Abstract
Purpose
Diexanthema copepods are ectoparasites on deep-sea isopods. This genus currently contains six species, all reported from the North Atlantic. Our study describes a new species of Diexanthema found on isopods from 7184 to 7186 m depth in the Kuril-Kamchatka Trench, northwestern Pacific.
Methods
We observed the copepod’s morphology, made camera-lucida drawings, and compared our species with congeners. We determined partial sequences for its 16S rRNA and 18S rRNA genes and constructed an 18S-based maximum-likelihood copepod tree to place it phylogenetically. We identified the host isopod species through morphology and cytochrome c oxidase subunit I (COI, cox1) and 18S sequences.
Results/Conclusion
We described the copepod as Diexanthema hakuhomaruae sp. nov. and identified its host as Eugerdella cf. kurabyssalis Golovan, 2015 (Desmosomatidae). This is the first Diexanthema copepod from the Pacific and also from hadal depths. Diexanthema hakuhomaruae most closely resembles D. bathydiaita Richie, 1975, parasitic on Nannoniscus sp. (Nannoniscidae) in the Atlantic, but differs from the latter in having a smooth body surface and leg 5 in the ventrolateral region of the urosome. In the 18S tree, D. hakuhomaruae was the sister group to the Rhizorhina clade, which is consistent with the morphology-based hypothesis that they are closely related.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nicothoidae is a family of parasitic copepods, with 22 genera and about 140 species [1]. Nicothoids utilize various crustaceans as hosts, including Ostracoda, Leptostraca, Decapoda, Amphipoda, Cumacea, Isopoda, Mysida, and Tanaidacea [2]. Nicothoid morphology is highly diverse. Rhizorhina copepods, for example, have an unsegmented spherical body, whereas Paranicothoe copepods have a multi-segmented body consisting of a prosome and urosome. Recent molecular phylogenetic analyses have called into question the monophyly of this family [3, 4].
The nicothoid genus Diexanthema is characterized by the following female features: a spherical prosome lacking a head process; an unsegmented urosome; antennules with two or fewer articles; maxillipeds absent; and caudal rami shorter than the urosome, or absent [1, 5,6,7]. It currently contains six named species, Diexanthema apoda Boxshall and Harrison, 1988, Diexanthema bathydiaita Richie, 1975, Diexanthema corrugatum Boxshall and Harrison, 1988, Diexanthema desistoma Richie, 1975, Diexanthema nudum Boxshall and Harrison, 1988, and Diexanthema ritchiei Boxshall and Harrison, 1988, all of which are parasitic on deep-sea isopods in the North Atlantic Ocean [8, 9] (Table 1). Boxshall and Lincoln [10] placed this genus in the “Rhizorhina group,” along with the genera Rhizorhina and Choniorhiza. The same authors proposed two other groups in Nicothoidae, the “Nicothoe group” and the “Sphaeronella group,” containing five and eight genera, respectively. The validity of this grouping has not yet been tested with molecular data.
In 2022, we collected an undescribed Diexanthema species parasitic on a desmosomatid isopod from the hadal zone in the Kuril-Kamchatka Trench, northwestern Pacific Ocean. This is the first Diexanthema copepod from outside the North Atlantic, and the first from hadal depths. Here we describe this species, provide partial sequences for multiple genes to aid future DNA barcoding, and infer its phylogenetic position in Siphonostomatoida based on an 18S-rRNA (18S) tree to test the validity of the “Rhizorhina group.”
Materials and Methods
Five copepods, each from a different host individual, were collected on 6 October 2022 during cruise KH-22-8 of R/V Hakuho-maru (Japan Agency for Marine-Earth Science and Technology; JAMSTEC), with small plankton nets attached inside an Agassiz trawl (cf. Figure 2B in Akiyama et al. [11]), at depths of 7184–7186 m. Copepods were attached to the pereonite-3 sternite or the basis of pereopods 3, 4, or 6 of the host isopods. They were photographed and then fixed and preserved in 80% ethanol.
Three copepods were detached from their host with chemically sharpened needles, and two were retained intact on the host for future non-destructive observation or molecular analysis. Two of the three detached copepods were used for morphological observations and one for DNA extraction (see below). The former were transferred through an ethanol series (70, 60, 50, 40, and 30% ethanol, each step for c. 5 min) at room temperature, mounted on cavity slides (T8-R004; Toshin Riko, Japan) in 30% ethanol, and observed with an Olympus BX53 microscope. Illustrations were prepared with Adobe Illustrator CS6 from draft line drawings made with a camera lucida. In copepods, body length (BL) was measured from the anterior to posterior ends of the body (prosome + urosome), and prosome width (PW) and urosome width (UW) at the widest portion of the prosome and urosome, respectively. In host isopods, BL (from the anterior edge of the cephalothorax to the tip of the pleotelson) and the pereonite-2 width (P2W) were measured. All measurements are presented in the text in micrometers unless noted otherwise. The specimens studied were deposited in the Invertebrate Collection of the Hokkaido University Museum (ICHUM), Sapporo, Japan, under catalog numbers ICHUM8451–ICHUM8455.
DNA was extracted from the whole body of one copepod and pereopod 1 of one host by using the NucleoSpin Tissue XS Kit (Macherey–Nagel, Germany). For the cytochrome c oxidase subunit I (COI) gene, PCR primers used for the amplification and cycle sequencing were LCO1490 and HCO2198 [12]. For the 18S rRNA gene, amplification primers were SR1 and SR12 [13], and six primers (18S-b3F, 18S-b4R, 18S-b5F, 18S-b6F, 18S-a6R, and 18S-b8F [14, 15]) were used in cycle sequencing. For the copepod 16S rRNA gene, the newly designed primers Copepod16S_F (CGCCTGTTTATCAAARACWY) and Copepod16S_R (TCGATTTGAACTCAAATCAWG) were used for amplification and cycle sequencing. PCR amplification conditions for COI with TaKaRa Ex Taq DNA polymerase (TaKaRa Bio, Japan) were as described by Munakata et al. [16]; those for 18S with KOD FX Neo (Toyobo, Japan) were as described by Okamoto and Kakui [17]; and those for 16S with KOD ONE PCR Master Mix (Toyobo) were 45 cycles of 98 °C for 10 s, 50 °C for 5 s, and 68 °C for 1 s. PCR products for 16S were separated on a 2% agarose gel, excised with a micro spatula, and purified with the MagExtractor PCR & Gel Clean Up Kit (Toyobo). All nucleotide sequences were determined with a BigDye Terminator Kit ver. 3.1 and a 3730 DNA Analyzer (Life Technologies, USA). Fragments were concatenated by using MEGA7 [18]. The sequences we determined were deposited in the International Nucleotide Sequence Database (INSD) through the DNA Data Bank of Japan.
The copepod 18S dataset for a phylogenetic analysis comprised the copepod dataset from Kakui and Munakata [4] and the one Diexanthema sequence we determined, representing 50 siphonostomatoid species and one outgroup taxon (Misophthriopsis okinawensis, a misophrioid). The sequences were aligned (1528 positions in the aligned dataset; see Online Resources 1 and 2) as described by Munakata et al. [19]; methods for selecting the optimal substitution model (GTR + F + R3), the maximum likelihood (ML) analysis, and drawing the tree were as described by Kakui and Shimada [20].
Results and Discussion
Host Identification
All host individuals were females with developing oostegites. An abbreviated description of their morphology is as follows. Pereonite 1 slightly longer than pereonite 2. Pleotelson with small posterolateral spines. Coxae I–II each with acute anterior projection (longer on coxa I than on coxa II). Pereopod 1 stout: ischium with five robust distodorsal, unequally bifid setae; carpus enlarged, ventral margin convex in proximal two thirds but straight in distal third, with minute distal, unequally bifid, distally setulate seta (UBDS) at base of penultimate seta, and row of four robust UBDS of irregular size. Operculum truncate, slightly concave distally.
According to these character states, our individuals were Eugerdella kurabyssalis Golovan, 2015, described from the Northwest Pacific Basin east of the Kuril-Kamchatka Trench [21] and later reported also from the Kuril-Kamchatka Trench [22]. Jennings et al. [22] molecularly detected Eugerdella cf. kurabyssalis Golovan, 2015 among Eugerdella specimens collected from the Kuril-Kamchatka Trench. It closely resembles E. kurabyssalis but shows minor differences from the latter, such as a smaller, less pronounced coxa [22], and has been collected from deeper depths than E. kurabyssalis (E. cf. kurabyssalis from 7081 to 7123 m; E. kurabyssalis from 4830 to 6051 m [21, 22]). In a BLAST search [23] of the public database, the partial COI sequence (LC741552; 655 bp long) we determined for one host individual (ICHUM8452) was most similar to the COI sequence from E. cf. kurabyssalis (MN179516; query cover 91%, identity score 99.83% [22]). Our isopods were collected from 7184 to 7186 m depth, similar to the depth range reported for E. cf. kurabyssalis. Although we could not judge whether the coxa in our specimens is smaller and less pronounced than in E. kurabyssalis, given the similarity in COI sequence and sampling depth, we concluded that the host isopods were E. cf. kurabyssalis.
Taxonomy
Diexanthema hakuhomaruae sp. nov.
Diagnosis (Females)
Body smooth, lacking furrows or minute hairs; anterior hood absent; caudal ramus present, with two spiniform setae (outer seta longer than inner); rod/horn-like antennule and antenna absent; semicircular irregular branching structure present; rootlet absent; mandible present; legs 1–4 absent; leg 5 present, small lobe shape, with three spiniform setae in ventrolateral region of urosome.
Type Host
Eugerdella cf. kurabyssalis Golovan, 2015 (Isopoda: Asellota: Desmosomatidae).
Type Locality
Station A5, Kuril-Kamchatka Trench axis, northwestern Pacific (41°14.024′ N 145°01.931′ E to 41°14.126′ N 145°01.108′ E), 7184–7186 m depth.
Attachment Sites
Pereopod basis and pereon sternite.
Material Examined
Holotype: female (ICHUM8451), BL 356, PW 346, UW 86, one vial containing extracted copepod and host (BL 1797, P2W 483). Paratypes: four females (ICHUM8452, BL 397, PW 359, UW 96, used in DNA extraction, one slide and one vial containing host [BL 1925, P2W 489]; ICHUM8453, BL 342, PW 326, UW 87, one vial containing extracted copepod and host [BL 1801, P2W 475]; ICHUM8454, BL 365, PW 385, UW 89, one vial containing copepod attached to host [BL 1984, P2W 499]; ICHUM8455, BL 400, PW 366, UW 95, one vial containing copepod attached to host [BL 1820, P2W 490]). All specimens were collected at the type locality on 6 October 2022 by R/V Hakuho-maru.
Representative DNA Sequences
One 16S (INSD accession number LC741550; 424 bp long) and one 18S (LC741551; 1762 bp long) sequences were determined from paratype female ICHUM8452. One COI (LC741552; 655 bp long, encoding 218 amino acids) and one 18S (LC741553; 2174 bp long) sequences were determined from host female ICHUM8452.
Etymology
The specific name (a noun in the genitive case) is from R/V Hakuho-maru, the vessel from which the type specimens were collected.
Description (Female, Based on the Holotype)
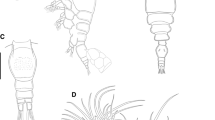
Body (Figs. 1, 2) transparent (white in ethanol), smooth, lacking furrows or minute hairs; prosome globular but slightly flattened dorsoventrally, containing bulging ovaries (Fig. 1b); anterior hood absent; urosome wide, oblong. BL/PW 1.03, BL/UW 4.14, PW/UW 4.02. Caudal ramus present, small lobe shaped, with two spiniform setae (outer seta longer than inner). Rod/horn-like antennule and antenna absent; semicircular irregular branching structure present posterior to oral sucker. Rootlet absent. Mandible present, styliform. Pore (maxillary gland pore?; arrow in Fig. 2e) present on ventrolateral swelling. Legs 1–4 absent. Leg 5 present, small lobe shaped, located in ventrolateral region of urosome, with three spiniform setae. Genital area without ornamentation; openings of seminal receptacles and genital apertures indistinct.
Male and Copepodid
Unknown.
Variation
One female paratype (ICHUM8453) observed in addition to the holotype showed all diagnostic characters seen in the holotype. Ranges (mean with standard deviation in parentheses) of BL, PW, UW, BL/PW, BL/UW, PW/UW for five individuals in the type series were 342–400 (372 ± 23), 326–385 (356 ± 20), 86–96 (91 ± 4), 0.95–1.11 (1.05 ± 0.05), 3.93–4.20 (4.10 ± 0.09), and 3.74–4.32 (3.94 ± 0.22), respectively.
Genetic Divergence and Phylogenetic Analysis
We attempted to determine COI, 16S, and 18S sequences for D. hakuhomaruae sp. nov. but were unable to PCR-amplify the COI region. Among copepod 16S sequences in the INSD database, one from Tripaphylus elongatus (C. B. Wilson, 1932) (as Paeon elongatus, FJ447423 [24]) was most similar to our 16S sequence in a BLAST search, but the query cover and identity score were low (46% and 76.73%, respectively). To date, no other nicothoid 16S sequences have been deposited in public databases [25].
In the ML tree (Fig. 3), D. hakuhomaruae sp. nov. formed a fully supported clade with the Rhizorhina clade. The other relationships were identical to those provided in Kakui and Munakata [4], except for minor differences in ultrafast bootstrap values.
Maximum-likelihood (ML) tree based on 18S sequences (1528 positions). Numbers near nodes are ultrafast bootstrap values. Black circles indicate 100% ultrafast bootstrap support. Clades containing more than two confamilial terminal taxa were collapsed (terminal triangles), except for those in Nicothoidae (shaded) and Asterocheridae. Group names follow Boxshall and Lincoln [10]. The scale bar indicates branch length in substitutions per site
Remarks
Diexanthema hakuhomaruae sp. nov. is the seventh species described in Diexanthema. Females lack rod/horn-like antennules, have mandibles, and lack legs 1–4, features shared with females of D. bathydiaita. The former differs from the latter in having a smooth body surface (body covered with minute hairs in D. bathydiaita) and leg 5 located ventrolaterally on the urosome (laterally in D. bathydiaita). Their host groups are different at the family level: Desmosomatidae for D. hakuhomaruae, Nannoniscidae for D. bathydiaita.
Diexanthema hakuhomaruae sp. nov. lacks rootlets observed in D. apoda. This species appears to use its oral sucker to attach to its host isopod.
Our 18S tree showed a close relationship between Diexanthema and Rhizorhina. This suggests that, although we lacked Choniorhiza sequences, the “Rhizorhina group” proposed by Boxshall and Lincoln [10] may reflect close phylogenetic relationships among its members.
Individuals of Diexanthema hakuhomaruae sp. nov. were, as with Rhizorhina individuals (KK unpublished data), easily deformed by changes in solution; for example, the body of one individual shrank when transferred into a 1:3:6 mixture of glycerin, absolute ethanol, and deionized water, making morphological observation difficult (transferred into 30% ethanol, it recovered its spherical shape). This suggests that differences in body form should be treated with caution in Diexanthema (and Rhizorhina) taxonomy. In addition, most appendages are strongly reduced or completely lacking in these nicothoid genera, and often few morphological differences are observable among congeners. As adopted in other parasitic groups [26], a “turbo taxonomy” [27] approach, i.e., providing concise morphological descriptions along with DNA-sequence and host data in establishing new species, may be advisable in the taxonomy of several character-poor nicothoid genera.
Conclusions
We described Diexanthema hakuhomaruae sp. nov. parasitic on the desmosomatid Eugerdella cf. kurabyssalis Golovan, 2015 collected from a hadal depth in the Kuril-Kamchatka Trench, northwestern Pacific. This species represents the first Diexanthema species from the Pacific, and the first from hadal depths. Our 18S-based tree confirmed a close relationship between Diexanthema and Rhizorhina, previously suggested by morphology. Continued molecular studies, with wider taxon sampling and additional molecular markers, should further elucidate the phylogenetic relationships among nicothoid copepods, with consequent advances in taxonomy.
Data Availability
The raw data (sampling locality; sampling date; museum deposition numbers, INSD accession numbers, and the depository for specimens) are included in the manuscript.
References
Walter TC, Boxshall G (2023) Nicothoidae Dana, 1852–1853. https://www.marinespecies.org/aphia.php?p=taxdetails&id=135530. Accessed 13 Mar 2023
Boxshall GA, Halsey SH (2004) An introduction to copepod diversity. The Ray Society, London
Kakui K (2016) Descriptions of two new species of Rhizorhina Hansen, 1892 (Copepoda: Siphonostomatoida: Nicothoidae) parasitic on tanaidacean crustaceans, with a note on their phylogenetic position. Syst Parasitol 93:57–68. https://doi.org/10.1007/s11230-015-9604-x
Kakui K, Munakata M (2023) A new Sphaeronella species (Copepoda: Siphonostomatoida: Nicothoidae) parasitic on Euphilomedes sp. (Ostracoda: Myodocopa: Philomedidae) from Hokkaido, Japan, with an 18S molecular phylogeny. Syst Parasitol 100:121–131. https://doi.org/10.1007/s11230-022-10075-z
Ohtsuka S, Boxshall GA, Harada S (2005) A new genus and species of nicothoid copepod (Crustacea: Copepoda: Siphonostomatoida) parasitic on the mysid Siriella okadai Ii from off Japan. Syst Parasitol 62:65–81. https://doi.org/10.1007/s11230-005-5483-x
Bamber RN, Boxshall GA (2006) A new genus and species of the Langitanainae (Crustacea: Peracarida: Tanaidacea: Tanaidae) bearing a new genus and species of nicothoid parasite (Crustacea: Copepoda: Siphonostomatoida: Nicothoidae) from the New Caledonia slope. Spec Divers 11:137–148. https://doi.org/10.12782/specdiv.11.137
Boyko CB (2009) Nomenclatural issues with Paranicothoe Carton, 1970 and Pseudonicothoe Avdeev & Avdeev, 1978 (Crustacea: Copepoda: Nicothoidae), with comments on the female isopod type specimen of Paranicothoe cladocera Carton, 1970. Proc Biol Soc Wash 122:206–211. https://doi.org/10.2988/08-49.1
Ritchie L (1975) A new genus and two new species of Choniostomatidae (Copepoda) parasitic on two deep sea isopods. Zool J Linn Soc 57:155–178. https://doi.org/10.1111/j.1096-3642.1975.tb01415.x
Boxshall GA, Harrison K (1988) New nicothoid copepods (Copepoda: Siphonostomatoida) from an amphipod and from deep-sea isopods. Bull Br Mus Nat Hist (Zool) 54:285–299
Boxshall GA, Lincoln RJ (1983) Some new parasitic copepods (Siphonostomatoida: Nicothoidae) from deep-sea asellote isopods. J Nat Hist 17:891–900. https://doi.org/10.1080/00222938300770701
Akiyama T, Shimomura M, Nakamura K (2008) Collection of deep-sea small arthropods: gears for collection and processing of samples on deck. TAXA 24:27–32. https://doi.org/10.19004/taxa.24.0_27. (in Japanese with English abstract)
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Nakayama T, Watanabe S, Mitsui K, Uchida H, Inouye I (1996) The phylogenetic relationship between the Chlamydomonadales and Chlorococcales inferred from 18SrDNA sequence data. Phycol Res 44:47–55. https://doi.org/10.1111/j.1440-1835.1996.tb00037.x
Kakui K, Katoh T, Hiruta SF, Kobayashi N, Kajihara H (2011) Molecular systematics of Tanaidacea (Crustacea: Peracarida) based on 18S sequence data, with an amendment of suborder/superfamily-level classification. Zool Sci 28:749–757. https://doi.org/10.2108/zsj.28.749
Kakui K, Fukuchi J, Shimada D (2021) First report of marine horsehair worms (Nematomorpha: Nectonema) parasitic in isopod crustaceans. Parasitol Res 120:2357–2362. https://doi.org/10.1007/s00436-021-07213-9
Munakata M, Tanaka H, Kakui K (2021) Heterocypris spadix sp. nov. (Crustacea: Ostracoda: Cypridoidea) from Japan, with information on its reproductive mode. Zool Sci 38:287–296. https://doi.org/10.2108/zs200127
Okamoto N, Kakui K (2022) Integrative taxonomy of Zeuxo (Crustacea: Peracarida: Tanaidacea) from Japan, with the description of a new species. Biologia 77:2497–2506. https://doi.org/10.1007/s11756-022-01121-8
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Munakata M, Tanaka H, Kakui K (2022) Taxonomy and natural history of Cavernocypris hokkaiensis sp. nov., the first ostracod reported from alpine streams in Japan. Zoosyst Evol 98:117–127. https://doi.org/10.3897/zse.98.80442
Kakui K, Shimada D (2022) Dive into the sea: first molecular phylogenetic evidence of host expansion from terrestrial/freshwater to marine organisms in Mermithidae (Nematoda: Mermithida). J Helminthol 96:e33. https://doi.org/10.1017/S0022149X22000256
Golovan OA (2015) Description of two ubiquitous species of Desmosomatidae (Isopoda: Asellota) from the Northwest Pacific Basin east of the Kuril-Kamchatka Trench. Zootaxa 4039:201–224. https://doi.org/10.11646/zootaxa.4039.2.1
Jennings RM, Golovan O, Brix S (2020) Integrative species delimitation of desmosomatid and nannoniscid isopods from the Kuril-Kamchatka trench, with description of a hadal species. Prog Oceanogr 182:102236. https://doi.org/10.1016/j.pocean.2019.102236
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Dippenaar SM (2009) Estimated molecular phylogenetic relationships of six siphonostomatoid families (Copepoda) symbiotic on elasmobranchs. Crustaceana 82:1547–1567. https://doi.org/10.1163/001121609X12511103974538
DDBJ (2022) DNA Data Bank Japan. https://www.ddbj.nig.ac.jp/ Accessed 29 Nov 2022
Summers MM, Al-Hakim II, Rouse GW (2014) Turbo-taxonomy: 21 new species of Myzostomida (Annelida). Zootaxa 3873:301–344. https://doi.org/10.11646/zootaxa.3873.4.1
Butcher BA, Smith MA, Sharkey MJ, Quicke DLJ (2012) A turbo-taxonomic study of Thai Aleiodes (Aleiodes) and Aleiodes (Arcaleiodes) (Hymenoptera: Braconidae: Rogadinae) based largely on COI barcoded specimens, with rapid descriptions of 179 new species. Zootaxa 3457:1–232. https://doi.org/10.11646/zootaxa.3457.1.1
Acknowledgements
We thank Shigeaki Kojima and Yasunori Kano for the opportunity to join R/V Hakuho-maru cruise KH-22-8, conducted as a part of the project “Comprehensive study of diversity and evolutionary mechanisms of deep-sea animals in trenches in the northwestern Pacific” supported by KAKENHI grant (JP19H00999) from the Japan Society for the Promotion of Science (JSPS); Captain Kazuhiko Kasuga and the crew of R/V Hakuho-maru, technicians from Marine Work Japan and MOL Marine & Engineering, and researchers aboard for support in collecting; Hiroshi Kajihara, Jamael Abato, and Tsuyoshi Takano for help in sorting samples; and Matthew H. Dick for reviewing the manuscript and editing the English. This study was supported in part by Atmosphere and Ocean Research Institute, The University of Tokyo, and by KAKENHI grants (JP19K06800 and JP22H02681) from JSPS.
Funding
This study was supported in part by the Atmosphere and Ocean Research Institute, The University of Tokyo, and by KAKENHI grants (JP19K06800 and JP22H02681) from the Japan Society for Promotion of Science (JSPS). The funding agencies had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
KK conceived and designed the study, made morphological observations on the copepods, and conducted the molecular analysis; MO made morphological observations on the isopods; KK, JF, and MO collected samples, wrote the manuscript, and read and approved the final draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is registered in ZooBank under https://zoobank.org/4A75D879-2540-46A8-AF47-97C424A48B63.
Supplementary Information
Below is the link to the electronic supplementary material.
11686_2023_676_MOESM1_ESM.fas
Supplementary file1 Aligned 18S sequences used in the maximum-likelihood analysis, with alignment-ambiguous sites retained (FAS 89 KB)
11686_2023_676_MOESM2_ESM.fas
Supplementary file2 Aligned 18S sequences used in the maximum-likelihood analysis, reduced to 1528 positions by removing alignment-ambiguous sites (FAS 78 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kakui, K., Fukuchi, J. & Ohta, M. Diexanthema hakuhomaruae sp. nov. (Copepoda: Siphonostomatoida: Nicothoidae) from the Hadal Zone in the Northwestern Pacific, with an 18S Molecular Phylogeny. Acta Parasit. 68, 413–419 (2023). https://doi.org/10.1007/s11686-023-00676-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-023-00676-z