Abstract
The crude protein content is critical to quality assessment when screening for nutrients, taste quality and commercial value. Thus, this study performed a non-destructive and rapid prediction of protein content in paddy based on line-scanning near-infrared hyperspectral imaging (1001–2300 nm) technology. Partial least squares regression (PLSR), principal component regression (PCR) and multiple linear regression (MLR) predictive models were established to evaluate protein content (5.5037–8.2543 g 100 g−1) in 100 intact paddy, and the models achieved high performance. Spectral pre-processing with De-trending to a certain extent could enhance the smoothness of the spectrum and reduce spectral noise effectively. Successive projection algorithm (SPA) was used to extract characteristic wavelengths to simplify the models. A set of 18 feature variables were selected from the original wavelength, and the SPA-PLSR model has the best performance to predict protein content in paddy. In addition, the simplified performed model with a higher value of coefficient of determination (R2) of R2C and R2P was 0.9078 and 0.8836, and the lower root mean square error (RMSE) of RMSEC and RMSEP was 0.0912 and 0.1675, respectively. The distribution maps of each sample protein contents in each pixel were obtained using the prediction model. Therefore, experimental results indicated the feasibility and possibility of a rapid and non-destructive hyperspectral imaging technique to detect the chemical component in paddy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paddy was the main essential natural source of carbohydrate (70–75%) [1], protein (4–18%) [2], fat [3] (1.5–2.5%), vitamin and mineral substance [4] for human nutrition, which has being significant staple food in the scale world. At present, the conventional paddy planning area in China was up to 245 million tons, whereas the hybrid paddy accounts for 200 million tons [2]. In particular, protein content in paddy determines the basis of pricing fluctuations and its commercial products. Thus, protein content was an important indicator of the internal paddy quality. This indicator has obvious significance on price, nutrition, cooking quality and taste of paddy and was essential that influence the growth of the food industry [5]. Rapid detection of protein content in paddy could promote the effective implementation of commercial and factory scale inspection, and the manufacturers could rapidly identify whether the characteristics of paddy meets their expectations. However, most of the traditional paddy protein content detection methods were destructive, cumbersome, time-consuming, inefficient and unable to achieve online detection [6]. Thus, a highly efficient, low-cost, effective and accurate food industry quality evaluation system is demand to develop.
In this study, a non-invasive, pollution-free, high-through put technique based on the hyperspectral imaging (HSI) technology was developed to investigate the chemical properties of internal substances and intact cereal [7]. HSI was a burgeoning technology that supplied the capacity to measure features across a far-ranging of wavebands simultaneously, thereby showing a particular representation of seeds or grains performance and internal material component interactions. The physical and chemical information about paddy could be extracted on account of the ability of HSI to collect both spectral and pictorial data [8]. Near-infrared (NIR) spectroscopy was connected to the molecular vibrations of hydrogen-containing functional groups with the specimen and is extensively used to predict major constituents of agriculture products. However, the conventional NIR spectrometers merely acquire spectral information on a patch of a spot area of the sample. Spectral information of intact paddy seeds was incapable of acquired, which could weaken the reliability and accuracy of prediction. Therefore, obtaining precise spectral information representing the whole paddy is necessary to improve the correction and stability of the predicted substance content [9].
HSI integrates NIR spectroscopy and digital imaging technique to provide a message on the spatial distribution of testing substance. In addition, HSI acquired the spectral (λ) and spatial (x, y) information and combined them into a 3D data matrix. A data cube contains hundreds to thousands of continuous images, narrow band spectrum, and 2D images of spectral message [10, 11]. In this work, chemical analysis method and NIR-HSI technology were combined to explore the protein content in paddy from different aspects. At present, the application of NIR-HSI technology is becoming increasingly widespread in non-destructive and efficient testing of quality and safety of agricultural product. For instance, NIR-HSI technology has shown a broad application prospects in fruits and vegetables [12, 13], agricultural products [14], dairy products [15], meat [16] and other fields. Although NIR detection methods perform well in predicting protein content in paddy, applying the combination of HSI and NIR spectroscopy in the analysis of paddy protein content remains limited. NIR-HIS technology with the advantages of contactless and rapid, and it has feasible superiority for homogeneity evaluation of whole paddy and other grains.
Accordingly, this research aimed to (1) determine protein content in paddy using traditional chemical methods combined with NIR-HSI technology; (2) evaluate the exploration performance of NIR-HSI technology based on three calibration models, namely, partial least squares regression (PLSR), principal component regression (PCA) and multiple linear regression (MLR); (3) select the characteristic variables from the NIR-HIS spectral region using successive projection algorithm (SPA) and interval variable iterative space shrinkage approach (iVISSA), iteratively retains informative variables (IRIV) and competitive adaptive reweighted sampling algorithm (CARS); (4) compare the selection variable subsets and prediction performance of different feature wavelength extraction algorithms and (5) apply calibration models combined with image processing algorithm to visualise the distribution of protein content in intact paddy.
Materials and methods
Paddy samples
A total of 100 varieties of paddy samples (52 kinds of japonica paddy, 34 kinds of indica paddy and 14 kinds of glutinous paddy) from different provinces (Shandong, Guizhou, Hubei, Jiangsu, Anhui, Hunan, Henan, Sichuan, etc.) in China were purchased. Then, the samples were ground to a uniform powder using a high-speed pulveriser (Deceleration Yili Industry and Trade Co. Ltd., model: QE-200., rotate speed: 28,000 r/min, power: 1200 w), placed in a sealed bag and stored under vacuum at room temperature. In each sample, 65 g of the original paddy samples (without ground) was randomly selected for HSI analyses. Each hyperspectral image contained 288 spectral bands. The paddy of each variety was ground and weighed to 0.5000 g of powder, and their protein content was determined using the AOAC 960.52 [17]. Stoichiometric methods were used to establish the relationship between the spectrum and actual protein content.
Hyperspectral imaging
In this research, the laboratory-based push broom NIR-HSI (901–2517 nm) system (Isuzu Optics Corp., Taiwan, China) was used, and the hyperspectral images of paddy samples were obtained under reflection mode [18]. The instruments line scanners that create a two-dimensional image of the sample by translating the sample on the objective table relative to the charge coupled device (CCD) camera. The light reflected by the samples could be acquired on the spatial spectral axes to obtain spatial and spectral information. The system includes a line scanning spectrometer (Specim, Spectral Imaging Ltd., Oulu, Finland), CCD camera (Andor, Ireland), two halogen lamps (Illumination Technologies Inc., New York, USA), a conveyor belt (Isuzu Optics Corp., Taiwan, China), a camera obscura (Schneider, German) and a computer (DELL, USA) for data acquisition and processing (Fig. 1).
Image and spectral processing
Figure 2 shows the schematic diagram of a corrected image of a hyperspectral image. In all of 288 white reference images and 288 black reference images were obtained to calibrate the captured images. The former was collected with a white board with a teflon surface having 99% reflectance under the same conditions as the sample acquisition process, whereas the latter was collected when the halogen lamp was turned off and the lens cover was covered. Then, the average values of the collected reference images were calculated; the samples were transmitted through the automatic conveyor belt, and the processed image information was collected on the stage. The sample images were calibrated as reflectivity images using the following Eq. (1) to improve the accuracy of spectral information and reduce the influence of light source and dark noise [19]
where RT, Iraw and Idark, Iwhite represented the corrected and raw hyperspectral images and the mean black and white reference images, respectively.
After calibrating the image, a circle of 60 × 60 pixels was selected on the image, and each sample was repeated several times. Afterwards, the average was calculated to automatically obtain the region of interests (ROI). All spectra of pixels in the ROI were extracted to obtain the average spectrum of each sample for the following data processing.
Optimal wavelength selection
The collected NIR hyperspectral data include hundreds of continuous bands. The optimal wavelength selection algorithm could simplify data processing, improve the calculation speed and the robustness of the model and remove irrelevant or redundant information bands of the sample [20]. In general, the selection of characteristic wavelengths was to select interval wavelengths from the full spectral region, and a high absolute coefficient was found in the established model [21]. Four spectral variable selection algorithms showed efficient performance, namely, SPA, IRIV, iVISSA and CARS, which were used to predict food quality by exploiting highly accurate calibration models.
In addition to non-information variables in the original spectral set, there were also exist some collinear or redundant variables. The purpose of SPA algorithm was to select the least redundant variables to resolve collinearity issue. SPA was a forward wavelength selection algorithm that began with a wavelength and after that merged a new wavelength in each iteration until reached the specified amount of N wavelengths [22]. IRIV was a characteristic wavelength selection algorithm on the basis of binary matrix shuffling filter (BMSF) [23]. This algorithm was used to split all wavelengths into strong information, weak information, non-information and interference wavelengths using model population analysis (MPA) and remove non-information and interference wavelengths with an iterative manner. Finally, the wavelength after backward elimination was selected as the characteristic wavelength [24]. The iVISSA was an extraction method in full wavelength range based on MPA and cross-validation of root mean square error (RMSECV). The algorithm combines the effect of wavelength combination and wavelength space compression [25]. The iVISSA-SPA algorithm was a mixture characteristic wavelength selection method. Firstly, iVISSA was selected to acquire a set of potential feature wavelengths for protein content. Then, SPA was used to select the most efficient subset of variables from the potential characteristic wavelengths to evaluate the validity of these remaining wavelengths. For CARS algorithm, the frequency of Monte Carlo sampling runs was 200, and fivefold cross-validation was used to confirm the optimal characteristic wavelength. All characteristic wavelength extraction algorithms were implemented in MATLABR2018b (Math Works, Natick, USA).
Multivariate data analysis
Model establishing was one of the paramount steps in multivariate data analysis. This investigate was based on three algorithms for multivariate calculation, namely, PLSR, PCR and MLR. The results of the three algorithms were compared to determine the best calibration model.
PLSR has been extensively applied in NIR-HSI technology and shown commendable advantages for strongly collinear spectral data. In this study, the PLSR model was used to establish a linear regression algorithm between the paddy spectral matrix (X) and reference protein content (Y) [26]. The PCR was a data dimensionality reduction model, which was normally used in HSI processing. The PCR reduced the redundant information by conversing strongly correlated variables into principal component variables with non-correlation [27]. Thus, for NIR hyperspectral images, the PCR model decreased the spectral dimension, where the number of principal components was seldom and the image variance was large. MLR was an alternative linear algorithm to analyse spectral information processing that built a model between multi-spectral variables and a sample variable by fitting a linear equation [26]. When the variables of the sample were greater than the spectral, the algorithms were considered suitable [28].
Visualisation of protein content distribution
The NIR-HSI technology could not only accurately detect the main nutrients in the samples, but also use image processing technology to present the spatial distribution of nutrients in the samples and the concentration gradient of different substances [29]. NIR hyperspectral imaging aimed to reveal the hidden message in the images and then qualitatively or quantitatively predict the characteristics of the measured samples. In this study, the PLSR model was utilised to visualise and map each pixel of a hyperspectral image as a chemical image to forecast protein content in paddy at the pixel level. The principle was achieved by calculating the spectrum of each pixel in the images, and the regression equation was established based on the PLSR model [30]. The generated images were called a visual image of the chemical composition, normally expressed on a linear colour scale [31].
Results and discussions
Spectral analysis
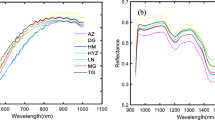
Figure 3 illustrates the spectral curves of 100 different paddy varieties after removing the wavelengths with excessive energy and distinct noise at the beginning and ending of the spectrum in the effective wavelength region of 1001–2300 nm (223 bands). The change in sample spectrum was primarily due to chemical components such as protein, starch, fat and moisture. In addition, the changes in spectral curves were mainly related to the overtones and combined vibrations of chemical molecular bonds, including C–H, O–H, N–H, C–O and S–H [32, 33]. The absorption peak at approximately 1221 nm was primarily related to the second overtone stretching of the C–H bond. The prominent peak occurred at about 1477 nm was ascribed to the combined action of N–H first overtone stretch and O–H first overtone stretch, respectively [34]. The bands at near 1762 nm were attributed to the C–H first overtone stretching. Another water-related absorption peak was observed at approximately 1941 nm, which was connected with O–H stretching and bending vibrations [35]. In addition, the characteristic peak of the protein was deemed to be concentrated at approximately 2185 nm [36]. Therefore, it is feasible to use NIR-HSI technology to determine protein content in paddy. It was clear that in the entire spectral region, the spectral curve trends of each variety were similar, but the reflectance intensity was different, which indicated that the internal substances of different paddy were basically the same, but the content of a single compound was different.
Rationality of sample division
It is very important to obtain the accuracy and stability of the model through reasonable dividing the samples into calibration set and prediction set. A high-performance sample set partitioning method could select calibration set samples to represent the statistical characteristics of the entire sample and the remaining samples [37]. The sample set partitioning based on joint x–y distance method (SPXY) algorithm could consider the spatial variability of x and y. SPXY algorithm has the advantage of covering a wide range of multi-dimensional spaces compared with the sample x-variable-based partitioning method [38]. As depicted in Table 1, through the SPXY algorithm, the 100 samples were divided into the calibration set and prediction set in a 2:1 ratio, resulting in 67 and 33 samples, respectively. The range of protein content in the calibration set (5.5037–8.2543 g 100 g−1) covers the prediction set (5.9102–8.1294 g 100 g−1), indicating that the division method was reasonable, which could improve the accuracy and robustness of the calibration model.
Modeling based on full wavelengths
In this research, a correction model was firstly established based on the full wavelength to remove irregularities and noise bands in the head and tail of the spectral region. PLSR and PCR calibration models were used to establish a quantitative relationship between the reflectance data matrix (X) acquired in the entire 223 wavelength range and protein content (Y) of the corresponding variables. An optimized model should have higher correlation coefficients and lower RMSE on the calibration and prediction sets.
Table 2 presents the statistical parameter results for the calibration set, cross-validation set and prediction set used to calculate the protein content model in paddy. In the PLSR and PCR models, R2C was 0.9289 and 0.9061, and RMSEC was 0.0800 and 0.0920, respectively in the calibration set. On the contrary, in the cross-validation set, R2CV was 0.7908 and 0.7641, and RMSECV was 0.1411 and 0.1490. In the prediction set, R2P was 0.8629 and 0.8574, and RMSEP was 0.1818 and 0.1854. The consequences demonstrated that the model estimate of the performance of PLSR and PCR algorithms was highly accurate. The NIR-HSI system can predict protein content in paddy with a full spectral range (1001–2300 nm) based on the reliability and stability of the acquired prediction model.
Characteristic wavelength selection and model establishing
This study had proven that the NIR-HSI system was suitable for predicting protein content in paddy with the whole wavelength range. However, there were 223 wavelength variables in the full NIR spectrum with an average interval of 6.28 nm for each wavelength, and the intervals were smaller. Small wavelength separations represented close or similar wavelength bands that carried repetitive information associated with the protein [19]. The full hyperspectral data disposing process was complicated and time-consuming, and the speed of the established model was limited, which was difficult to convert into online detection. Therefore, extracting the characteristic wavelengths instead of the limited number of full wavelengths was necessary to establish the calibration model and obtain similar accuracy and stability. In the current research, SPA, IRIV, iVISSA-SPA and CARS algorithms were applied to select the characteristic wavelengths that were strongly correlated with protein content.
Characteristic wavelength selection by SPA
The SPA algorithm can efficiently solve data redundancy and covariance reduction. The theory of the SPA used RMSE value to analyse the performance of the established model. The accuracy and stability of the model are high under low RMSE. Figure 4a shows the curve of the RMSE value obtained by the SPA with the increase of the selected variables. With the increase of the number of selected variables, the RMSE value began to drop sharply whereas it still exist a certain fluctuation until the number of selected variables was 18 (marked with a red box). The RMSE (0.1680) reached the minimum value and then showed a gradual upward trend. However, the specific number of extracted variables was incapable clearly known. The congruent relationship between characteristic wavelengths and variable index was shown in Fig. 4b to clearly demonstrate the results of extracting characteristic wavelengths by SPA. The position of the red box in the figure represents the characteristic variables of protein. Figure 4b shows 18 valid variables (1063, 1107, 1158, 1296, 1396, 1421, 1477, 1507, 1732, 1797, 1826, 1861, 2030, 2052, 2101, 2164, 2255 and 2300 nm) selected from 223 spectral variables, accounting for 8.07% of the total wavelengths. In this study, the optimal wavelengths selected based on the SPA contained the information most relevant to the protein.
Characteristic wavelength selection by iVISSA-SPA
In this investigation, the iVISSA was a wavelength interval spectral region selection method. The iVISSA selected 77 characteristic wavelengths, accounting for 34.5% of the total number of wavelengths (Fig. 5a). The number of characteristic wavelengths selected using this approach was numerous, and the interval distance amongst wavelengths was micro and similar bands carry similar message. Therefore, optimising effective variables for dimension decomposition of obtained data is necessary to avoid redundant information amongst adjacent bands, thereby improving the operation speed and efficiency of the model. Although the model based on the characteristic variables selected by the iVISSA algorithm exhibits good performance, the wavelength variables still need to be further optimised to adapt to future online detection technologies [39]. Therefore, on the basis of the iVISSA, the SPA was used to further screen the characteristic wavelengths [39,40,41], and a combination of the iVISSA–SPA was proposed to select the optimal characteristic variables to determine protein content in paddy. Figure 5b depicted the RMSE curve of different varieties of paddy subsets of variables after combining the iVISSA–SPA. The red box represented the number of optimal variables (16), and the corresponding RMSE was small (0.1820). Figure 5c illustrated that the corresponding characteristic wavelengths of the variables and protein were 1001, 1063, 1095, 1408, 1427, 1593, 1690, 1803, 1815, 2030, 2058, 2190, 2215, 2251, 2265 and 2300 nm, accounting for 7.17% of the total wavelengths. The selected optimal variables contained almost the overall spectral region, reducing data redundancy and preserving key information.
Characteristic wavelength selection by IRIV
The IRIV algorithm eliminated non-information variables and weak information variables through multiple iterations and retained effective information variables as characteristic wavelengths. After a few several rounds of iterations, the number of remainder variables correspondingly decreased. The refined evaluation of backward elimination strategies had good performance due to interaction amongst variables [23, 42, 43]. Figure 6a illustrated the process of the changes in the number of remaining wavelengths with the increase of iterations rounds. In the wavelength range of 1001–2300 nm, the IRIV algorithm was used to conduct a total of eight iterations round. During the first three iterations round, the number of wavelengths decreased sharply from 223 to 43 on account of many irrelevant information wavelengths were eliminated and then decreased slowly in the subsequent round. This result was remained stable at the 7th iterations round, followed by backward elimination of eight variables. Figure 6b shown 20 selected variables at 1013, 1038, 1045, 1408, 1678, 1684, 1690, 2014, 2030, 2036, 2052, 2058, 2063, 2068, 2074, 2215, 2246, 2255, 2295 and 2300 nm, accounting for 8.97% of the total wavelength.
Characteristic wavelength selection by CARS
On the basis of the calibration set, the full spectrum data and protein content were associated for the important information as the CARS algorithm to extract the feature variables. The number of Monte Carlo sampling runs was set to 200, and fivefold cross-validation was used to predict the performance of the model [10, 11]. Figure 7 shows the change related to selected wavelengths, RMSECV value and regression coefficient path with the increase of the number of sampling runs. Figure 7a clearly shows that in the initial stage, the variables with useless or redundant information were quickly screened out, and then the number of variables reduced steadily. These two steps represented the rough and precise screening in the exponentially decreasing function (EDF) process, respectively. After enforced eliminating variables based on the EDF, the algorithm further filters variables in a competitive manner. Figure 7b shows the fivefold RMSECV value decreasing at samplings runs 1–100 because non-information variables were eliminated. Nevertheless, in the subsequent process, RMSECV started to increase due to the elimination of some relevant wavelengths. The selected optimal wavelengths correspond to the minimum fivefold RMSECV of 0.1221. Figure 7c shows the regression coefficient path of each variable under different sampling runs. At the blue * position, the number of sampling runs was 100, and the minimum RMSECV value was the best subset. The CARS algorithm results show that 21 characteristic wavelengths (1296, 1352, 1358, 1581, 1587, 1593, 1678, 1690, 1696, 1732, 2014, 2019, 2052, 2058, 2068,2074, 2195, 2215, 2246, 2260 and 2300 nm); Fig. 7d were selected from 223 wavelengths, accounting for 9.42% of the total wavelengths.
Model construction and comparison
This paper established the prediction model (PLSR, PCR and MLR) for protein content in paddy based on the full wavelength and selected characteristic wavelengths, respectively. As indicates in Fig. 8, the modeling schemes of different algorithms could made great predictions for 100 varieties of paddy. The selected characteristic wavelength algorithm has high performance on account of the ability to simplify the model by removing extraneous variables compared with the full spectrum model. And further improves the stability and accuracy of the model. The PLSR model possessed higher capability and robustness than PCR and MLR prediction models when considering full spectrum and extracted feature spectrum. In the case of full-spectrum PLSR and PCR models, R2C, R2CV and R2P were 0.9289, 0.9061; 0.7908, 0.7642 and 0.8629, 0.8574, respectively, and RMSEC, RMSECV and RMSEP were 0.0800, 0.0920; 0.1411, 0.1490 and 0.1818, 0.1854, respectively. The comparison results shown that the PLSR model with 18 characteristic wavelengths based on the SPA was the optimal model, which could be better predicting the protein content in paddy. The performance of SPA-PLSR was the best with R2C = 0.9087, RMSEC = 0.0912, R2CV = 0.8188, RMSECV = 0.1331, R2P = 0.8836, and RMSEP = 0.1675 based on the prediction model. Although the prediction results of the PLSR model employing wavelengths selected by iVISSA-SPA, IRIV and CARS were not as great as that of SPA-PLSR, they were inside the acceptable limits. Figure 9 was a scatter plot of predicted and actual protein content in paddy based on the SPA-PLSR, SPA-PCR and SPA-MLR model. The solid line represented the regression line for the ideal correlation between actual and predicted values of protein. The samples points were closely distributed near the regression line, demonstrating that the prediction performance of the model was better. The model was based on extracting the characteristic wavelength, which could reduce complexity and improve accuracy. From the perspective of the accuracy of the prediction model and spectral information, the characteristic wavelengths extraction method adopted in this research was stable and reliable.
Visualisation of the distribution of protein content
The HSI technology can simultaneously provide spectral and image information of the samples. The primary preponderance of hyperspectral images was that each pixel contains ample spectral information, which could be regarded as a prediction dataset. The optimal model was selected by the characteristic wavelength algorithm to visualise protein content in paddy, and the pixel of the ROI in the hyperspectral image of samples was transformed to predict protein content. The established optimal SPA-PLSR model was transferred to each pixel of the image to construct a visualisation distribution map and predict protein content of all pixels in paddy samples. Protein content predicted by the optimal model was represented by a linear colour bar (Fig. 10). In the visualisation pixels in hyperspectral images with similar spectral characteristics had an analogical colour value (protein content). In the visualisation distribution map, the red area represents high protein content, and the purple area represents low protein content. With the increase of protein content, the colour gradually changed from dark-blue to dark-red. Protein content of different paddy varieties could be clearly differentiated by colour, and the distribution was uneven. The HSI technology could achieve visualisation in the arbitrarily area of paddy, directly characterise the distribution of protein content in paddy and then aid the comprehensive evaluation of paddy quality.
Conclusions
This study aims to investigate the application of HSI technology in the 1001–2300 nm spectral range to predict protein content of paddy and to compare the performance of multiple regression models. In improving the prediction performance, PLSR, PCR and MLR regression models were constructed, whereas the comprehensive performance of the PLSR model had a better performance. Compared with the full spectrum and iVISSA-SPA, IRIV and CARS algorithms, the PLSR model established by the SPA selected 17 characteristic wavelengths from 223 wavelengths, indicating its high predictive ability. The optimal model constructed based on SPA-PLSR showed that R2P and RMSEP were 0.8836 and 0.1675, respectively. In conclusion, it was feasible to predict protein content in paddy using the NIR-HSI technology, which is a non-destructive, rapid and accurate method. This research could provide ponderable reference for non-destructive and fast detection of chemical components such as protein in grains. Future work will select representative wavelengths by using varieties of paddy in different regions and years to improve the performance and stability of the model.
Data availability
The authors will supply the relevant data in response to reasonable requests.
References
C.J. Bergman, 9—rice end-use quality analysis, in Rice, 4th edn., ed. by J. Bao (AACC International Press, St Paul, 2019), pp.273–337. https://doi.org/10.1016/B978-0-12-811508-4.00009-5
T.B. Bagchi, S. Sharma, K. Chattopadhyay, Development of NIRS models to predict protein and amylose content of brown rice and proximate compositions of rice bran. Food Chem. 191, 21–27 (2016). https://doi.org/10.1016/j.foodchem.2015.05.038
D. Mridha, P.C. Gorain, M. Joardar, A. Das, S. Majumder, A. De, N.R. Chowdhury, U. Lama, R. Pal, T. Roychowdhury, Rice grain arsenic and nutritional content during post harvesting to cooking: a review on arsenic bioavailability and bioaccessibility in humans. Food Res. Int. 154, 111042 (2022). https://doi.org/10.1016/j.foodres.2022.111042
N. Hussain, D.-W. Sun, H. Pu, Classical and emerging non-destructive technologies for safety and quality evaluation of cereals: a review of recent applications. Trends Food Sci. Technol. 91, 598–608 (2019). https://doi.org/10.1016/j.tifs.2019.07.018
C. Maione, B.L. Batista, A.D. Campiglia, F. Barbosa, R.M. Barbosa, Classification of geographic origin of rice by data mining and inductively coupled plasma mass spectrometry. Comput. Electron. Agric. 121, 101–107 (2016). https://doi.org/10.1016/j.compag.2015.11.009
H. Mæhre, L. Dalheim, G.K. Edvinsen, E.O. Elvevoll, I.J. Jensen, Protein determination-method matters. Foods 7(1), 5 (2018). https://doi.org/10.3390/foods7010005
S. Srivastava, H.N. Mishra, Detection of insect damaged rice grains using visible and near infrared hyperspectral imaging technique. Chemom. Intell. Lab. Syst. 221, 104489 (2022). https://doi.org/10.1016/j.chemolab.2021.104489
P. Mishra, M.S.M. Asaari, A. Herrero-Langreo, S. Lohumi, B. Diezma, P. Scheunders, Close range hyperspectral imaging of plants: a review. Biosyst. Eng. 164, 49–67 (2017). https://doi.org/10.1016/j.biosystemseng.2017.09.009
M. Krimmer, C. Farber, D. Kurouski, Rapid and noninvasive typing and assessment of nutrient content of maize kernels using a handheld Raman spectrometer. ACS Omega 4(15), 16330–16335 (2019). https://doi.org/10.1021/acsomega.9b01661
C. Liu, W. Huang, G. Yang, Q. Wang, J. Li, L. Chen, Determination of starch content in single kernel using near-infrared hyperspectral images from two sides of corn seeds. Infrared Phys. Technol. 110, 103462 (2020). https://doi.org/10.1016/j.infrared.2020.103462
H. Liu, B. Bruning, T. Garnett, B. Berger, Hyperspectral imaging and 3D technologies for plant phenotyping: from satellite to close-range sensing. Comput. Electron. Agric. 175, 105621 (2020). https://doi.org/10.1016/j.compag.2020.105621
Y. Ji, L. Sun, Y. Li, J. Li, S. Liu, X. Xie, Y. Xu, Non-destructive classification of defective potatoes based on hyperspectral imaging and support vector machine. Infrared Phys. Technol. 99, 71–79 (2019). https://doi.org/10.1016/j.infrared.2019.04.007
Y. Xia, W. Huang, S. Fan, J. Li, L. Chen, Effect of spectral measurement orientation on online prediction of soluble solids content of apple using Vis/NIR diffuse reflectance. Infrared Phys. Technol. 97, 467–477 (2019). https://doi.org/10.1016/j.infrared.2019.01.012
Y. Long, W. Huang, Q. Wang, S. Fan, X. Tian, Integration of textural and spectral features of Raman hyperspectral imaging for quantitative determination of a single maize kernel mildew coupled with chemometrics. Food Chem. 372, 131246 (2022). https://doi.org/10.1016/j.foodchem.2021.131246
W.H. Su, D.W. Sun, Fourier transform infrared and raman and hyperspectral imaging techniques for quality determinations of powdery foods: a review. Compr. Rev. Food Sci. Food Saf. 17(1), 104–122 (2018). https://doi.org/10.1111/1541-4337.12314
W. Cheng, D.-W. Sun, H. Pu, Q. Wei, Characterization of myofibrils cold structural deformation degrees of frozen pork using hyperspectral imaging coupled with spectral angle mapping algorithm. Food Chem. 239, 1001–1008 (2018). https://doi.org/10.1016/j.foodchem.2017.07.011
C. Ma, Z. Ren, Z. Zhang, J. Du, C. Jin, X. Yin, Development of simplified models for nondestructive testing of rice (with husk) protein content using hyperspectral imaging technology. Vib. Spectrosc. 114, 103230 (2021). https://doi.org/10.1016/j.vibspec.2021.103230
J.-L. Xu, C. Riccioli, D.-W. Sun, Efficient integration of particle analysis in hyperspectral imaging for rapid assessment of oxidative degradation in salmon fillet. J. Food Eng. 169, 259–271 (2016). https://doi.org/10.1016/j.jfoodeng.2015.08.015
J. Ma, J.-H. Cheng, D.-W. Sun, D. Liu, Mapping changes in sarcoplasmatic and myofibrillar proteins in boiled pork using hyperspectral imaging with spectral processing methods. LWT 110, 338–345 (2019). https://doi.org/10.1016/j.lwt.2019.04.095
G. Xuan, C. Gao, Y. Shao, X. Wang, Y. Wang, K. Wang, Maturity determination at harvest and spatial assessment of moisture content in okra using Vis-NIR hyperspectral imaging. Postharvest Biol. Technol. 180, 111597 (2021). https://doi.org/10.1016/j.postharvbio.2021.111597
W.-H. Su, S. Bakalis, D.-W. Sun, Chemometrics in tandem with near infrared (NIR) hyperspectral imaging and Fourier transform mid infrared (FT-MIR) microspectroscopy for variety identification and cooking loss determination of sweet potato. Biosyst. Eng. 180, 70–86 (2019). https://doi.org/10.1016/j.biosystemseng.2019.01.005
J. Li, H. Zhang, B. Zhan, Y. Zhang, R. Li, J. Li, Nondestructive firmness measurement of the multiple cultivars of pears by Vis-NIR spectroscopy coupled with multivariate calibration analysis and MC-UVE-SPA method. Infrared Phys. Technol. 104, 103154 (2020). https://doi.org/10.1016/j.infrared.2019.103154
Y.-H. Yun, W.-T. Wang, M.-L. Tan, Y.-Z. Liang, H.-D. Li, D.-S. Cao, H.-M. Lu, Q.-S. Xu, A strategy that iteratively retains informative variables for selecting optimal variable subset in multivariate calibration. Anal. Chim. Acta 807, 36–43 (2014). https://doi.org/10.1016/j.aca.2013.11.032
K. Yao, J. Sun, C. Chen, M. Xu, X. Zhou, Y. Cao, Y. Tian, Non-destructive detection of egg qualities based on hyperspectral imaging. J. Food Eng. 325, 111024 (2022). https://doi.org/10.1016/j.jfoodeng.2022.111024
P. Zhang, Z. Xu, Q. Wang, S. Fan, W. Cheng, H. Wang, Y. Wu, A novel variable selection method based on combined moving window and intelligent optimization algorithm for variable selection in chemical modeling. Spectrochim. Acta A 246, 118986 (2021). https://doi.org/10.1016/j.saa.2020.118986
J.-L. Xu, C. Riccioli, D.-W. Sun, Development of an alternative technique for rapid and accurate determination of fish caloric density based on hyperspectral imaging. J. Food Eng. 190, 185–194 (2016). https://doi.org/10.1016/j.jfoodeng.2016.06.007
T. Lilo, C.L.M. Morais, K.M. Ashton, C. Davis, T.P. Dawson, F.L. Martin, J. Alder, G. Roberts, A. Ray, N. Gurusinghe, Raman hyperspectral imaging coupled to three-dimensional discriminant analysis: classification of meningiomas brain tumour grades. Spectrochim. Acta A 273, 121018 (2022). https://doi.org/10.1016/j.saa.2022.121018
B. Zhang, B. Gu, G. Tian, J. Zhou, J. Huang, Y. Xiong, Challenges and solutions of optical-based nondestructive quality inspection for robotic fruit and vegetable grading systems: a technical review. Trends Food Sci. Technol. 81, 213–231 (2018). https://doi.org/10.1016/j.tifs.2018.09.018
H.-D. Yu, L.-W. Qing, D.-T. Yan, G. Xia, C. Zhang, Y.-H. Yun, W. Zhang, Hyperspectral imaging in combination with data fusion for rapid evaluation of tilapia fillet freshness. Food Chem. 348, 129129 (2021). https://doi.org/10.1016/j.foodchem.2021.129129
Y.-Y. Pu, D.-W. Sun, Vis–NIR hyperspectral imaging in visualizing moisture distribution of mango slices during microwave-vacuum drying. Food Chem. 188, 271–278 (2015). https://doi.org/10.1016/j.foodchem.2015.04.120
W.H. Su, D.W. Sun, Multispectral imaging for plant food quality analysis and visualization. Compr. Rev. Food Sci. Food Saf. 17(1), 220–239 (2018). https://doi.org/10.1111/1541-4337.12317
V. Wiedemair, D. Mair, C. Held, C.W. Huck, Investigations into the use of handheld near-infrared spectrometer and novel semi-automated data analysis for the determination of protein content in different cultivars of Panicum miliaceum L. Talanta 205, 120115 (2019). https://doi.org/10.1016/j.talanta.2019.120115
G. Yang, Q. Wang, C. Liu, X. Wang, S. Fan, W. Huang, Rapid and visual detection of the main chemical compositions in maize seeds based on Raman hyperspectral imaging. Spectrochim. Acta A 200, 186–194 (2018). https://doi.org/10.1016/j.saa.2018.04.026
E. Lancelot, D. Bertrand, M. Hanafi, B. Jaillais, Near-infrared hyperspectral imaging for following imbibition of single wheat kernel sections. Vib. Spectrosc. 92, 46–53 (2017). https://doi.org/10.1016/j.vibspec.2017.05.001
X. Li, F. Feng, R. Gao, L. Wang, Y. Qian, C. Li, G. Zhou, Application of near infrared reflectance (NIR) spectroscopy to identify potential PSE meat. J. Sci. Food Agric. 96(9), 3148–3156 (2016). https://doi.org/10.1002/jsfa.7493
L.C.R. Silva, G.S. Folli, L.P. Santos, I.H.A.S. Barros, B.G. Oliveira, F.T. Borghi, I.P. Santos, P.R. Filgueiras, W. Romão, Quantification of beef, pork, and chicken in ground meat using a portable NIR spectrometer. Vib. Spectrosc. 111, 103158 (2020). https://doi.org/10.1016/j.vibspec.2020.103158
J. Chen, S. Zhu, G. Zhao, Rapid determination of total protein and wet gluten in commercial wheat flour using siSVR-NIR. Food Chem. 221, 1939–1946 (2017). https://doi.org/10.1016/j.foodchem.2016.11.155
F. Hu, M. Zhou, P. Yan, D. Li, W. Lai, S. Zhu, Y. Wang, Selection of characteristic wavelengths using SPA for laser induced fluorescence spectroscopy of mine water inrush. Spectrochim. Acta A 219, 367–374 (2019). https://doi.org/10.1016/j.saa.2019.04.045
J. Zhang, Y. Ma, G. Liu, N. Fan, Y. Li, Y. Sun, Rapid evaluation of texture parameters of Tan mutton using hyperspectral imaging with optimization algorithms. Food Control 135, 108815 (2022). https://doi.org/10.1016/j.foodcont.2022.108815
T. An, S. Yu, W. Huang, G. Li, X. Tian, S. Fan, C. Dong, C. Zhao, Robustness and accuracy evaluation of moisture prediction model for black tea withering process using hyperspectral imaging. Spectrochim. Acta A 269, 120791 (2022). https://doi.org/10.1016/j.saa.2021.120791
R. Yuan, M. Guo, C. Li, S. Chen, G. Liu, J. He, G. Wan, N. Fan, Detection of early bruises in jujubes based on reflectance, absorbance and Kubelka-Munk spectral data. Postharvest Biol. Technol. 185, 111810 (2022). https://doi.org/10.1016/j.postharvbio.2021.111810
Y. Li, B. Ma, C. Li, G. Yu, Accurate prediction of soluble solid content in dried Hami jujube using SWIR hyperspectral imaging with comparative analysis of models. Comput. Electron. Agric. 193, 106655 (2022). https://doi.org/10.1016/j.compag.2021.106655
Y. Xu, H. Zhang, C. Zhang, C. Wu, J. Li, Y. Xia, S. Fan, Rapid prediction and visualization of moisture content in single cucumber (Cucumis sativus L.) seed using hyperspectral imaging technology. Infrared Phys. Technol. 102, 103034 (2019). https://doi.org/10.1016/j.infrared.2019.103034
Funding
This work was supported by the National Natural Science Foundation of China (32171910), Shandong Natural Science Foundation (ZR2020MC085), Major Scientific and Technological Innovation Project of Shandong Province (2019JZZY010734).
Author information
Authors and Affiliations
Contributions
JZ: performed the experiments, data curation and writing the original manuscript; ZG, SW and ZR: contributed to the software, conception and methodology of this study; MY and SZ: investigation, resources; XY, JD and CM: funding acquisition, supervision, project administration and writing—review and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Guo, Z., Ren, Z. et al. Variable selection methods to determine protein content in paddy using near-infrared hyperspectral imaging. Food Measure 17, 4506–4519 (2023). https://doi.org/10.1007/s11694-023-01964-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-023-01964-y