Abstract
Recently, multi-task based feature selection methods have been used in multi-modality based classification of Alzheimer’s disease (AD) and its prodromal stage, i.e., mild cognitive impairment (MCI). However, in traditional multi-task feature selection methods, some useful discriminative information among subjects is usually not well mined for further improving the subsequent classification performance. Accordingly, in this paper, we propose a discriminative multi-task feature selection method to select the most discriminative features for multi-modality based classification of AD/MCI. Specifically, for each modality, we train a linear regression model using the corresponding modality of data, and further enforce the group-sparsity regularization on weights of those regression models for joint selection of common features across multiple modalities. Furthermore, we propose a discriminative regularization term based on the intra-class and inter-class Laplacian matrices to better use the discriminative information among subjects. To evaluate our proposed method, we perform extensive experiments on 202 subjects, including 51 AD patients, 99 MCI patients, and 52 healthy controls (HC), from the baseline MRI and FDG-PET image data of the Alzheimer’s Disease Neuroimaging Initiative (ADNI). The experimental results show that our proposed method not only improves the classification performance, but also has potential to discover the disease-related biomarkers useful for diagnosis of disease, along with the comparison to several state-of-the-art methods for multi-modality based AD/MCI classification.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
As the common form of dementia worldwide, Alzheimer’s disease (AD) is a primary neurodegenerative brain disease occurring in elderly people. It was first described by a German psychiatrist and neuropathologist Alois Alzheimer in 1906 and was named after him (Berchtold and Cotman 1998). It was reported that there were 26.6 million AD patients in the world in 2006 (Berchtold and Cotman 1998). Also, it is predicted that 1 in 85 people will be affected by AD by 2050 (Brookmeyer et al. 2007). There is a prodromal state between normal aging and AD, called mild cognitive impairment (MCI). Most individuals with MCI will eventually progress to dementia within 5 years (Gauthier et al. 2006). There is no cure for AD and no treatment to reverse or halt its progression. Therefore, accurate diagnosis of AD and MCI is very important to delay the disease progression. However, since the change of AD-related brain is prior to the symptom of AD, it is critical to detect those changes for early diagnosis of AD. Recently, neuroimaging technology is increasingly used to identify such abnormal changes in the early stage of AD (Cheng et al. 2012; Petersen et al. 1999; Sui et al. 2012; Ye et al. 2011; Zhang et al. 2011).
Early studies on AD/MCI classification mainly focus on using a single modality of biomarker, such as magnetic resonance imaging (MRI) (De Leon et al. 2007; Fan et al. 2008; McEvoy et al. 2009), fluorodeoxyglucose positron emission tomography (FDG-PET) (Higdon et al. 2004; Morris et al. 2001; De Santi et al. 2001), cerebrospinal fluid (CSF) (Mattsson et al. 2009; Shaw et al. 2009), etc. However, in those studies, some useful complementary information across different modalities of biomarkers is ignored, which is helpful for further improving the accuracy of classification. Recently, several researchers have explored to combine multiple modalities of biomarkers (Apostolova et al. 2010; Fjell et al. 2010; Landau et al. 2010; Walhovd et al. 2010; Jie et al. 2013). For instance, Hinrichs et al. (Hinrichs et al. 2009) combined two modalities, i.e., MRI and PET, for classification of AD. Bouwman et al. (Bouwman et al. 2007) proposed to combine two modalities of MRI and CSF to identify MCI patients from healthy controls (HC). Fellgiebel et al. (Fellgiebel et al. 2007) used PET and CSF to predict cognitive deterioration in MCI. Zhang et al. (Zhang et al. 2011) combined three modalities, i.e., MRI, FDG-PET and CSF, to classify AD/MCI from HC. Gray et al. (Gray et al. 2013) used four modalities, i.e., MRI, FDG-PET, CSF and genetic information, for AD classification. These existing studies have suggested that different modalities of biomarkers can provide the inherently complementary information that can improve accuracy in disease diagnosis when used together (Apostolova et al. 2010; Fjell et al. 2010; Landau et al. 2010; Walhovd et al. 2010; Foster et al. 2007).
In multi-modality based classification methods, traditional feature selection approaches, such as the least absolute shrinkage and selection operator (Lasso) and t-test, are often performed to help select the disease-related brain features for training a good learning model (Tibshirani 1996; Wee et al. 2012). However, one main disadvantage of those feature selection methods is that they usually ignore the inherent relatedness among features from different modalities. Recently, multi-modality based feature selection methods have been proposed to overcome this problem. For example, Huang et al. (Huang et al. 2011) presented a sparse composite linear discrimination analysis to recognized AD-related ROIs from multi-modality data. Liu et al. (Liu et al. 2014) proposed a multi-task based feature selection with each task corresponding to a learning model using individual modality of data and embedding inter-modality information into multi-task learning model for AD classification. Gray et al. (Gray et al. 2013) constructed a multi-modality classification framework based on pairwise similarity measures which come from random forest classifiers for the classification between AD/MCI and HC. However, in those methods, some useful discriminative information, such as the distribution information of intra-class and inter-class subjects, is not well mined, which may affect the final classification performance.
To address that problem, in this paper, we propose a new discriminative multi-task feature selection (DMTFS) model, which considers both the inherent relations among multi-modality data and the distribution information of intra-class subjects (i.e., subjects from the same class) and inter-class subjects (i.e., subjects from different classes) from each modality. Specifically, we first formulate feature selection on multi-modality data as multi-task learning problem with each task corresponding to a learning problem on individual modality. Then, two regularized terms are included into the proposed DMTFS model. Specifically, the first term is the group-sparsity regularizer (Ng and Abugharbieh 2011; Yuan and Lin 2006), which ensures only a small number of common brain region-specific features to be jointly selected from multi-modality data. Furthermore, we introduce a new Laplacian regularization term into the proposed objective function, which preserves the compactness of intra-class subjects and the separability of inter-class subjects, and hence induces the more discriminative features. Finally, we adopt the multi-kernel support vector machine (SVM) technique to fuse multi-modality data for performing classification of AD/MCI. To evaluate the proposed method, a series of experiments are performed on the baseline MRI and FDG-PET image data of 202 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), which includes 51 AD patients, 99 MCI patients, and 52 HC. The experimental results show the superiority of our proposed method, in comparison with the existing multi-modality based methods.
Methods
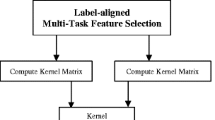
Figure 1 shows the overview of our proposed framework, which contains three major steps, i.e., image pre-processing and feature extraction, discriminative multi-task feature selection, and multi-kernel SVM classification. In this section, before giving the detailed descriptions of these steps, we will first introduce the subjects used in this study.
Subjects
The dataset we used in this study is obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.adni-info.org). ADNI is a non-profit organization which was founded in 2003 by the National Institute of Biomedical Imaging and Bioengineering. Many researchers of institutions work together to achieve this organization. The ADNI is committed to evaluate the progression of early Alzheimer’s disease, i.e., MCI, by combining some technology such as magnetic resonance imaging (MRI), fluorodeoxyglucose positron emission tomography (FDG-PET), Cerebrospinal fluid (CSF) and other clinical diagnosis, which greatly improve the efficiency of diagnosis, and save the time of treatment and the cost to the patients.
Following our previous works (Zhang et al. 2011, 2012), we evaluate the proposed method on the baseline MRI and FDG-PET data of 202 ADNI subjects, which contain 51 AD patients, 99 MCI patients (including 43 MCI converters (MCI-C) and 56 MCI non-converters (MCI-NC)), as well as 52 healthy controls (HC). Specifically, we build multiple binary classifiers to confirm the classification performance of our proposed method, including AD vs. HC, MCI vs. HC, and MCI-C vs. MCI-NC.
Image Pre-processing and feature extraction
The same image pre-processing as in (Zhang et al. 2011, 2012) is performed for all MRI and PET images, including anterior commissure (AC) - posterior commissure (PC) correction, skull-stripping, removal of cerebellum, and segmentation of structural MR images into three different tissues: grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF). With atlas warping, we can partition each subject image into 93 regions of interests (ROIs). For each of the 93 ROIs, we compute the GM tissue volume from the subject’s MRI image. For PET image, we first rigidly align it with its respective MRI image of the same subject, and then compute the average value of PET signals in each ROI. Therefore, for each subject, we can finally obtain totally 93 features from MRI image and another 93 features from PET image.
Discriminative Multi-Task Feature Selection (DMTFS)
Before deriving our proposed discriminative multi-task feature selection (DMTFS) method, we first briefly introduce the traditional multi-task feature selection (MTFS) model (Zhang et al. 2012). Suppose X m = [x m1 , …, x m i , …, x m N ]T ∈ R N × d as training subjects from the m -th modality (i.e., task), and Y = [y 1, …, y i , …, y N ]T ∈ R N represents the corresponding response vector from all training subjects, where d and N are the numbers of features and training subjects, respectively. Here, x m i is a feature vector of the i -th subject from the m -th modality, and y i ∈ {+1, − 1} is the response class label (i.e., patient or healthy control). In addition, w m ∈ R d represents the weight vector of linear function for the m -th task, and W = [w 1, …, w m, …, w M] ∈ R d × M denotes the weight matrix including all w m. Then, the MTFS model is to optimize the following objective function:
where M is the number of modalities, ‖W‖2,1 = ∑ d j = 1 ‖w j ‖2 s the l 2,1 -norm of weight matrix which calculates the sum of l 2 -norm of w j (Yuan and Lin 2006), and w j is the j-th row of W which represents the weight vector of the j -th feature across M tasks. Here, the l 2,1 -norm is adopted to enforce the group sparsity on the weight matrix, i.e., encouraging a number of rows in the weight matrix being zero. The first term in Eq. (1) is the empirical loss function, which measures the error between predicted value obtained from learning model and the true value. λ is a regularization parameter which balances the relative importance of both terms. The larger λ value means the more zero rows appear in the weight matrix, i.e., few of features are preserved.
In MTFS model, a linear function (i.e., f(x) = w T x) was used to map the data from the original high-dimensional feature space to one-dimensional space. This model only focuses on the relationship between label and subject, and thus ignores the distribution information of subjects from each modality, such as the compactness of intra-class subjects and the separability of inter-class subjects. This kind of information may help induce the more discriminative features and thus further improve the classification performance. Figure 2 illustrates an example. Here each color denotes a class, and the points with the same color denote that they come from the same class. The arrows with green color denote that green points (which are intra-class nearest neighbors) should be closer to the central green point in the new feature space. Also, the arrows with purple color denote that purple points (which are inter-class nearest neighbors) should be far away from the central green point in the new feature space. Intuitively, Fig. 2 shows that intra-class samples should be closer while inter-class samples should be far away in the new feature space.
To address this problem, inspired by some recent works (Cai et al. 2007; Xue et al. 2009), we propose a new discriminative regularization term to preserve the distribution information of subjects. To be specific, in each modality, for each subject x m i , we first seek its k nearest neighbors, i.e., n(x m i ) = {x m,1 i , x m,2 i , …, x m,k i }, and define two disjoint subject subsets as follows:
where n w (x m i ) includes the neighbors that have the same label with the subject x m i , and n b (x m i ) contains the neighbors having different labels with the subject x m i . Then, to discover discriminative structure and geometrical information of the data, we construct two graphs, i.e., intra-class graph G m w and inter-class graph G m b , with each subject as a node for both graphs. Let Z m w and Z m b denote the weight matrices of G m w and G m b , respectively. We define:
Then, to preserve the discriminative and structural information of two graphs during linear mapping, we introduce a new discriminative regularization term as:
Where
and
Here, L m w = D m w − Z m w and L m b = D m b − Z m b represent intra-class and inter-class Laplacian matrices for the m-th modality, respectively. D m w,ii = ∑ N j = 1 Z m w,ij and D m b,ii = ∑ N j = 1 Z m b,ij are the corresponding diagonal matrices. σ is a positive constant which controls the relative importance of both terms.
With the regularizer in Eq. (6), our proposed discriminative multi-task feature selection model (DMTFS) has the following objective function:
where λ and σ are positive constants whose values can be determined via inner cross-validation on the training data. Below, we give an algorithm to solve the optimization problem in Eq. (9).
Optimization algorithm
In our study, we use the Accelerated Proximal Gradient (APG) technique (Chen et al. 2009; Liu 2999) to solve the optimization problem in Eq. (9). Specifically, we first separate the objective function in Eq. (9) into a non-smooth part as:
and a smooth one as:
Then, the function h(W) + g(W) can be approximately expressed by the following function:
where ‖ ⋅ ‖F denotes the Frobenius norm, h(W k ) represents the gradient of h(W) at the point W k in the k -th iteration process, and 〈W − W k , ∇h(W k )〉 denotes the inter product of matrixes which equals to Tr((W − W k )T∇h(W k )). Here, n represents the iteration step size, whose value can be decided by using line search.
Finally, the iterative process of APG algorithm can be interpreted as follows:
Where \( {\boldsymbol{P}}_{\boldsymbol{k}}={\boldsymbol{W}}_{\boldsymbol{k}}-\frac{1}{n}\nabla h\left({\boldsymbol{W}}_{\boldsymbol{k}}\right) \). According to (Chen et al. 2009; Liu 2999), we can decompose the problem in Eq. (13) into d separate sub-problems, for which the analytical solutions can be easily obtained. Also, from (Chen et al. 2009; Liu 2999), instead of performing gradient descent based on W k , we can compute the following formulation as:
where \( {\alpha}_k=\frac{\left(1-{\tau}_{k-1}\right){\tau}_k}{\tau_{k-1}} \) and \( {\tau}_k=\frac{2}{k+3} \).
Multi-kernel SVM classification
After selecting the discriminative and common features (i.e., brain regions) across multiple modalities, we then use the multi-kernel SVM method proposed in (Zhang et al. 2011) for final classification of AD/MCI from healthy controls. Specifically, based on the features obtained from the proposed method, we compute a linear kernel across different subjects for each modality and then use the following function to integrate the multiple kernels:
where K m(x m i , x m j ) represents the kernel function over the m -th modality between the sample x i and x j , and α m ≥ 0 is a weight parameter with constraint of ∑ m α m = 1. Here, we find the optimal values of α m by using a coarse-grid search on the training subjects with range from 0 to 1 and the interval value of 0.1. Finally, the LIBSVM toolbox (Chang and Lin 2011) is adopted to perform SVM with the mixed kernel defined in Eq. (15).
Results
Classification performance
In this paper, we adopt 10-fold cross-validation to evaluate the classification performance. Specifically, we divide the whole samples into 10 parts, leaving one part for testing and the remaining parts as training data in each cross-validation. This process is repeated for 10 independent times to avoid the bias in random partition of samples. Four performance measures, including classification accuracy (ACC) measuring the proportion of subjects correctly classified among the whole subjects, sensitivity (SEN) measuring the proportion of AD or MCI patients correctly classified, specificity (SPE) measuring the proportion of healthy controls correctly classified, and the area under receiver operating characteristic (ROC) curve (AUC), are used to evaluate the classification performance of different classification methods.
We compare our proposed DMTFS method with several other methods, including multi-task feature selection method (denoted as MTFS) (Zhang et al. 2012), and multi-modal classification method proposed in (Zhang et al. 2011) using the least absolution shrinkage and selection operator (Lasso) as feature selection (denoted as MML). For further comparison, we also concatenate the MRI and PET features into a long feature vector, followed by the sequential forward floating selection (SFFS) (Pudil et al. 1994) for feature selection, and then using the standard SVM for classification. Table 1 lists the comparison of different methods for AD/MCI classifications. Figure 3 plots the ROC curves of different methods.
From Table 1 and Fig. 3, we can see that our proposed method outperforms the other methods in all performance measures for both AD and MCI classifications. Specifically, our method achieves the classification accuracies of 95.92 % and 82.13 % for AD vs. HC and MCI vs. HC, respectively, while the best accuracies of other methods are only 92.07 % and 74.17 %, respectively. In addition, our method achieves high AUC values of 0.97 and 0.82 for AD vs. HC and MCI vs. HC, respectively, showing better diagnostic power than the other methods for AD/MCI classifications.
On the other hand, we also perform experiments on classifying MCI converters (MCI-C) from MCI non-converters (MCI-NC), with the corresponding results shown in Table 2 and Fig. 4. As can be seen from Table 2 and Fig. 4, our proposed method achieves better classification performances than other methods for MCI-C vs. MCI-NC classification. Specifically, our proposed method achieves a classification accuracy of 71.12 % for MCI-C vs. MCI-NC classification, which is nearly 10 % higher than the best result by other methods.
In addition, we perform significance test on the classification performances between our proposed method and other compared methods by using the standard paired \( t \)test under the significance level of 95 %. Table 3 shows the results of \( t \)test between our method and any other method. As we can see from Table 3, for all the three classification tasks, i.e., AD vs. HC, MCI vs. HC, and MCI-C vs. MCI-NC, our proposed method is significantly better than other compared methods, which again shows the advantages of our proposed method.
The most discriminative brain regions
Because of the importance of the brain region-related disease in early diagnosis, we besides reporting classification performances, and also investigate the top selected features (i.e., brain regions) by our proposed DMTFS method. To be specific, since the selected features in each cross-validation are not the same, we select those features as the most discriminative features which have the highest occurrence frequency in every cross-validation folds.
Figure 5 plots the top 15 selected brain regions for MCI vs. HC classification. As can be seen from Fig. 5, our method can effectively identify those disease-related brain regions such as hippocampal, amygdala, precuneus, and temporal pole, which have been reported to be relevant with AD in the previous studies (Dai et al. 2009; Del Sole et al. 2008; Misra et al. 2009; Solodkin et al. 2013; Van Hoesen and Hyman 1990; Wang et al. 2012). For example, hippocampus are located in the temporal lobe of the brain, which are the role of the memory and spatial navigation. The Hippocampi are the first damaged regions in AD, showing loss of memory and spatial orientation. Hyman BT et al. (Hyman et al. 1984) also mentioned that the focal pattern of pathology isolates the hippocampal may induce to damage of memory in AD. Amygdala is the subcortical central of the limbic system, and has the function of regulating visceral sensation and producing emotions. Many researchers have found that the important role of amygdala in AD patients (Knafo et al. 2009; Poulin et al. 2011). Such as Knafo et al. (Knafo et al. 2009) mentioned that individuals who have AD with a significant shrinkage of amygdala, and extensive gliosis. In addition, precuneus (Del Sole et al. 2008; Karas et al. 2007) and temporal pole (Nobili et al. 2008) also show significant abnormalities in AD.
Discussion
In this paper, we propose a new discriminative multi-task feature selection method for AD vs. HC, MCI vs. HC, and MCI-C vs. MCI-NC classifications. Experimental results demonstrate that our proposed method achieves better classification performances and also identifies more discriminative features, compared with the existing multi-modality based methods. Specifically, our proposed method achieves an accuracy of 95.92 % for classification between AD and HC, a high accuracy of 82.13 % for the classification between MCI and HC, and a high accuracy of 71.12 % for classification between MCI-C vs. MCI-NC.
Multi-modality based classification
Since different modalities may provide complementary information for diagnosis of AD (Apostolova et al. 2010; Landau et al. 2010), a lot of recent studies have investigated combining multi-modalities of data for AD diagnosis, showing improved classification performances (Walhovd et al. 2010; Bouwman et al. 2007; Wee et al. 2012; Ye et al. 2008; Davatzikos et al. 2011). For more comparisons, Table 4 lists the comparison between our proposed method and several other state-of-the-art methods for multi-modality based AD/MCI classification. For example, Huang et al. (Huang et al. 2011) proposed the sparse composite linear discriminant analysis (SCLDA) model performed on MRI and PET modalities of data, achieving the accuracy of 94.30 % for AD classification. Gray et al. (Gray et al. 2013) used four modalities (including MRI, PET, CSF and genetic) of data and achieved the accuracies of 89.00 %, 74.60 % and 58.00 % for classifying AD, MCI and MCI-C, respectively. Liu et al. (Liu et al. 2014) used two modalities including MRI and PET and achieved the accuracies of 94.40 %, 78.80 % and 67.80 % for classifying AD, MCI and MCI-C, respectively. As we can see from Table 4, our proposed method consistently outperforms the other state-of-the-art methods for multi-modality based classifications of AD, MCI, and MCI-C.
Effect of parameters
In our proposed model, there are two regularization terms including the group-sparsity regularizer and the discriminative regularizer. Accordingly, two regularization parameters (i.e., λ and σ) are used to balance the contributions of different terms. More specifically, λ is used to control the group sparsity of the model, and σ is used to balance the relative importance between the intra-class Laplacian matrix and the inter-class Laplacian matrix. Figure 6 gives the classification accuracies of our proposed method under different values of the parameter λ. For comparison, we also give the classification results of standard MTFS method (i.e., without the discriminative regularization term). Also, it is worth noting that when λ = 0, no feature selection step is performed, i.e., all features are used for subsequent classification. In addition, we also test different values for the parameter σ, ranging from 0 to 1 at a step size of 0.1, with a fixed λ value, as shown in Fig. 7.
As we can see from Fig. 6, under all values of λ, our proposed method significantly outperforms the MTFS method on all three classification tasks (i.e., AD vs. HC, MCI vs. HC and MCI-C vs. MCI-NC), which again shows the advantage of our method by introducing the discriminative regularization term based on the intra-class and inter-class Laplacian matrices. On the other hand, Fig. 7 indicates that the corresponding curves w.r.t different values of σ are very smooth on all the three classification tasks, showing a good robustness, i.e., insensitive to the values of σ.
Comparsion with single model methods
Here, to estimate the effect of combining multi-modality image data and provide a more comprehensive comparison of the result from the proposed model, we further perform two experiments, that are (1) using only MRI modality, and (2) using only PET modality. It’s worth noting that our proposed model can also be used in single-modality case, where our model degrades into discriminative single-task (modality) feature selection followed by SVM classification. With corresponding results shown in Table 5. As can be seen from Table 5, using multi-modalities (i.e., MRI + PET) achieves significantly better performances than only using single modality (MRI or PET).
Comparison with other feature selection methods
In order to further show the superiority of our proposed method, we compare it with other popular feature selection methods including RelieF (Kira and Rendell 1992) and Elastic Net (Zou and Hastie 2005). For fair comparison, we use the same classifier (i.e., multi-kernel SVM) after performing feature selection using RelieF, Elastic Net and our proposed method. Table 6 gives the classification accuracies of different feature selection methods for AD vs. HC, MCI vs. HC and MCI-C vs. MCI-NC, respectively. As we can see from Table 6, our proposed method always achieves the best classification accuracies in all the three classification tasks, compared to RelieF and Elastic Net. In particular, our proposed method exceeds nearly 10 percentage points than other two compared methods in the classification accuracy of MCI-C vs. MCI-NC. This result again validates the efficacy of our proposed method.
Limitations
The current study is limited by the following two factors. First, in this paper, we use two modalities, i.e., MRI and PET, for AD/MCI classification. However, there exist other modalities (e.g., CSF and APOE) which may also contain commentary information for further improving the classification performance. Second, we only consider two class classification problems (i.e., AD vs. HC, MCI vs. HC and MCI-C vs. MCI-NC), while did not test our proposed method for multi-class classification. In the future, we will address the above limitations to further improve the classification performance.
Conclusion
This paper proposed a discriminative multi-task feature selection method for classification of AD/MCI. Different from the existing multi-modality based feature selection methods, our proposed method explores both the distribution information of intra-class subjects and inter-class subjects. Experimental results on the ADNI dataset show that our proposed method not only improves the classification performance, but also has potential to discover the disease-related biomarkers useful for diagnosis of disease, in comparison with the state-of-the-art multi-modality based methods.
References
Apostolova, L. G., Hwang, K. S., Andrawis, J. P., Green, A. E., Babakchanian, S., Morra, J. H., et al. (2010). 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiology of Aging, 31, 1284–1303.
Berchtold, N. C., & Cotman, C. W. (1998). Evolution in the conceptualization of dementia and Alzheimer’s disease: greco-roman period to the 1960s. Neurobiology of Aging, 19, 173–189.
Bouwman, F., Schoonenboom, S., van Der Flier, W., Van Elk, E., Kok, A., Barkhof, F., et al. (2007). CSF biomarkers and medial temporal lobe atrophy predict dementia in mild cognitive impairment. Neurobiology of Aging, 28, 1070–1074.
Brookmeyer, R., Johnson, E., Ziegler-Graham, K., & Arrighi, H. M. (2007). Forecasting the global burden of Alzheimer’s disease. Alzheimer’s & Dementia, 3, 186–191.
Cai, D., He, X., Zhou, K., Han, J., Bao, H. (2007). Locality Sensitive Discriminant Analysis, in IJCAI, pp. 708–713.
Chang, C.-C., & Lin, C.-J. (2011). LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST), 2, 27.
Chen, X., Pan, W., Kwok, J.T., Carbonell, J.G. (2009). Accelerated gradient method for multi-task sparse learning problem. in Data Mining, 2009. ICDM’09. Ninth IEEE International Conference on, pp. 746–751.
Cheng, B., Zhang, D., Shen, D. (2012). Domain transfer learning for MCI conversion prediction, in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2012, ed: Springer, pp. 82–90.
Dai, W., Lopez, O. L., Carmichael, O. T., Becker, J. T., Kuller, L. H., & Gach, H. M. (2009). Mild cognitive impairment and Alzheimer disease: patterns of altered cerebral blood flow at MR imaging 1. Radiology, 250, 856–866.
Davatzikos, C., Bhatt, P., Shaw, L. M., Batmanghelich, K. N., & Trojanowski, J. Q. (2011). Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology of Aging, 32, 2322. e19–2322. e27.
De Leon, M., Mosconi, L., Li, J., De Santi, S., Yao, Y., Tsui, W., et al. (2007). Longitudinal CSF isoprostane and MRI atrophy in the progression to AD. Journal of Neurology, 254, 1666–1675.
De Santi, S., de Leon, M. J., Rusinek, H., Convit, A., Tarshish, C. Y., Roche, A., et al. (2001). Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiology of Aging, 22, 529–539.
Del Sole, A., Clerici, F., Chiti, A., Lecchi, M., Mariani, C., Maggiore, L., et al. (2008). Individual cerebral metabolic deficits in Alzheimer’s disease and amnestic mild cognitive impairment: an FDG PET study. European Journal of Nuclear Medicine and Molecular Imaging, 35, 1357–1366.
Fan, Y., Batmanghelich, N., Clark, C. M., & Davatzikos, C. (2008). Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage, 39, 1731–1743.
Fellgiebel, A., Scheurich, A., Bartenstein, P., & Müller, M. J. (2007). FDG-PET and CSF phospho-tau for prediction of cognitive decline in mild cognitive impairment. Psychiatry Research: Neuroimaging, 155, 167–171.
Fjell, A. M., Walhovd, K. B., Fennema-Notestine, C., McEvoy, L. K., Hagler, D. J., Holland, D., et al. (2010). CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer’s disease. The Journal of Neuroscience, 30, 2088–2101.
Foster, N. L., Heidebrink, J. L., Clark, C. M., Jagust, W. J., Arnold, S. E., Barbas, N. R., et al. (2007). FDG-PET improves accuracy in distinguishing frontotemporal dementia and Alzheimer’s disease. Brain, 130, 2616–2635.
Gauthier, S., Reisberg, B., Zaudig, M., Petersen, R. C., Ritchie, K., Broich, K., et al. (2006). Mild cognitive impairment. The Lancet, 367, 1262–1270.
Gray, K. R., Aljabar, P., Heckemann, R. A., Hammers, A., & Rueckert, D. (2013). Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. NeuroImage, 65, 167–175.
Higdon, R., Foster, N. L., Koeppe, R. A., DeCarli, C. S., Jagust, W. J., Clark, C. M., et al. (2004). A comparison of classification methods for differentiating fronto‐temporal dementia from Alzheimer’s disease using FDG‐PET imaging. Statistics in Medicine, 23, 315–326.
Hinrichs, C., Singh, V., Mukherjee, L., Xu, G., Chung, M. K., & Johnson, S. C. (2009). Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. NeuroImage, 48, 138–149.
Huang, S., Li, J., Ye, J., Wu, T., Chen, K., Fleisher, A. et al., (2011). Identifying Alzheimer’s Disease-Related Brain Regions from Multi-Modality Neuroimaging Data using Sparse Composite Linear Discrimination Analysis. in Advances in Neural Information Processing Systems, pp. 1431–1439.
Hyman, B. T., Van Hoesen, G. W., Damasio, A. R., & Barnes, C. L. (1984). Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science, 225, 1168–1170.
Jie, B., Zhang, D., Cheng, B., Shen, D. (2013). Manifold regularized multi-task feature selection for multi-modality classification in Alzheimer’s disease, in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013, ed: Springer, pp. 275–283.
Karas, G., Scheltens, P., Rombouts, S., van Schijndel, R., Klein, M., Jones, B., et al. (2007). Precuneus atrophy in early-onset Alzheimer’s disease: a morphometric structural MRI study. Neuroradiology, 49, 967–976.
Kira, K., & Rendell, L.A. (1992). The feature selection problem: Traditional methods and a new algorithm. in AAAI, pp. 129–134.
Knafo, S., Venero, C., Merino‐Serrais, P., Fernaud‐Espinosa, I., Gonzalez‐Soriano, J., Ferrer, I., et al. (2009). Morphological alterations to neurons of the amygdala and impaired fear conditioning in a transgenic mouse model of Alzheimer’s disease. The Journal of Pathology, 219, 41–51.
Landau, S., Harvey, D., Madison, C., Reiman, E., Foster, N., Aisen, P., et al. (2010). Comparing predictors of conversion and decline in mild cognitive impairment. Neurology, 75, 230–238.
Liu, J., & Ye, J.(2010). Efficient l1/lq norm regularization. arXiv preprint arXiv:1009.4766.
Liu, F., Wee, C.-Y., Chen, H., & Shen, D. (2014). Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s Disease and mild cognitive impairment identification. NeuroImage, 84, 466–475.
Mattsson, N., Zetterberg, H., Hansson, O., Andreasen, N., Parnetti, L., Jonsson, M., et al. (2009). CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA, 302, 385–393.
McEvoy, L. K., Fennema-Notestine, C., Roddey, J. C., Hagler, D. J., Jr., Holland, D., Karow, D. S., et al. (2009). Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment 1. Radiology, 251, 195–205.
Misra, C., Fan, Y., & Davatzikos, C. (2009). Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. NeuroImage, 44, 1415–1422.
Morris, J. C., Storandt, M., Miller, J. P., McKeel, D. W., Price, J. L., Rubin, E. H., et al. (2001). Mild cognitive impairment represents early-stage Alzheimer disease. Archives of Neurology, 58, 397–405.
Ng, B., & Abugharbieh, R. (2011). Generalized sparse regularization with application to fMRI brain decoding. in Information Processing in Medical Imaging, pp. 612–623.
Nobili, F., Salmaso, D., Morbelli, S., Girtler, N., Piccardo, A., Brugnolo, A., et al. (2008). Principal component analysis of FDG PET in amnestic MCI. European Journal of Nuclear Medicine and Molecular Imaging, 35, 2191–2202.
Petersen, R. C., Smith, G. E., Waring, S. C., Ivnik, R. J., Tangalos, E. G., & Kokmen, E. (1999). Mild cognitive impairment: clinical characterization and outcome. Archives of Neurology, 56, 303–308.
Poulin, S. P., Dautoff, R., Morris, J. C., Barrett, L. F., Dickerson, B. C., & A. s. D. N. Initiative. (2011). Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Research: Neuroimaging, 194, 7–13.
Pudil, P., Novovičová, J., & Kittler, J. (1994). Floating search methods in feature selection. Pattern Recognition Letters, 15, 1119–1125.
Shaw, L. M., Vanderstichele, H., Knapik‐Czajka, M., Clark, C. M., Aisen, P. S., Petersen, R. C., et al. (2009). Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Annals of Neurology, 65, 403–413.
Solodkin, A., Chen, E. E., Hoesen, G. W., Heimer, L., Shereen, A., Kruggel, F., et al. (2013). In vivo parahippocampal white matter pathology as a biomarker of disease progression to Alzheimer’s disease. Journal of Comparative Neurology, 521, 4300–4317.
Sui, J., Adali, T., Yu, Q., Chen, J., & Calhoun, V. D. (2012). A review of multivariate methods for multimodal fusion of brain imaging data. Journal of Neuroscience Methods, 204, 68–81.
Tibshirani, R. (1996). Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological), pp. 267–288.
Van Hoesen, G. W., & Hyman, B. T. (1990). Hippocampal formation: anatomy and the patterns of pathology in Alzheimer’s disease. Progress in Brain Research, 83, 445–457.
Walhovd, K., Fjell, A., Dale, A., McEvoy, L., Brewer, J., Karow, D., et al. (2010). Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiology of Aging, 31, 1107–1121.
Wang, C., Stebbins, G. T., Medina, D. A., Shah, R. C., Bammer, R., & Moseley, M. E. (2012). Atrophy and dysfunction of parahippocampal white matter in mild Alzheimer’s disease. Neurobiology of Aging, 33, 43–52.
Wee, C.-Y., Yap, P.-T., Zhang, D., Denny, K., Browndyke, J. N., Potter, G. G., et al. (2012). Identification of MCI individuals using structural and functional connectivity networks. NeuroImage, 59, 2045–2056.
Xue, H., Chen, S., & Yang, Q. (2009). Discriminatively regularized least-squares classification. Pattern Recognition, 42, 93–104.
Ye, J., Chen, K., Wu, T., Li, J., Zhao, Z., Patel, R. et al. (2008). Heterogeneous data fusion for alzheimer’s disease study. in Proceedings of the 14th ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 1025–1033.
Ye, J., Wu, T., Li, J., & Chen, K. (2011). Machine learning approaches for the neuroimaging study of Alzheimer’s disease. Computer, 44, 99–101.
Yuan, M., & Lin, Y. (2006). Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society, Series B (Statistical Methodology), 68, 49–67.
Zhang, D., Wang, Y., Zhou, L., Yuan, H., & Shen, D. (2011). Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage, 55, 856–867.
Zhang, D., Shen, D., & A. s. D. N. Initiative. (2012). Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage, 59, 895–907.
Zou, H., & Hastie, T. (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society, Series B (Statistical Methodology), 67, 301–320.
Acknowledgments
This work is supported in part by National Natural Science Foundation of China (Nos. 61422204, 61473149), the Jiangsu Natural Science Foundation for Distinguished Young Scholar (No. BK20130034), the Specialized Research Fund for the Doctoral Program of Higher Education (No. 20123218110009), the NUAA Fundamental Research Funds (No. NE2013105), and NIH grants (EB006733, EB008374, EB009634, and AG041721).
For this project, the dataset we collected and used was provided by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation and the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., with participation from the U.S. Food and Drug Administration. What’s more, Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The Northern California Institute for Education and Research is the grantee organization, as well as the Alzheimer’s Disease Cooperative Study at the University of California, San Diego coordinate the study. ADNI data are disseminated by the Laboratory for Neuron Imaging at the University of California, Los Angeles.
Conflicts of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Additional information
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: www.loni.ucla.edu/ADNI/Collaboration/ADNI_Authorship_list.pdf
Rights and permissions
About this article
Cite this article
Ye, T., Zu, C., Jie, B. et al. Discriminative multi-task feature selection for multi-modality classification of Alzheimer’s disease. Brain Imaging and Behavior 10, 739–749 (2016). https://doi.org/10.1007/s11682-015-9437-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-015-9437-x