potential of these CNC strains to serve as hosts for antibiotic-resistance genes.
Additionally, the histidine decarboxylase (hdc) gene, involved in the histamine synthesis, the most important biogenic amine from the toxicological and hygienic aspects in dry-sausages [80], was absent in all the strains studied here. Some authors have suggested that the use of decarboxylase-negative starters and the low water activity might reduce the biogenic amines formation in dry-fermented sausages [81]. Further studies should be performed to search for genes involved in the synthesis of other biogenic amines, such as tyramine and cadaverine.
The strains that showed the best technological and safety characteristics, summarized in Table 1, were selected to formulate the UNQ-MFS starter: L. sakei UNQLs16 (MK478379), as representant of the LAB group, and M. caseolyticus UNQMca2 (MK478377), S. saprophyticus UNQSscb2 (MK478378), Rothia sp. UNQRcb1o (MK478385), S. equorum UNQSeco12 (MK478384) and S. xylosus UNQSxco16 (MK478387), as representatives of CNC group.
UNQ-MFS was compared with a commercial starter (C-MFS), evaluating two meat batches (each with one of the starters) on a pilot scale (INTI, Argentina). Both fermentative processes were followed through physicochemical (pH and weight loss), microbiological (counts), and molecular (implantation analysis) parameters.
The LAB’s main role is to reduce the pH of the matrix, by consumption of sugars, which leads to the production of some acid products (mainly lactic acid) [4,5,6]. This is consistent with the fast LAB growth and the decrease in pH observed at the beginning of fermentation process. The subsequent restoration of pH observed is related to proteolytic activity. Proteolysis is initially carried out by endogenous exopeptidases, whose activity is favored at low pH. In the advanced stages of the fermentation process, proteolysis is mainly due to microbial enzymes, especially attributed to CNC metabolism. Thus, it increases the bioavailability of small peptides, free amino acids, and basic compounds such as ammonium, all of which raise pH [15, 19, 82]. Furthermore, the increase in free amino acids and non-protein nitrogen has been related to other effects, such as favoring the drying process during sausage maturation [5, 83]. In relation to this, in both meat batches studied, a gradual decrease in weight was observed, reaching final values of around 30%. Additionally, both the decrease in pH at the beginning of the process and the loss of weight are important parameters related to the safety of the final product, since under such conditions, pathogenic and spoiling microorganisms are not capable of growth [4,5,6, 84]. In this sense, no counts of the Gram-negative bacteria and companions were obtained from the 7th day of the process for both meat batches which contributes to the safety of the final products.
For comparative purposes, the texture profile of the final products resulting from both fermentation starters (UNQ and Commercial) was analyzed. Since the physicochemical characteristics that are relevant for the development of the texture profile, such as pH and drying [7, 18, 19, 77, 78], were similar for both fermentation meat batches studied, and considering that the elaboration, ripening, and ingredients used for each meat batch were the same, we could attribute the differences found for the parameters of chewiness and hardness to a differential microbial metabolic activity introduced by the fermentation starters used. In addition to the pH and drying, the other factor that greatly contributes to the development of the texture profile, are the proteolytic activity and the formation of low molecular weight molecules [20,21,22,23]. In addition, the mold observed on the surface of the sausages seems to come from the work area since fungal starters were not used and both batches of meat were dried in the same place. However, the batch inoculated with the UNQ-MFS developed more whitish fungal colonization than the one inoculated with the C-MFS. Therefore, we could infer that the metabolites produced by the microorganisms present in the UNQ-MFS may have favored the growth of a better fungal microflora. However, further studies should be carried out in the future to address these aspects.
Other authors have compared autochthonous and commercial meat fermentation starters that included different species of LAB and CNC, using a similar inoculum (approximately 107 CFU/g), and their results partially agree with ours. Franciosa et al. [88] did not find significant differences in pH between the commercial and autochthonous batches. In addition, they carried out sensory analysis, determining a consumer preference for autochthonous batches compared to the commercial one, attributed to significant differences in parameters such as tenderness and firmness. Although a sensory analysis was not included in the present work, we found significant differences in texture parameters between the commercial and autochthonous batches, that could influence the consumer’s perception. On the other hand, Frece et al. [89] studied the same autochthonous and commercial starters in different fermented meat products and demonstrated significant differences between autochthonous and commercial starters in some of them, in terms of pH and the drying process. Their autochthonous starter cultures also yielded better results in the organoleptic evaluation.
The implantation analysis of LAB strains from the UNQ-MFS starter was performed by RAPD-PCR, like in previous studies [85,86,87,88,89], and the implantation of L. sakei UNQLs16 strain was confirmed. Its electrophoretic profile was the only one repeated over time, suggesting the ability of L. sakei UNQLs16 to lead the fermentation process, and that no other native LAB (from raw ingredients or working surfaces) was able to be implanted. Regarding the CNC strains included in the UNQ-MFS starter, they were not able to implant in the meat matrix, since none of the electrophoretic profiles observed matched those corresponding to inoculated CNC strains. Nor did we find any native electrophoretic profiles (native strains from the raw materials) that were repeated over time. The failure of CNC inoculated strains to dominate the process may be due to an insufficient inoculum concentration, since they were inoculated in a 1–5 ratio with respect to L. sakei UNQLs16. It is also possible that the low water activity and the reduced pH have limited the growth of this microbial group. This finding correlate to the decrease in CNC counts observed in the meat batch inoculated with UNQ-MFS. Other authors have shown a successful CNC implantation in fermentation processes of dry-sausages, the most relevant species being S. equorum, S. saprophyticus and S. xylosus [28,29,30, 88]. Interestingly, Frece et al. [89] showed the presence of an inoculated S. carnosus after 180 days of storage. These authors speculated that the autochthonous starter cultures applied (S. carnosus and L. plantarum) prolonged the shelf-life of the final product.
On the other hand, the implantation of the strains from C-MFS was also evaluated. Profiles of isolates obtained from meat batch inoculated with C-MFS did not show implantation or dominance of any of the strains obtained from it; however, a repeated profile was found throughout time. We believe that the implanted strain in this meat batch could come from some of the raw elements used in the formulation, such as meat, spices, utensils, etc. Alternatively, the strain could have been found at the state of viable-non-cultivable (VNC) at the time of isolation from the C-MFS and been reactivated during the fermentation process.
Conclusions
We proved the feasibility of formulating a native starter, able of successfully carrying out a pilot scale fermentative meat process, using autochthonous strains obtained from artisanal dry-sausages. These traditional products, which have obtained the quality certificate of Geographical Indication in Argentina, are both culturally and economically relevant food products. The autochthonous L. sakei UNQLs16 strain was able to implant and dominate the fermentation process, resulting in dry-fermented sausages with similar characteristics to those from which it was isolated. On the other hand, the need to adjust the CNC inoculum in future assays was evidenced, given that the CNC strains did not survive throughout the fermentative process. This adjustment could favor the implantation of these strains, which would be beneficial because of their relevance during fermentation of meat products. Although none of the strains obtained from C-MFS seems to have been implanted in the meat batch used for comparative purposes, the physicochemical and microbiological characteristics of sausages show that the fermentation process occurred spontaneously, probably due to the microbiota having been present in or on the raw materials or equipment used. We can conclude that L. sakei UNQLs16 has adapted better to the specific meat matrix and to the manufacturing process compared to the commercial starter. This autochthonous strain has survived until the end, leading the fermentation process, and is, therefore, a strong candidate to be included in a technological transference to the productive sector as an MFS. We believe that the design of a fermentative starter with native strains is not only relevant for the regional producers, but also to expand the knowledge and to increase the number and diversity of available strains capable of leading the fermentative processes, contributing to the safety and quality of fermented foodstuffs.
Data Availability
This work is part of undergraduate thesis, which has not yet been uploaded to our institutional repository.
References
F. Villani, A. Casaburi, C. Pennacchia, L. Filosa, F. Russo, D. Ercolini, Appl. Environ. Microbiol. 73(17), 5453–5463 (2007). https://doi.org/10.1128/AEM.01072-07
C. Fontana, P.S. Cocconcelli, G. Vignolo, Int. J. Food Microbiol. 103(2), 131–142 (2005). https://doi.org/10.1016/j.ijfoodmicro.2004.11.046
Y. Liu, Z. Wan, K.W. Yohannes, Q. Yu, Z. Yang, H. Li, J. Liu, J. Wang, Front. Microbiol. 11, 611260 (2021). https://doi.org/10.3389/fmicb.2020.611260
A. Najjari, M. Boumaiza, S. Jaballah, A. Boudabous, H.I. Ouzari, Food Sci. Nutr. 8(8), 4172–4184 (2020). https://doi.org/10.1002/fsn3.1711
C.E. dos Santos Cruxen, G.D. Funck, L. Haubert, G. da Silva Dannenberg, J. de Lima Marques, F.C. Chaves, W.P. da Silva, Â.M. Fiorentini, Food Res. Int. 122, 371–382 (2019). https://doi.org/10.1016/j.foodres.2019.04.018
M.S. Ammor, B. Mayo, Meat Sci. 76(1), 138–146 (2007). https://doi.org/10.1016/j.meatsci.2006.10.022
E.H. Drosinos, S. Paramithiotis, G. Kolovos, I. Tsikouras, I. Metaxopoulos, Food Microbiol. 24(3), 260–270 (2007). https://doi.org/10.1016/j.fm.2006.05.001
H. Wang, J. Xu, Q. Liu, Q. Chen, F. Sun, B. Kong, Food Chem. 386, 132830 (2022). https://doi.org/10.1016/j.foodchem.2022.132830
D. Yu, M.-q Feng, J. Sun, Food Control 123, 107743 (2021). https://doi.org/10.1016/j.foodcont.2020.107743
C.E. dos Santos Cruxen, G.D. Funck, G. da Silva Dannenberg, L. Haubert, J. de Lima Marques, I.S. Kroning, F.C. Chaves, W.P. da Silva, Â.M. Fiorentini, LWT 86, 538–543 (2017). https://doi.org/10.1016/j.lwt.2017.08.045
F. Ravyts, L. Steen, O. Goemaere, H. Paelinck, L. De Vuyst, F. Leroy, Food Microbiol. 27(7), 945–954 (2010). https://doi.org/10.1016/j.fm.2010.05.030
F. Leroy, J. Verluyten, L. De Vuyst, Int. J. Food Microbiol. 106(3), 270–285 (2006). https://doi.org/10.1016/j.ijfoodmicro.2005.06.027
M. Kieliszek, K. Pobiega, K. Piwowarek, A.M. Kot, Molecules 26(7), 1858 (2021). https://doi.org/10.3390/molecules26071858
A. Waśko, M. Kieliszek, Z. Targoński, Prep. Biochem. Biotechnol. 42(5), 476–488 (2012). https://doi.org/10.1080/10826068.2012.656869
Y. Xiao, Y. Liu, C. Chen, T. Xie, P. Li, Food Res. Int. 135, 109247 (2020). https://doi.org/10.1016/j.foodres.2020.109247
Y. Huang, H. Li, T. Huang, F. Li, J. Sun, Food Chem. 149, 31–39 (2014). https://doi.org/10.1016/j.foodchem.2013.10.081
M. Gómez, J.M. Lorenzo, Meat Sci. 95(3), 658–666 (2013). https://doi.org/10.1016/j.meatsci.2013.06.005
Q. Chen, B. Kong, Q. Han, Q. Liu, L. Xu, Meat Sci. 121, 196–206 (2016). https://doi.org/10.1016/j.meatsci.2016.06.012
J.M.A. Aro, P. Nyam-Osor, K. Tsuji, K.-i Shimada, M. Fukushima, M. Sekikawa, Food Chem. 119(1), 279–285 (2010). https://doi.org/10.1016/j.foodchem.2009.06.025
P. Ikonić, M. Jokanović, L. Petrović, T. Tasić, S. Škaljac, B. Šojić, N. Džinić, V. Tomović, J. Tomić, B. Danilović, Int. J. Food Prop. 19(9), 1924–1937 (2016). https://doi.org/10.1080/10942912.2015.1089280
M. Spaziani, M. Del Torre, M.L. Stecchini, Meat Sci. 81(1), 77–85 (2009). https://doi.org/10.1016/j.meatsci.2008.06.017
L.C. Roseiro, C. Santos, M. Sol, M. Borges, M. Anjos, H. Gonçalves, A. Carvalho, Meat Sci. 79(4), 784–794 (2008). https://doi.org/10.1016/j.meatsci.2007.11.012
Ü. Dalmış, A. Soyer, Meat Sci. 80(2), 345–354 (2008). https://doi.org/10.1016/j.meatsci.2007.12.022
P. Kumar, M. Chatli, A.K. Verma, N. Mehta, O. Malav, D. Kumar, N. Sharma, Crit. Rev. Food Sci. Nutr. 57(13), 2844–2856 (2017). https://doi.org/10.1080/10408398.2015.1074533
M. Zagorec, M.-C. Champomier-Vergès, Microorganisms 5(3), 56 (2017). https://doi.org/10.3390/microorganisms5030056
M.S. Mainar, D.A. Stavropoulou, F. Leroy, Int. J. Food Microbiol. 247, 24–37 (2017). https://doi.org/10.1016/j.ijfoodmicro.2016.05.021
V. Pisacane, M.L. Callegari, E. Puglisi, G. Dallolio, A. Rebecchi, Int. J. Food Microbiol. 207, 57–65 (2015). https://doi.org/10.1016/j.ijfoodmicro.2015.04.029
R. Talon, S. Leroy, Meat Sci. 89(3), 303–309 (2011). https://doi.org/10.1016/j.meatsci.2011.04.029
D.A. Stavropoulou, E. Van Reckem, S. De Smet, L. De Vuyst, F. Leroy, Int. J. Food Microbiol. 274, 52–59 (2018). https://doi.org/10.1016/j.ijfoodmicro.2018.03.008
L. Cocolin, P. Dolci, K. Rantsiou, Meat Sci. 89(3), 296–302 (2011). https://doi.org/10.1016/j.meatsci.2011.04.011
D.A. Stavropoulou, P. Filippou, S. De Smet, L. De Vuyst, F. Leroy, Food Microbiol. 76, 180–188 (2018). https://doi.org/10.1016/j.fm.2018.05.006
R. Talon, S. Leroy, I. Lebert, Meat Sci. 77(1), 55–62 (2007). https://doi.org/10.1016/j.meatsci.2007.04.023
D. Bassi, E. Puglisi, P.S. Cocconcelli, Curr. Opin. Food Sci. 2, 118–122 (2015). https://doi.org/10.1016/j.cofs.2015.03.002
A. Zinnai, F. Venturi, M. Quartacci, G. Andrich, Ital. J. Food Sci. 23(1), 80 (2011)
A. Zinnai, F. Venturi, C. Sanmartin, M.F. Quartacci, G. Andrich, J. Biosci. Bioeng. 115(1), 43–49 (2013). https://doi.org/10.1016/j.jbiosc.2012.08.008
F. Pasini, F. Soglia, M. Petracci, M.F. Caboni, S. Marziali, C. Montanari, F. Gardini, L. Grazia, G. Tabanelli, Nutrients 10(10), 1497 (2018). https://doi.org/10.3390/nu10101497
F. Gardini, Y. Özogul, G. Suzzi, G. Tabanelli, F. Özogul, Front. Microbiol. 7, 1218 (2016). https://doi.org/10.3389/fmicb.2016.01218
K. Neffe-Skocińska, A. Okoń, D. Zielińska, P. Szymański, B. Sionek, D. Kołożyn-Krajewska, Appl. Sci. 10(12), 4311 (2020). https://doi.org/10.3390/app10124311
F.G. Pavli, A.A. Argyri, N.G. Chorianopoulos, G.-J.E. Nychas, C.C. Tassou, LWT 118, 108810 (2020)
A. Marcobal, B. de las Rivas, M.V. Moreno-Arribas, R. Munoz, J. Food Prot. 68(4), 874–878 (2005). https://doi.org/10.4315/0362-028X-68.4.874
C. Le Jeune, A. Lonvaud-Funel, B. Ten Brink, H. Hofstra, J. Van der Vossen, J. Appl. Bacteriol. 78(3), 316–326 (1995)
M.J. Fraqueza, Int. J. Food Microbiol. 212, 76–88 (2015). https://doi.org/10.1016/j.ijfoodmicro.2015.04.035
World Health Organization, in Antimicrobial resistance: global report on surveillance (World Health Organization, 2014)
B.Q. Salazar, Nacameh 11(2), 33–49 (2017)
G.S. Vila, G.N. Pose, J.A. Segura, V. Ludemann (2016), http://hdl.handle.net/11336/123084
R.S. Canel, J.R. Wagner, S.A. Stenglein, V. Ludemann, Int. J. Food Microbiol. 164(1), 81–86 (2013). https://doi.org/10.1016/j.ijfoodmicro.2013.03.022
Dirección Nacional de Alimentos y Bebidas. Ministerio de Producción y Trabajo, Presidencia de la Nación. Indicación Geográfica, Salame Típico de Colonia Caroya. https://alimentosargentinos.magyp.gob.ar/HomeAlimentos/IGeo/productos_reg/SalameCaroya/Salame_Tipico_Colonia_Caroya.pdf. Accessed 12 Oct 2023
A. Moavro, S. Stenglein, L. Delfederico, J. Wagner, V. Ludemann, LWT 110, 255–261 (2019). https://doi.org/10.1016/j.lwt.2019.04.074
O.R.D. Santa, R.E.F. de Macedo, H.S.D. Santa, C.M. Zanette, R.J.S. de Freitas, N.N. Terra, Food Sci. Technol. 34, 780–786 (2014). https://doi.org/10.1590/1678-457X.6467
Libro Farmacopea Argentina 7ª Ed. Comisión Permanente para la Farmacopea Argentina; Administración Nacional de Medicamentos, Alimentos y Tecnología Médica (ANTMAT); Ministerio de Salud
B. Bravo-Ferrada, L. Delfederico, A. Hollmann, V.D. La Hens, Y. Curilén, A. Caballero, L. Semorile, Int. J. Microbiol. Res. 3(1), 48 (2011). https://doi.org/10.9735/0975-5276.3.1.48-55
A.M. Rodas, S. Ferrer, I. Pardo, Syst. Appl. Microbiol. 26(3), 412–422 (2003). https://doi.org/10.1078/072320203322497446
P. Cocconcelli, D. Porro, S. Galandini, L. Senini, Lett. Appl. Microbiol. 21(6), 376–379 (1995). https://doi.org/10.1111/j.1472-765X.1995.tb01085.x
J. Stenlid, J.-O. Karlsson, N. Högberg, Mycol. Res. 98(1), 57–63 (1994). https://doi.org/10.1016/S0953-7562(09)80337-7
N. Akopyanz, N.O. Bukanov, T.U. Westblom, S. Kresovich, D.E. Berg, Nucleic Acids Res. 20(19), 5137–5142 (1992). https://doi.org/10.1093/nar/20.19.5137
L. Delfederico, A. Hollmann, M. Martínez, N.G. Iglesias, G. De Antoni, L. Semorile, J. Dairy Res. 73(1), 20–27 (2006). https://doi.org/10.1017/S0022029905001408
M.Y. Farhat, G. Thomas, C. Cunard, E. Cole, A. Myers, P. Ramwell, J. Pharmacol Exp. Ther. 254(1), 289–293 (1990)
J. McFarland, J. Am. Med. Assoc. 49(14), 1176–1178 (1907). https://doi.org/10.1001/jama.1907.25320140022001f
Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing (2020), chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/, https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf. Accessed 16 Nov 2022
European Food Safety Authority, EFSA J. 6(7), 732 (2008)
A.A. Miles, S. Misra, J. Irwin, Epidemiol. Infect. 38(6), 732–749 (1938). https://doi.org/10.1017/S002217240001158X
E. Plaza Reina, Estudio de actualización de mermas de producto, para mejorar la rentabilidad de alimentos Lacali SA (Thesis, Universidad Autónoma de Occidente, Colombia, 2013)
T. Aymerich, B. Martin, M. Garriga, M. Vidal-Carou, S. Bover-Cid, M. Hugas, J. Appl. Microbiol. 100(1), 40–49 (2006). https://doi.org/10.1111/j.1365-2672.2005.02772.x
Di Rienzo, J. A., Casanoves, F., Balzarini, M. G., Gonzalez, L., Tablada, M., & Robledo, C. W. InfoStat Versión; Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Córdoba, Argentina. 2017. (program website: https://www.infostat.com.ar/)
Y. Wu, C. Cui, W. Sun, B. Yang, M. Zhao, J. Food Process Eng 32(6), 844–854 (2009). https://doi.org/10.1111/j.1745-4530.2008.00249.x
S. Mazhar, K.N. Kilcawley, C. Hill, O. McAuliffe, Front. Microbiol. 11, 1533 (2020). https://doi.org/10.3389/fmicb.2020.01533
Y. Hu, Y. Li, X.-a Li, H. Zhang, Q. Chen, B. Kong, LWT 154, 112723 (2022). https://doi.org/10.1016/j.lwt.2021.112723
Y. Sanz, F. Toldrá, Appl. Environ. Microbiol. 68(4), 1980–1987 (2002). https://doi.org/10.1128/AEM.68.4.1980-1987.2002
C. Devirgiliis, P. Zinno, G. Perozzi, Front. Microbiol. 4, 301 (2013). https://doi.org/10.3389/fmicb.2013.00301
B.M. Marshall, D.J. Ochieng, S.B. Levy, Microbe 4(5), 231–238 (2009)
D. Gevers, M. Danielsen, G. Huys, J. Swings, Appl. Environ. Microbiol. 69(2), 1270–1275 (2003). https://doi.org/10.1128/AEM.69.2.1270-1275.2003
A.S. Hummel, C. Hertel, W.H. Holzapfel, C.M. Franz, Appl. Environ. Microbiol. 73(3), 730–739 (2007). https://doi.org/10.1128/AEM.02105-06
I. Klare, C. Konstabel, G. Werner, G. Huys, V. Vankerckhoven, G. Kahlmeter, B. Hildebrandt, S. Müller-Bertling, W. Witte, H. Goossens, J. Antimicrob. Chemother. 59(5), 900–912 (2007). https://doi.org/10.1093/jac/dkm035
S. Federici, F. Ciarrocchi, R. Campana, E. Ciandrini, G. Blasi, W. Baffone, Meat Sci. 98(4), 575–584 (2014). https://doi.org/10.1016/j.meatsci.2014.05.019
É. Laslo, É. György, A. Czikó, Acta Univ. Sapientiae Aliment. 12, 54–69 (2019)
M. Danielsen, A. Wind, Int. J. Food Microbiol. 82(1), 1–11 (2003). https://doi.org/10.1016/S0168-1605(02)00254-4
S. Zhang, J.-H. Oh, L.M. Alexander, M. Özçam, J.-P. van Pijkeren, J. Bacteriol. (2018). https://doi.org/10.1128/jb.00607-17
D.J. Das, A. Shankar, J.B. Johnson, S. Thomas, Nutrition 69, 110567 (2020). https://doi.org/10.1016/j.nut.2019.110567
M. Nawaz, J. Wang, A. Zhou, C. Ma, X. Wu, J.E. Moore, B. Cherie Millar, J. Xu, Curr. Microbiol. 62(3), 1081–1089 (2011). https://doi.org/10.1007/s00284-010-9856-2
P. Ikonic, M. Jokanovic, T. Peulic, N. Cucevic, Z. Tomicic, S. Skaljac, M. Ivic, Evolution of amino acids and biogenic amines in traditional dry-fermented sausage Sjenički sudžuk during processing, in IOP Conference Series: Earth and Environmental Science (IOP Publishing, 2019)
R. González-Tenorio, B. Fonseca, I. Caro, A. Fernández-Diez, V. Kuri, S. Soto, J. Mateo, Meat Sci. 94(3), 369–375 (2013). https://doi.org/10.1016/j.meatsci.2013.03.017
J.M. Lorenzo, M. Gómez, S. Fonseca, Food Control 46, 382–389 (2014). https://doi.org/10.1016/j.foodcont.2014.05.025
P. Ikonić, T. Tasić, L. Petrović, S. Škaljac, M. Jokanović, A. Mandić, B. Ikonić, Food Control 30(1), 69–75 (2013). https://doi.org/10.1016/j.foodcont.2012.06.021
R. Talon, S. Leroy-Sétrin, S. Fadda, Research Advances in the Quality of Meat and Meat Products (Research Signpost, Trivandrum, 2002), pp.175–191
S. Hernández-Macias, N. Ferrer-Bustins, O. Comas-Basté, A. Jofré, M. Latorre-Moratalla, S. Bover-Cid, M. del Carmen Vidal-Carou, Foods 10(8), 1916 (2021). https://doi.org/10.3390/foods10081916
L. Perea-Sanz, J.J. López-Díez, C. Belloch, M. Flores, Meat Sci. 164, 108103 (2020). https://doi.org/10.1016/j.meatsci.2020.108103
R. Rubio, A. Jofré, T. Aymerich, M.D. Guàrdia, M. Garriga, Meat Sci. 96(2), 937–942 (2014). https://doi.org/10.1016/j.meatsci.2013.09.008
I. Franciosa, I. Ferrocino, M.R. Corvaglia, M. Giordano, M. Coton, J. Mounier, K. Rantsiou, L. Cocolin, Food Res. Int. 162, 112007 (2022). https://doi.org/10.1016/j.foodres.2022.112007
J. Frece, D. Kovačević, S. Kazazić, J. Mrvčić, N. Vahčić, D. Ježek, M. Hruškar, I. Babić, K. Markov, Food Technol. Biotechnol. 52(3), 307–316 (2014). https://hrcak.srce.hr/126177
Acknowledgements
We are grateful to Andrea Guillade to read and revised English, redaction, and cohesion.
Funding
This research was funded by Universidad Nacional de Quilmes (Programa Microbiología Molecular Básica y Aplicada), Grant Number 990/19; by the Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CICPBA; project PIT-AP-BA), Grant Number 173/16; by Agencia Nacional de Promoción Científica y Técnica (ANPCyT): PICT 2019, Grant Number 0008 and PICT aplicado 2021, Grant Number 00013. LS and DV are members of the Research Career of CIC-BA, and GR is a fellow of the National Scientific and Technical Research Council Argentina (CONICET), Argentina.
Author information
Authors and Affiliations
Contributions
LD and LCS contributed to the conception and design of the study. GAR and JF performed the statistical analysis. JF and DVH performed the isolation, identification, typing and the technological properties assays. MS performed the meat batches preparation and the physicochemical characteristics assays. GAR performed the assays of the strains safety-related properties, the formulation of the UNQ-MFS and inoculum preparation, microbial monitoring assays, the design of the figures and tables, and wrote the first draft of the manuscript. GAR and LD wrote sections of the manuscript. All authors contribute to manuscript revision, read, and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rivas, G.A., Francioni, J., Sánchez, M. et al. An indigenous bacterial consortium from Argentinean traditional dry sausages as a pilot-scale fermentation starter. Food Measure 18, 516–528 (2024). https://doi.org/10.1007/s11694-023-02189-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-023-02189-9