Abstract
Nectonema, the only horsehair worm (Nematomorpha) genus found in marine environments, was previously known to be parasitic only in decapod crustaceans. We report Nectonema sp. as the first record of a marine nematomorph parasitic in isopod crustaceans. This is also the third record of marine nematomorphs from the North Pacific. Six infected isopods (Natatolana japonensis) collected from 1425 m of depth in the Sea of Japan each contained one to seven (mean 2.33) nematomorphs in the body cavity in the pereon. There was no correlation between the host body length and number of parasites. For Nectonema sp., we describe and illustrate morphological features of the parasitic juvenile stage and present nucleotide sequences for the cytochrome c oxidase subunit I gene (COI or cox1; 451 nt), 18S rRNA gene (1777 nt), and region spanning the internal transcribed spacer 1 (ITS1) and the 28S rRNA gene including the 5.8S rRNA gene and ITS2 (1218 nt in total). In an 18S maximum-likelihood tree that included 24 nematomorph species, Nectonema sp. grouped with N. agile from the northwestern Atlantic; the 18S gene from these two taxa was divergent by 11.8% K2P distance, suggesting that they are different species. Nectonema species may have a broader range of host groups than previously suspected, but may have been previously misidentified as nematode parasites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nectonema is the only nematomorph genus found in marine environments. There are five species described in this genus to date: Nectonema agile Verrill, 1879 (type species) from the Atlantic Ocean and Mediterranean and Black Seas; Nectonema melanocephalum Nierstrasz, 1907 from Indonesia; Nectonema munidae Brinkmann, 1930 from Bergen and Norway; Nectonema svensksundi Bock, 1913 from Spitzbergen, Norway; and Nectonema zealandica Poinar and Brockerhoff, 2001 from New Zealand (Schmidt-Rhaesa et al. 2013). The occurrence records of this group in the North Pacific have been limited, with only two reports from Japan: Oku et al. (1983) and Yoshida (2016) reported unidentified Nectonema individuals from the brachyuran Erimacrus isenbeckii (Brandt, 1848) and the anomuran Pagurus brachiomastus (Thallwitz, 1891), respectively.
The life cycle of nematomorphs contains a larval stage, a parasitic juvenile stage, and a free-living adult stage (Hanelt and Janovy 2004). In Nectonema, juveniles have so far been reported only from crustaceans in the order Decapoda (shrimps, hermit crabs, crabs, etc.), among which more than 27 species have been reported as hosts (Schmidt-Rhaesa et al. 2013). In a single host species, Nectonema individuals at various developmental stages can be found (Huus 1932; Schmidt-Rhaesa 1996).
Here, we report the first record of Nectonema (as Nectonema sp.) parasitic in a species in the order Isopoda, a morphologically diverse crustacean group with more than 10,000 described species. This is also the third record of marine nematomorphs from the North Pacific. We describe the morphology of this nematomorph and present nucleotide sequences for its cytochrome c oxidase subunit I (COI) gene, 18S rRNA (18S) gene, and ITS cluster, including the 3´ region of internal transcribed spacer 1 (ITS1), the 5.8S rRNA gene, ITS2, and the 5´ region of the 28S rRNA gene.
Materials and methods
Isopods were collected from plastic jars that contained cut sardine bait and had been placed in baited traps (Saito et al. 2014: Fig. 1J) during a cruise of R/V Soyo-maru (National Research Institute of Fisheries Science, Japan); the traps were recovered on 16 July 2014 at station Kago-4 (40°00.59′ N 135°57.63′ E), 1425 m depth, Sea of Japan. The isopods were picked from the jars by Ken Fujimoto and kept alive at 4 °C until 30 July. Infected individuals were anesthetized with 35‰ MgCl2 and dissected to extract nematomorphs. One infected isopod and one extracted nematomorph were photographed live. Isopods were fixed and preserved in ethanol; nematomorphs removed from them were fixed in DESS solution (Yoder et al. 2006) or 99% ethanol.
Body length (BL) was measured from the anterior edge of the cephalothorax to the tip of the pleotelson for the isopods, and from the anterior to posterior tip of the body for the nematomorphs; the cephalothorax width (CW) of the isopods was measured at the widest portion of the cephalothorax.
Isopods were dissected with needles under a Nikon SMZ1500 stereomicroscope; detached appendages were mounted on glass slides in glycerin and observed with a Nikon E600 microscope. Two nematomorphs in DESS solution were transferred into a 1:3:6 mixture of glycerin, absolute ethanol, and deionized water and placed in a thermostatic chamber at 40 °C for two days, after which they were mounted on glass slides in glycerin and observed with an Olympus BX51 microscope. Illustrations of nematomorphs were prepared with Inkscape 1.0 from digital micrograph images. Morphological terminology for Nectonema here follows Poinar and Brockerhoff (2001).
Total DNA was extracted from part of the body from each of four nematomorphs by using a NucleoSpin Tissue XS Kit (TaKaRa Bio, Japan). Primers used for PCR and sequencing are listed in Table 1. PCR amplification conditions for the ITS cluster and COI with TaKaRa Ex Taq DNA polymerase (TaKaRa Bio) were 94 °C for 1 min; 35 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 1 min; and 72 °C for 2 min. Conditions for 18S amplification with KOD FX Neo (Toyobo, Japan) were 94 °C for 1 min; 45 cycles of 98 °C for 10 s, 65 °C for 30 s, and 68 °C for 75 s; and 68 °C for 3 min. PCR products for 18S were separated on a 2% agarose gel, excised with a micro spatula, and purified with a MagExtractor PCR & Gel Clean Up Kit (Toyobo). All nucleotide sequences were determined by direct sequencing with a BigDye Terminator Kit ver. 3.1 and a 3730 DNA Analyzer (Life Technologies, USA). Fragments were concatenated by using MEGA7 (Kumar et al. 2016).
The 18S dataset for a phylogenetic analysis included our 18S sequence from Nectonema sp. and 25 sequences from 23 nematomorph species and two outgroup taxa (a nematode and a tardigrade) taken from public databases (DDBJ 2021; Online Resource 1). Methods for alignment of all 18S sequences (1806 positions in the aligned dataset; Online Resource 2) and selection of the optimal substitution model (GTR + I + G) were as described by Homma et al. (2020). The Kimura (1980) 2-parameter (K2P) distance between the aligned Nectonema sequences was calculated with MEGA7. A maximum likelihood (ML) analysis was conducted in RAxML-NG (Kozlov et al. 2019), with nodal support values obtained by analysis of 1000 bootstrap pseudoreplicates. The ML tree was drawn by using FigTree v1.4.4 (Rambaut 2021).
Our nematomorph specimens were deposited in the Invertebrate Collection of the Hokkaido University Museum (ICHUM), Sapporo (ICHUM-6178–6191); host isopods were deposited in the Seto Marine Biological Laboratory (SMBL-V0598–0603). The sequences we determined were deposited in the International Nucleotide Sequence Database through the DNA Data Bank of Japan, under the accession numbers LC605980–605988.
Results and discussion
The six infected isopods we observed (three males, BL 14.7–15.8 mm; three females, BL 13.5–16.9 mm) were identified as Natatolana japonensis (Richardson, 1904) in the suborder Cymothoida (Fig. 1). All infected individuals harbored nematomorphs internally in the cavity of the pereon. The nematomorph infections ranged from one to seven individuals per host (mean intensity = 2.33) (Table 2). The prevalence of infection is unknown because we lack data on the total number of isopods in the baited traps. No clear correlation was detected between host size (BL) and the number of parasites (N) (N = 1.51 × BL – 20.6; R2 = 0.543).
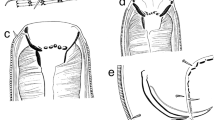
The nematomorphs ranged in BL from 23.7 mm to 94.0 mm (mean, 58.3 mm; N = 12 intact individuals; Online Resource 3). An abbreviated description of individual ICHUM-6181 (Fig. 2) is as follows. Anterior and posterior ends rounded. Cuticle colorless. Epidermis with single cell layer. Cuticular natatory bristles not observed. Cephalic papillae absent. Septum distinct. Anterior chamber translucent, with ca. 10 giant cells. Anterior region not pigmented. Mouth opening at anterior apex. Oral cavity with eight (?) bifurcated hooks protruding from cuticular wall. Sclerotized proboscis with two anterior scissor-shaped spines. Pharynx sclerotized, connecting with intestine posterior to septum. Body filled with mesenchyme between septum and subposterior region. Gonads not observed. Subposterior region with posterior opening (gonopore?) surrounded by elongate epidermal cells.
Nectonema sp. (ICHUM-6181). Anterior (a, b, e, f) and posterior (c, d) portions and sclerotized proboscis with two anterior scissor-shaped spines (b’) in glycerin, as microphotographs (a, c, e, f) and line drawings (b, b’, d). Arrowheads in e indicate giant cells. Abbreviations: ac, anterior chamber; bh, bifurcated hook; cu, cuticle; du, duct (gonoduct?); ep, epidermis; gc, giant cell; in, intestine; me, mesenchyme; mo, mouth opening; mu, muscle; ph, pharynx; po, posterior opening (gonopore?); pr, proboscis; se, septum; sp, scissor-shaped spine
COI and ITS-cluster sequences were determined from four nematomorphs (ICHUM-6182–6184, 6188). The four COI sequences (accession numbers LC605980–605983; 451 nt long, translating to 150 amino acids) were identical, as were the four ITS-cluster sequences (accession numbers LC605984–605987; 1218 nt long). No Nectonema COI and ITS-cluster sequences have previously been deposited in public databases (DDBJ 2021). The 18S sequences (accession numbers LC605988, 605989) determined from two nematomorphs (ICHUM-6184, 6188; 1777 nt long) were identical, and were 11.8% divergent in K2P distance from the only Nectonema 18S sequence available in public databases (N. agile; Bleidorn et al. 2002). In the ML tree for 18S (Fig. 3), Nectonema sp. and N. agile formed a clade with 100% bootstrap support, which was the sister group to a Gordiida clade having 87% bootstrap support; the relationships within Gordiida differed from those in previous studies (e.g., Bleidorn et al. 2002; Tobias et al. 2017), which may have resulted from the difference in the dataset used.
Morphology-based species identification is difficult in Nectonema, and Schmidt-Rhaesa (2005) noted that the five known species may have been described based on specimens at different developmental stages, making the descriptions not directly comparable. It should be noted that our Nectonema samples differ from juveniles of three species (N. agile, N. munidae, and N. zealandica; no juveniles have been reported in the other two species) in having ca. 10 giant cells and lacking cephalic papillae. Molecular identification is currently unavailable for this group because no molecular markers have been determined for all named Nectonema species. Identification of our material to species was not possible, as no adult specimens were obtained.
On the basis of the 11.8% K2P divergence, Nectonema sp. is likely not conspecific with N. agile. Furthermore, its occurrence in an isopod indicates that Nectonema may use other, undetected groups besides Decopoda for hosts. In the parasitic stage, nematomorphs resemble nematodes, and infections of unusual hosts in the past might have been misidentified as nematode infections. To understand true diversity of marine nematomorphs, host surveys targeting non-decapods as well as decapods and integrative taxonomic approaches will be necessary.
References
Bleidorn C, Schmidt-Rhaesa A, Garey JR (2002) Systematic relationships of Nematomorpha based on molecular and morphological data. Invertebr Biol 121:357–364
Bock S (1913) Zur Kenntnis von Nectonema und dessen systematischer Stellung. Zool Bidrag Uppsala 2:1–28
Brandt JF (1848) Vorläufige Bemerkungen über eine neue, eigenthümliche, der Fauna Russlands angehörige Gattung oder Untergattung von Krabben (Crustacea Brachyura) aus der Edwards’schen Abtheilung der Corysten. Bull Cl Phys Math Acad Imp Sci Saint-Pétersbourg 7:177–180
Brinkmann A (1930) Über Nectonema munidae n. sp. Bergen Mus Årbok 9:1–15
DDBJ (2021) DNA Data Bank of Japan. https://www.ddbj.nig.ac.jp/index.html. Accessed 14 Feb 2021
Hanelt B, Janovy J Jr (2004) Untying a Gordian knot: the domestication and laboratory maintenance of a Gordian worm, Paragordius varius (Nematomorpha: Gordiida). J Nat Hist 38:939–950
Homma R, Uyeno D, Kakui K (2020) Integrative taxonomy of Pseudolepeophtheirus longicauda (Crustacea: Copepoda: Caligidae) parasitic on Platichthys stellatus (Actinopterygii: Pleuronectidae). Parasitol Int 78:102135
Huus J (1932) Über die Begattung bei Nectonema munidae Br. und über den Fund der Larve von dieser Art. Zool Anz 97:33–37
Kakui K, Katoh T, Hiruta SF, Kobayashi N, Kajihara H (2011) Molecular systematics of Tanaidacea (Crustacea: Peracarida) based on 18S sequence data, with an amendment of suborder/superfamily-level classification. Zool Sci 28:749–757
Kakui K, Shimada D (2017) A new species of Tanaopsis (Crustacea: Tanaidacea) from Japan, with remarks on the functions of serial ridges and grooves on the appendages. Zootaxa 4282:324–336
Kanzaki N, Futai K (2002) A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within the xylophilus group. Nematology 4:35–41
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Nakayama T, Watanabe S, Mitsui K, Uchida H, Inouye I (1996) The phylogenetic relationship between the Chlamydomonadales and Chlorococcales inferred from 18SrDNA sequence data. Phycological Res 44:47–55
Nierstrasz HF (1907) Die Nematomorpha Der Siboga-Expedition. Siboga Exped 20:1–21
Oku Y, Fukumoto S, Ohbayashi M, Koike M (1983) A marine horsehair worm, Nectonema sp., parasitizing atelecyclid crab, Erimacrus isenbeckii, from Hokkaido, Japan. Jpn J Vet Res 31:65–69
Poinar G Jr, Brockerhoff AM (2001) Nectonema zealandica n. sp. (Nematomorpha: Nectonematoidea) parasitising the purple rock crab Hemigrapsus edwardsi (Brachyura: Decapoda) in New Zealand, with notes on the prevalence of infection and host defence reactions. Syst Parasitol 50:149–157
Rambaut A (2021) FigTree v1.4.4. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 14 Feb 2021
Richardson H (1904) Contributions to the natural history of the Isopoda. Proc US Natl Mus 27:1–89
Saito H, Hasegawa K, Kogure Y, Yosho I, Ueda Y, Fujita T (2014) An outline of “Research on Deep-sea Fauna of the Sea of Japan, 2009–2013.” Natl Mus Nat Sci Monogr 44:1–22
Schmidt-Rhaesa A (1996) Ultrastructure of the anterior end in three ontogenetic stages of Nectonema munidae (Nematomorpha). Acta Zool 77:267–278
Schmidt-Rhaesa A (2005) Nematomorpha (horse-hair worms). In: Rohde K (ed) Marine parasitology. CSIRO Publishing, Collingwood, pp 213–216
Schmidt-Rhaesa A, Pohle G, Gaudette J, Burdett-Coutts V (2013) Lobster (Homarus americanus), a new host for marine horsehair worms (Nectonema agile, Nematomorpha). J Mar Biol Assoc UK 93:631–633
Thallwitz J (1891) Decapoden-Studien, insbesondere basirt auf A. B. Meyer’s Sammlungen im Ostindischen Archipel, nebst einer Aufzählung der Decapoden und Stomatopoden des Dresdener Museums. Abh Ber K Zool Anthr Ethn Mus Dresden 3:1–56
Tobias ZJC, Yadav AK, Schmidt-Rhaesa A, Poulin R (2017) Intra- and interspecific genetic diversity of New Zealand hairworms (Nematomorpha). Parasitology 144:1026–1040
Verrill AE (1879) Notice of recent additions to the marine Invertebrata, of the northeastern coast of America, with descriptions of new genera and species and critical remarks on others. Proc US Natl Mus 3:356–405
Yoder M, Tandingan de Ley I, King IW, Mundo-Ocampo M, Mann J, Blaxter M, Poiras L, de Ley P (2006) DESS: a versatile solution for preserving morphology and extractable DNA of nematodes. Nematology 8:367–376
Yoshida R (2016) Akkeshi wan nai ni okeru oogata koukakurui kisei seibutsu sou no haaku to bunruigakuteki kenkyu [Taxonomic study and faunal survey of parasites in large crustaceans in Akkeshi Bay]. http://akkeshi-bekanbeushi.com/josei/report/report_h27/03yoshida.pdf. Accessed 18 May 2021 [in Japanese]
Zhu X, Gasser RB, Podolska M, Chilton NB (1998) Characterisation of anisakid nematodes with zoonotic potential by nuclear ribosomal DNA sequences. Int J Parasitol 28:1911–1921
Acknowledgements
We thank Ken Fujimoto, Shiro Sawadaishi, and the crew of R/V Soyo-maru for collecting specimens and providing facilities; Matthew H. Dick for reviewing the manuscript and editing our English; and two anonymous reviewers for improving this manuscript with their comments.
Funding
This research did not receive any specific grant support from public, commercial, or non-profit funding agencies.
Author information
Authors and Affiliations
Contributions
KK conceived and designed the study, collected samples, and conducted the molecular analysis. JF identified the host isopods. KK and DS made morphological observations of the nematomorphs. DS made drawings. KK wrote the first draft of the manuscript, and all authors commented on the first draft and read and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: David Bruce Conn
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kakui, K., Fukuchi, J. & Shimada, D. First report of marine horsehair worms (Nematomorpha: Nectonema) parasitic in isopod crustaceans. Parasitol Res 120, 2357–2362 (2021). https://doi.org/10.1007/s00436-021-07213-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-021-07213-9