Abstract
Well-established targeted technologies to engineer genomes such as zinc-finger nuclease-based editing (ZFN), transcription activator-like effector nuclease-based editing (TALEN), and clustered regularly interspaced short palindromic repeats and associated protein system-based editing (CRISPR/Cas) are proving to advance basic and applied research in numerous plant species. Compared with systems using ZFNs and TALENs, the most recently developed CRISPR/Cas system is more efficient due to its use of an RNA-guided nuclease to generate double-strand DNA breaks. To accelerate the applications of these technologies, we provide here a detailed overview of these systems, highlight the strengths and weaknesses of each, summarize research advances made with these technologies in model and crop plants, and discuss their applications in plant functional genomics. Such targeted approaches for genetically modifying plants will benefit agricultural production in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
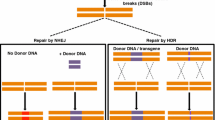
Targeted genome engineering is an advanced approach for scientists to efficiently alter DNA in plant cells by inserting foreign DNA into the genomic DNA at specific locations (Weeks et al. 2016; Weinthal et al. 2010; Xie et al. 2015). In this type of genome editing, sequence-specific nucleases are used to induce double-strand breaks (DSBs) in DNA, and the repair of the breaks achieves the desired sequence modification through nonhomologous end joining (NHEJ), as well as homologous recombination (HR). NHEJ can effectively create a gene knockout by causing frame shift mutations in the coding region of a gene (Sternberg et al. 2014; Urnov et al. 2010; Weinthal et al. 2010). HR can achieve precise gene modifications by repairing the DNA damage with DNA sequences of homology to the DNA sequence near the DSB (Sauer et al. 2016; Sprink et al. 2015; Tovkach et al. 2009; Urnov et al. 2010). Different technologies have been developed using synthetic nucleases (Jinek et al. 2012) and other restriction enzymes to induce site-specific DSBs for plant genome editing (Sternberg et al. 2014; Urnov et al. 2010; Weinthal et al. 2010; Xie and Yang 2013).
Zinc-finger nucleases (ZFNs), the first nucleases used for genome editing in plants, represented a breakthrough in genome modification because this approach exploits protein engineering to cut arbitrary DNA sequences (Puchta and Fauser 2013; Weinthal et al. 2010; Wood et al. 2011). ZFNs are engineered fusion proteins comprising DNA-binding zinc-finger motifs and the restriction enzyme FokI endonuclease (Chandrasegaran and Carroll 2015; Urnov et al. 2010; Weinthal et al. 2010). In ZFNs, 18–24-bp DNA sequences are recognized by two ZFN units strung together and a DSB is generated by the FokI dimer that was reconstituted by the pair of ZFNs (Puchta and Fauser 2013; Weinthal et al. 2010; Wood et al. 2011). ZFNs have been used for gene targeting to edit genomes of plant species such as Arabidopsis, tobacco, potato, maize, and soybean (Anand et al. 2013; Butler et al. 2015; Hansen et al. 2012; Petolino 2015; Urnov et al. 2010; Weeks et al. 2016). A study of molecular mechanisms on DSB formation through cleavage of 3-finger ZFNs demonstrated that the paired DNA binding sites are located at a specific genomic locus in plants (Chandrasegaran and Carroll 2015). Combining FokI endonuclease and zinc finger proteins (ZFPs) in ZFNs provides a general way to generate a site-specific double-strand break (DSB) in the plant genome (Puchta and Fauser 2013; Weinthal et al. 2010). The approaches using ZFN assembly and validation have great potential application for plant genome engineering through gene targeting (Puchta and Fauser 2013).
Transcription activator-like effector nuclease (TALEN)-based editing, another of the most commonly used techniques for plant genome engineering (Ainley et al. 2013; Durai et al. 2005; Jabalameli et al. 2015; Petolino 2015; Puchta and Fauser 2013; Ul Ain et al. 2015; Urnov et al. 2010; Weeks et al. 2016; Weinthal et al. 2010), is based on the function of bacterial transcription activator-like effectors (TALEs), that can be fused to the FokI cleavage domain. TALENs are hybrid proteins consisting of a genetically modified DNA-binding domain fused to the FokI nonspecific nuclease domain. The sequence repeats in TALEs are composed of 33–35 amino acids, with repeat-variable di-residues at positions 12 and 13 to determine pairing with the target DNA sequence (Sprink et al. 2015; Wood et al. 2011; Yan et al. 2013; Zhang et al. 2015b). For example, the DNA-binding domain derived from the Xanthomonas transcriptional activator-like effector (TALE) protein has been used to fuse with the functional domain of the FokI endonuclease in the application of TALENs in plants (Osakabe and Osakabe 2015; Shan et al. 2015; Sprink et al. 2015; Wood et al. 2011; Yan et al. 2013; Zhang et al. 2015b). Efficient TALENs have been used for successful genome editing in different plant species including Brachypodium, rice, tobacco, and wheat (Johnson et al. 2013; Kumar and Jain 2015; Lee et al. 2016; Osakabe and Osakabe 2015). Different genetic methods have been developed to facilitate the assembly of repeat arrays (Chen and Gao 2014; Clasen et al. 2016; Johnson et al. 2013; Kumar and Jain 2015; Lee et al. 2016; Osakabe and Osakabe 2015). For example, a free designable DNA binding domain can be used in TALENs to exploit alternative approaches in plants (Petolino 2015; Puchta and Fauser 2013). ZFNs and TALENs have been successfully used for genome modification in more than 40 different plant species and cell types for genome engineering, indicating their potential application of plant genetic engineering (Petolino 2015; Puchta and Fauser 2013; Wood et al. 2011).
Among different genome editing technologies, the type II clustered regularly interspaced short palindromic repeat and the associated protein system (CRISPR/Cas) is the most powerful tool for revealing gene function and for engineering genomes in plants (Barrangou 2015; Barrangou and Marraffini 2014; Bondy-Denomy and Davidson 2014). The CRISPR/Cas system, which is based on the application of RNA-guided engineered nucleases, are derived from the adaptive function of the immune system of bacteria (Streptococcus pyogenes) and of archaea, where cells cleave invading nucleic acid viruses in a sequence-dependent manner to protect themselves (Barrangou 2015; Barrangou and Marraffini 2014; Bondy-Denomy and Davidson 2014). At the proximal DNA end of the CRISPR locus, spacers integrate into a region between two adjacent repeats to generate immunity (Carter and Wiedenheft 2015; Charpentier and Marraffini 2014; Chylinski et al. 2014). Compared to ZFNs and TALENs, CRISPR/Cas9 is a more efficient tool because the RNA-guided nucleases provide targeted DSBs in cellular genomes (Charpentier and Marraffini 2014; Cong and Zhang 2015; Dow et al. 2015). Currently, the CRISPR/Cas9 system has been applied in a large number of plant species including Arabidopsis, tobacco, rice, tomato, and popular (de Lange et al. 2014; Duan et al. 2016; Endo et al. 2015; Gao et al. 2015; Jinkerson and Jonikas 2015; Mikami et al. 2015a, b; Schaeffer and Nakata 2015; Vazquez-Vilar et al. 2016; Zhang et al. 2014, 2016b). In the CRISPR/Cas system, the CRISPR RNA (crRNA) and the trans-encoded CRISPR RNA (tracrRNA) fused to generate a chimeric guide RNA (sgRNA) to cleave a specific DNA target region (Endo et al. 2015; Mikami et al. 2015a, b; Ren et al. 2014; Vazquez-Vilar et al. 2016). The Cas9 protein and sgRNA form an RNA-guided endonuclease to modify targeted DNA sequence in the plant genome (Jinek et al. 2012, 2013, 2014). In sgRNA-Cas9 complex, the chimeric guide RNA determines the catalytic action at a site-specific manner (Fig. 1). Unlike ZFNs and TALENs genome editing which uses protein motifs to recognize DNA sequences, CRISPR–Cas9 uses RNA–DNA recognition to edit genomic DNA sequence (Chandrasegaran and Carroll 2015).
Mechanism for targeted genetic modification by sequence-specific zinc-finger nucleases (ZFNs) through homologous recombination (HR) and non-homologous end joining (NHEJ) DNA repair. Two ZFN arrays are fused to the catalytic domain of the FokI endonuclease (gray) and bound to their target DNA site. Each monomer consists of three zinc fingers (green) that make base-specific contacts. A ZFN heterodimer binds to its specific DNA target. These ZFNs dimerize at the spacer region of the DNA target sequence, leading to the cleavage of the target DNA via a double-strand break (DSB). Upon dimerization, a DSB is created. To recognize the particular 9-bp target sequence, the sequential combination of three ZFs corresponding to each triplet nucleotide sequences of the target would be required. The repair of ZFN-induced DNA DSB via homologous recombination can be utilized for gene correction or gene insertion of a few nucleotides whereas the NHEJ-mediated repair pathway can be used for gene disruption. Total target DNA sequences are typically 18–36-bp. Each ellipsis represents a zinc finger domain that recognizes specific triplet nucleotides
In this review, we provide an overview of the function and types of these major genome engineering technologies in plants (Jinek et al. 2012), highlighting the use of ZFNS, TALENS, and CRISPR/Cas9 by addressing the current approaches to design efficient system and to decrease any off-target effects. We summarize potential applications of these technologies for genome engineering in plants to improve crop traits, virus resistance, and stress tolerance, and emphasize their use in model plant and crop species (Table 1).
ZFN-based genome engineering
ZFNs achieve sequence-specific cleavage for reverse genetics in plants (Urnov et al. 2010) and can be used to modify plant genomes via repair of a DSB in a targeted genomic region (Petolino 2015; Weinthal et al. 2010). ZFPs provide a flexible framework for creating custom ZFN protocols with new sequence-specificities (Durai et al. 2005). The expression of ZFNs can induce genomic DSBs to drive the replacement of target gene sequences with foreign DNA as site-specific mutagens in plant species for research in plant genetics and biotechnology (Weinthal et al. 2010). ZFNs can facilitate gene targeting, additions, deletions, and inactivation with low efficiency and reproducibility (Mahfouz et al. 2011). Various research groups are developing ZFNs that will enable both site-specific mutagenesis and controlled foreign DNA integration in living plant cells (Petolino 2015). The induction of DSBs at specific genomic locations by ZFNs results in genetic changes including gene deletion and site-specific mutagenesis in living plant cells (Tzfira et al. 2012). ZFN-mediated genome editing is becoming applicable technology in crop species for rapidly engineering crop trait such as and increasing quality, productivity and abiotic stress tolerance (Ainley et al. 2013; Jia and Wang 2014).
In the ZFN system, a single nuclease is sufficient to knock out the function of target genes through the frameshifts in the coding exon (Gupta et al. 2013). The development of ZFNs as a tool to generate site-specific genomic DSBs for genome modification and gene-targeting applications in plant cells (Marton et al. 2013; Petolino 2015; Weinthal et al. 2010). As the use of ZFNs in genome editing has increased, factors that affect the efficiency of ZFNs that recognize two adjacent unique DNA sequences have been investigated (Petolino 2015; Urnov et al. 2010). One subunit of ZFNs can generate a pair of hybrid nucleases to recognize the specific DNA region and to stimulate gene targeting in multiple cell types (Yan et al. 2013).
ZFNs have been developed that can repress transcription, bind methylated DNA or act as fluorescent chromatin probes (Scott et al. 2014). ZFNs are nuclease-based precision genome editing technique (Fig. 1) that is enabling efficient gene editing including target gene disruption, DNA fragment insertion, repair, genetic modification, and gene deletion (Steentoft et al. 2014). Targeted genome editing using ZFNs has the potential to improve plant molecular breeding by providing the approaches to modify genomes in a predictable manner (Bortesi and Fischer 2015). Investigation on the molecular mechanism of cleavage by ZFNs demonstrated that the paired binding sites is a unique genomic DNA region in cells and that a ZFN-induced DSB stimulates homologous recombination (Chandrasegaran and Carroll 2015). ZFNs have been used to knock down gene expression (Hutter et al. 2015), to cut specific DNA sequence (Droz-Georget Lathion et al. 2015), and to remove essential splicing sequences (Ousterout et al. 2015). ZFNs mediate targeted genetic modification by increasing the DNA mutation rate via induction of DSBs at a predetermined genomic DNA region (Petersen and Niemann 2015b). ZFN-mediated DSB generation at a specific genomic DNA region followed by NHEJ repair can lead to gene-specific mutations via nucleotide base-pair insertions or deletions (Petolino 2015). Gene mutations can also be corrected using ZFNs (Qin and Gao 2016).
ZFN-based genome engineering in model plants
The technology of ZFNs has been used for targeted genome engineering in model plant species including Arabidopsis (Puchta and Fauser 2013; Urnov et al. 2010) and tobacco (Mahfouz et al. 2011; Marton et al. 2010; Puchta and Fauser 2013). ZFN-expressing transgenic plant systems have been used to produce mutants by transferring the gene directly into the target cells. Tovkach et al. (2009) have developed in planta activity assays to confirm the activity of ZFNs in plant cells. The assays are based on introduction of a mutated uidA- and ZFN-expressing cassettes into target plants cells that offer cloning flexibility and simple assembly of the tested ZFNs in plants (Tovkach et al. 2009).
Marton et al. (2010) reported a nontransgenic technology for ZFN delivery using a novel tobacco rattle virus (TRV)-based expression system to introduce ZFNs into a different tissues and cells of intact plants (Marton et al. 2010). ZFN-mediated targeted mutagenesis was detected in newly developed infected tissues of tobacco (Nicotiana tabacum). The stability of ZFN-induced genetic changes in the next generation was confirmed by sequence analysis, indicating a viable alternative to the production of ZFN-mediated genetic modification while avoiding direct transformation of plants. The frequency of targeted DNA integration via homologous recombination established ZFNs as a feasible technique for higher plants including tobacco and Arabidopsis so that targeted genomic DNA modifications will become routine for plant species including crop plants (Puchta and Fauser 2013).
ZFN-based genome engineering in crop plants
ZFNs-based genome engineering has been successfully applied in crop plants including maize and potato (Belhaj et al. 2013; Butler et al. 2015; Petolino 2015; Weeks et al. 2016). Weeks et al. (2016) documented the progress and future uses of designer nucleases with model plant species and to engineer genes and genomes in economically important crop species for enhanced food production. Ainley et al. (2013) reported the combination of high-efficiency targeted genome editing driven by engineered ZFNs with trait-stacking in crop plants and illustrated the utility of the nuclease-driven genome editing technology in maize. They also used microparticle bombardment to deliver donor DNA and the corresponding ZFN expression construct to immature embryos, demonstrating a simple, facile and rapid technology for trait-stacking (Ainley et al. 2013; Puchta and Fauser 2013). ZFNs have also been used in potato (Solanum tuberosum) for targeted mutagenesis via nonhomologous end joining (Butler et al. 2015).
Although ZFNs opened the door to custom nuclease-targeted genome engineering in the late 1990s, it has not been widely adopted for plants because designing multi-zinc-finger modules is very difficult, the interactions between amino acid residues and base pairs of the target DNA sequence are complicated, and the assembly of specific DNA-binding proteins for each target gene is laborious for plants (Belhaj et al. 2013; Petolino 2015; Urnov et al. 2010). An in-depth understanding of the interactions between single amino acids in the zinc-finger module with each base pair of the target DNA sequence is required to facilitate the design of accurate zinc-finger modules. In addition, off-target cleavage as a result of nonspecific binding of zinc-finger motifs can be a problem (Durai et al. 2005; Puchta and Fauser 2013; Weinthal et al. 2010; Wood et al. 2011; Yan et al. 2013). Moreover, the costs are high for the modest efficacy in many applications in plants (Blackburn et al. 2013).
TALEN-based genome engineering
TALENs are useful tools for performing targeted genomic DNA editing in plants and cultured plant cells (Fig. 2) (Marton et al. 2013; Reyon et al. 2012), providing the ability to delete chromosomal segments, study the function of large noncoding sequences, screen genomes, disrupt genes, and develop new crop traits (Chen and Gao 2014; Gupta et al. 2013; Jia and Wang 2014; Nemudryi et al. 2014; Shan et al. 2014). Luo et al. (2014) reported that a pair of transcription activator-like effector nucleases (TALENs) improved genome editing, compared to the use of zinc finger nucleases and thus increase gene targeting efficiency in vitro and in vivo.
Mechanism for targeted genetic modification by the transcription activator-like effector nucleases through homologous recombination (HR) and non-homologous end joining (NHEJ) DNA repair. DNA sequence recognition and site-specific DSB formations by transcription activator-like effector nuclease. Two transcription activator-like effector nucleases (TALENs) bound to their target site. Repeat variable di-residues in each repeat in the transcription activator-like (TAL) effector array (gray) make a base-specific contact with the target DNA. The TAL effector repeat arrays are fused to endonuclease FokI to form hybrid TALE nucleases for gene editing purposes. A meganuclease binds to a DNA target. The catalytic domain also determines base specificity. TALENs create specific DSBs in target sequences of DNA around 30–40-bp. The resulted DNA double-strand break then undergoes homologous recombination or error-prone NHEJ DNA repair during which deletions or insertions of a few nucleotides often occur. Each ellipse represents a zinc finger (ZF) domain that recognizes the specific nucleotide
Targeted gene editing with TALENs has become increasingly popular since these powerful tools have greatly facilitated the generation of plant models for basic research and harbor an enormous potential for applications in genetics (Mussolino et al. 2015). Recent advances in TALENs have benefited current molecular biology research (Osakabe and Osakabe 2015) and molecular breeding (Redel and Prather 2015; Bortesi and Fischer 2015). Applications of TALENs are expanding in research, medicine, and biotechnology, and specific sequences can be efficiently inserted, removed or changed in cultured plant cells (Lee et al. 2016). They can also be used for targeted crop protection strategies in molecular breeding programs for disease resistance (Nejat et al. 2016). TALENs are being used for genome editing and diverse biotechnological applications in model plants and various crop plants (Kumar and Jain 2015).
TALEN-based genome engineering in model plants
In a study on biofuel production, TALENs were used to efficiently edit the genome of the yeast Saccharomyces cerevisiae to induce overproduction of fatty acids by simultaneously editing the FAA1 and FAA4 genes encoding acyl-CoA synthetase (Aouida et al. 2015). Thus, TALENs can be an excellent tool for metabolic engineering.
TALENs have been developed for genome editing in Arabidopsis for germline-modification by stably integrated transgenes into plant genome. Using TALEN technology, Forner et al. (2015) observed very high phenotype frequencies in the T2 generation and demonstrated that constitutive TALEN expression did not cause additional off-target effects (Forner et al. 2015). TALENs can result in high-frequency targeted-gene mutagenesis, although a rapid method to determine the cleavage efficiency is still needed in plants. Based on cleavage-dependent luciferase gene correction, Johnson et al. (2013) reported an assay that consists of coinfiltrating Nicotiana benthamiana leaves with two Agrobacterium tumefaciens strains. Johnson et al. (2013) detected cleavage by transcription activator-like effector nucleases in the Arabidopsis CRUCIFERIN3 gene (Johnson et al. 2013).
TALENs enable facile editing of genomes for genome engineering in cells of different plant species. Baltes et al. (2014) used geminivirus-based replicons for transient expression of TALENs and delivery of DNA repair templates in tobacco. In addition to generation of the nuclease-mediated DNA double-strand breaks, increased frequency of gene targeting was achieved by application of the repair template and of the geminivirus replication initiator proteins. Baltes et al. (2014) demonstrated the feasibility of using geminivirus replicons to generate plants with genomic DNA modification in plants (Baltes et al. 2014).
TALENs-based genome engineering in crop plants
TALENs open up new opportunities for targeted genome engineering in crop plants to obtain agronomically important traits. Budhagatapalli et al. (2015) demonstrated the feasibility of genome editing by TALENs resulting in a predicted alteration of gene function in barley. As a result of cobombardment of leaf epidermis, Budhagatapalli et al. (2015) detected yellow fluorescent protein accumulation in about three of 100 mutated cells, indicating that a readily screenable marker system could be useful and important for genome editing (Budhagatapalli et al. 2015). In rice, Ma et al. (2015) developed a TALENs-based method for genome editing, and Shan et al. (2015) engineered TALENs to target the OsBADH2 gene. Shan et al. (2015) also obtained TALENs-based mutagenesis by simultaneously introducing three different pairs of TALENs to target three different rice genes in rice cells via bombardment. Wang et al. (2015) replaced a single base in situ in the rice OsEPSPS gene by co-transformation with TALEN and chimeric RNA–DNA oligonucleotides including RNA–DNA or DNA–RNA strand composition and obtained one mutant showing target base substitution. They also obtained 16 mutants with based deletions of different lengths. Molecular analysis of the mutants demonstrated that the induced mutations were stably passed to the next generation.
In Zea mays, Liang et al. (2014) designed five TALENs targeting four genes, (ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4) and obtained 23.1% targeting efficiency in protoplasts, and 13.3–39.1% targeting efficiency in transgenic plants. The TALENs induced targeted mutations in Z. mays protoplasts at an efficiency of 9.1%, indicating that TALENs are suitable for genome editing in maize (Liang et al. 2014). TALENs have also been used to knockout the VInv gene in potato (Solanum tuberosum) variety Ranger Russet. Clasen et al. (2016) obtained 11 modified plant lines that contain TALEN DNA insertions in the genome of potato, indicating that TALENs can be used to quickly improve traits in potato plants. TALENs-mediated targeted mutagenesis has been also reported in potato via nonhomologous end joining (Butler et al. 2015).
Compared to ZFNs, TALENs have fewer off-target effects due to the longer target recognition site (Chandrasegaran and Carroll 2015; Duda et al. 2014; Durai et al. 2005; Petolino 2015; Weinthal et al. 2010; Wood et al. 2011; Wright et al. 2014; Yan et al. 2013). However, the construction of multiple repeat sequences is a challenging task. Although computer programs are available for efficient TALE design and target prediction and TALE libraries are also available for mammalian systems, this technology requires sophisticated molecular design and assembly of individual DNA-binding proteins for every DNA target sequence. Knowledge of nuclease-specific features, as well as their specificities and mutation signatures, is critical for choosing the most appropriate genome editing tools for specific applications (Kim and Kim 2014). Yan et al. (2013) demonstrated that the increased size of the DNA recognition sequences in TALENs is not essential to achieve higher efficiency or specificity for gene targeting in cells (Yan et al. 2013). Although TALENs-based genome modifications are an important technology for genetic engineering in crop plants (Puchta and Fauser 2013), they have also been shown to cut at off-target sites with mutagenic consequences (Chandrasegaran and Carroll 2015).
CRISPR/Cas9-based genome engineering
The CRISPR/Cas9 system is a simple, powerful, highly efficient tool for genome engineering because it can precisely modify endogenous genes for biological studies and disease resistance (Fig. 3). It can provide at least a 10-fold increase in efficiency over previously published genome editing technologies for studying gene function (Bassett et al. 2013). Because the repair of the on- and off-target cleavage results in additional DNA sequence modification including insertions, deletions and point mutations, CRISPR/Cas9 systems need to be carefully designed for different plant species to avoid potential off-target cleavage sites (Cradick et al. 2013). The CRISPR/Cas9 system has been used to edit the genome and block gene expression and thus may be a powerful tool to protect against pathogens (Ebina et al. 2013). The high efficiency and heritability of CRISPR/Cas9-based mutagenesis, combined with the ease and limited off-target effects make the system very useful for in vivo studies (Hruscha et al. 2013).
Mechanism for targeted genetic modification by the Cas9/sgRNA complex through homologous recombination (HR) and non-homologous end joining (NHEJ) DNA repair. Cas9 interacts with the DNA strand, and the single guide RNA (sgRNA) hybridizes with the specific 20-nt sequence of the targeted gene, leading to cleavage of the targeted DNA. The target DNA is properly oriented in the active site of Cas9 through the PAM (tandem guanosine nucleotides) binding site. The interaction allows separate nuclease domains of Cas9 to independently cleave each strand of the target DNA sequence at a point 3-nt upstream of the PAM site. The resultant DNA double-strand break then undergoes HR or error-prone NHEJ DNA repair during which deletions or insertions of a few nucleotides often occur. crRNA, CRISPR RNA; HNH, His-Asn-His endonuclease; RuvC, Escherichia coli ruv endonuclease family member C, PAM, the protospacer adjacent motif
The CRISPR/Cas9 genome engineering system is also efficient, specific, and flexible for gene correction. Schwank et al. (2013) used the CRISPR/Cas9 system to correct a specific gene locus by homologous recombination in cultured cells of human patients; the corrected allele is expressed and fully functional. This study provided proof of concept that a gene can be corrected by homologous recombination in cells with a single-gene hereditary defect (Schwank et al. 2013). In addition, Auer et al. (2014) reported CRISPR/Cas9-mediated knock-in of DNA cassettes into the genome of zebrafish (Auer et al. 2014).
The CRISPR/Cas9 systems facilitate directed mutagenesis of specific loci in the genome and thus can greatly speed up analysis of gene function (Waaijers et al. 2013). The CRISPR/Cas9 system has been used to generate targeted mutations in model organisms (Bassett and Liu 2014; Bassett et al. 2014). By targeting a constitutive exon of the AGO1 gene, Bassett et al. (2014) achieved a homozygous mutation in up to 82% of cells. The system is also useful for transcriptional regulation (de Lange et al. 2014). The CRISPR/Cas9 system is capable of targeting multiple genomic sites in one shot (Petersen and Niemann 2015b). Recent investigations demonstrated that the CRISPR/Cas9 system could function as a sequence-specific nuclease in plants, providing new opportunities for generating genetically modified plants (Petersen and Niemann 2015a). The CRISPR/Cas9 system has quickly become the preferred genome-editing tool of plant scientists for proof of concept and functional studies in model systems. Such studies have led to the development of multiplexing for inducing multiple cleavage and site-specific transgene insertion. With conceptual CRISPR/Cas9 studies, plant molecular biologists are beginning to apply this genome editing technology for improving crops (Schaeffer and Nakata 2015) and generating novel mutant phenotypes (van Tol and van der Zaal 2014).
CRISPR/Cas9-based genome engineering in model plants
The CRISPR/Cas9 system for targeted mutagenesis has been applied in the model plants Arabidopsis thaliana and Nicotiana tabacum using transient expression systems and transgenic plants. Jiang et al. (2014) demonstrated that the CRISPR/Cas9 system has promise for facile editing of GFP gene in Arabidopsis. Gao et al. (2015) obtained insertion and deletion mutations in NtPDS and NtPDR6 of N. tabacum at frequencies of 16.2–20.3% using the CRISPR/Cas9 system.
The CRISPR/Cas9 system also offers unparalleled power for elucidating plant defense mechanisms and developing novel options for pest management (Gurr and You 2015). It has also been used for targeted genome modification in different plant species to obtain molecular immunity against DNA viruses. For example Ali et al. (2015b) designed a CRISPR/Cas9 system to introduce genomic mutations into the targeted TYLCV DNA sequence in N. benthamiana plants.
Targeted DNA sequence modification of plant genome is very important for elucidating gene functions for basic and applied plant research (Schaeffer and Nakata 2015; Zhang et al. 2014). The CRISPR/Cas9-based genome editing technology enables deletion of a large imprinted lncRNA and chromosome deletions (Han et al. 2014; He et al. 2015; Hemphill et al. 2015; Schaeffer and Nakata 2015). Vazquez-Vilar et al. (2016) described the adaptation of the RNA-guided Cas9 system to a modular DNA construction framework for increased use in synthetic plant biology in N. benthamiana (Vazquez-Vilar et al. 2016). The detailed procedure to design, construct, and examine dual gRNAs for plant codon-optimized Cas9 (pcoCas9)-mediated genome modification using A. thaliana and N. benthamiana protoplasts as experimental systems is valuable and provides opportunities to apply the CRISPR/Cas9 system for genome editing in whole plants (Ali et al. 2015a).
CRISPR/Cas9-based genome engineering in crop plants
Mikami et al. (2015a) evaluated mutation frequency in rice and identified the best Cas9/gRNA expression cassette for targeted mutagenesis in rice calli (Mikami et al. 2015a). Cas9 expression level and mutation frequency were positively correlated, and a prolonged tissue culture period increases the chance of inducing de novo mutations in nonmutated cells. An endogenous tRNA-processing system has been engineered as a simple platform to boost the multiplex editing capability of the CRISPR/Cas9 system in rice. Xie et al. (2015) demonstrated that synthetic genes with tandemly arrayed tRNA–gRNA architecture were successfully processed into gRNAs with the desired 5′ targeted sequences in vivo to achieve multiplex genome modification in transgenic rice plants (Xie et al. 2015).
The CRISPR/Cas9 system was also used to induce targeted mutations in OsRAV2 of rice and provides a better understanding of the salt response of OsRAVs and the molecular regulatory mechanisms of plant genes under salt stress (Duan et al. 2016). The mutations induced using the CRISPR/Cas9 system occurred very early in the development of the shoot apical meristem and were stably transmitted to the T1 and T2 populations in a Mendelian fashion (Zhang et al. 2016b). Xu et al. (2015) described the CRISPR/Cas9-mediated genome editing of four rice genes. A high frequency of mutagenesis was achieved in T0 generations, and the mutations in T1 lines were stably transmitted to next generations, indicating a standard pattern of germline transmission (Xu et al. 2015). The CRISPR/Cas9 system also efficiently induced targeted gene editing in 11 target genes of two rice subspecies (Zhang et al. 2014). The gene mutations were passed to the next generation without any detectable new mutations. To validate the applicability of the CRISPR/Cas9 system to target mutagenesis of paralogous genes in rice, Endo et al. (2015) designed a single-guide RNA (sgRNA) that recognized 20-bp sequences of the target CDKB2 gene. Targeted mutations in four CDK genes in plants derived from Cas9/sgRNA-transformed calli revealed that single, double, and triple mutants of CDKA2, CDKB1 and CDKB2 can be created by a single sgRNA (Endo et al. 2015).
To develop a toolkit for additional plant selectable markers, gRNA modules, and gRNA expression, Ito et al. (2015) developed a CRISPR/Cas9 binary vector set that is based on the pGreen backbone and a gRNA module vector set for multiplex genome editing in tomato plants. The toolkit has been used in maize protoplasts and transgenic maize lines and was highly efficient and specific, and the induced mutations were inherited by the next generation, indicating stable expression of the CRISPR/Cas9 system in maize (Svitashev et al. 2015).
The CRISPR/Cas9 system was developed that efficiently and precisely mutates genomic DNA sequences in soybean by knocking-out a GFP transgene and modifying nine endogenous loci in 95% of 88 hairy-root transgenic events analyzed (Jacobs et al. 2015). Du et al. (2016) developed a CRISPR/Cas9 system for two soybean genome-editing targets, GmPDS11 and GmPDS18, with a single targeting efficiency range of 11.7–18.1%. They also achieved a targeting efficiency of 12.5% in a double-mutation gene-targeting experiment.
The CRISPR/Cas9 system has been used to develop virus resistance in cucumber (Cucumis sativus L.) by disrupting the protein synthesis function of the recessive gene eIF4E (Chandrasekaran et al. 2016). The Cas9/sgRNA constructs targeted the N- and C-terminus of the eIF4E gene. Such CRISPR/Cas9 technology is expected to be applicable for generating resistance in other crop plants.
The CRISPR/Cas9 system, with the Cauliflower mosaic virus 35S promoter to express Cas9, has been used for targeted editing of the genome of the liverwort Marchantia polymorpha, inducing stable mutants (Sugano et al. 2014). The CRISPR/Cas9 system is also efficient for editing the gene of phytoene desaturase (PDS) that is a key enzyme in carotenoid biosynthesis in a hybrid Petunia: transgenic shoot lines with an albino phenotype accounted for 55.6–87.5% of the total regenerated plants (Zhang et al. 2016a).
CRISPR/Cas9-based genome engineering in woody plants
Although the CRISPR/Cas system has been successfully used to edit the genome of various model and crop plant species, it has been rarely used in woody plants. Recently, Fan et al. (2015) designed four gRNAs to target the phytoene desaturase gene 8 (PtoPDS) in Populus tomentosa after Agrobacterium-mediated transformation for editing. Of the 59 mutant phenotypes detected, 30 were homozygous, indicating the first successful stable transformation of a woody plant using the CRISPR/Cas system (Fan et al. 2015). Experimental evidence also demonstrated the sensitivity of CRISPR/Cas9 to allelic heterozygosity. Improvements of the CRISPR/Cas9 system for multiplex genome editing would accelerate not only basic molecular genetics research but also applied plant improvement (Tsai and Xue 2015).
Conclusion
Because of their efficiency and versatility, the genome engineering platforms ZFN, TALEN, and the CRISPR/Cas9 are promising tools for targeted genetic modifications in plants, investigations of functional genomics and improvement of crop traits. The advantages of the CRISPR–Cas9 system are the ease of RNA design and high efficiency. Although these genome-editing technologies enable scientists to modify the genome, they also may mutate off-target sites. Therefore, the major challenges in targeted genetic modification in plants in the future years are (1) improving the efficacy, specificity, and delivery; (2) increasing the ability to initiate, maintain, and regenerate plant cell and tissue cultures to increase the proportion of mutated cells and the number of plants containing heritable mutations; (3) analyzing risk versus benefit for the platforms (Chandrasegaran and Carroll 2015); (4) expanding the availability of crop-specific vectors and improving transformation protocols of crop plants; and (5) manipulating large genomic DNA regions, modifying transgene-free genomic DNA, and discovering novel gene functions.
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol 204:273–281
Ainley WM, Sastry-Dent L, Welter ME, Murray MG, Zeitler B, Amora R, Corbin DR, Miles RR, Arnold NL, Strange TL, Simpson MA, Cao Z, Carroll C, Pawelczak KS, Blue R, West K, Rowland LM, Perkins D, Samuel P, Dewes CM, Shen L, Sriram S, Evans SL, Rebar EJ, Zhang L, Gregory PD, Urnov FD, Webb SR, Petolino JF (2013) Trait stacking via targeted genome editing. Plant Biotechnol J 11:1126–1134
Ali Z, Abul-Faraj A, Piatek M, Mahfouz MM (2015a) Activity and specificity of TRV-mediated gene editing in plants. Plant Signal Behav 10:e1044191
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM (2015b) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16:238
Anand P, Schug A, Wenzel W (2013) Structure based design of protein linkers for zinc finger nuclease. FEBS Lett 587:3231–3235
Aouida M, Li L, Mahjoub A, Alshareef S, Ali Z, Piatek A, Mahfouz MM (2015) Transcription activator-like effector nucleases mediated metabolic engineering for enhanced fatty acids production in Saccharomyces cerevisiae. J Biosci Bioeng 120:364–371
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163
Barrangou R (2015) Diversity of CRISPR–Cas immune systems and molecular machines. Genome Biol 16:247
Barrangou R, Marraffini LA (2014) CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54:234–244
Bassett AR, Liu JL (2014) CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics 41:7–19
Bassett AR, Tibbit C, Ponting CP, Liu JL (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4:220–228
Bassett AR, Tibbit C, Ponting CP, Liu JL (2014) Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol Open 3:42–49
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39
Blackburn PR, Campbell JM, Clark KJ, Ekker SC (2013) The CRISPR system-keeping zebrafish gene targeting fresh. Zebrafish 10:116–118
Boehm CR, Ueda M, Nishimura Y, Shikanai T, Haseloff J (2016) A cyan fluorescent reporter expressed from the chloroplast genome of Marchantia polymorpha. Plant Cell Physiol 57:291–299
Bondy-Denomy J, Davidson AR (2014) To acquire or resist: the complex biological effects of CRISPR–Cas systems. Trends Microbiol 22:218–225
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Budhagatapalli N, Rutten T, Gurushidze M, Kumlehn J, Hensel G (2015) Targeted modification of gene function exploiting homology-directed repair of TALEN-mediated double-strand breaks in barley. G3 (Bethesda) 5:1857–1863
Butler NM, Atkins PA, Voytas DF, Douches DS (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE 10:e0144591
Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W (2015) CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE 10:e0136064
Carter J, Wiedenheft B (2015) SnapShot: CRISPR-RNA-guided adaptive immune systems. Cell 163(260–260):e261
Chandrasegaran S, Carroll D (2015) Origins of programmable nucleases for genome engineering. J Mol Biol 428:963–989
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17:1140–1153
Charpentier E, Marraffini LA (2014) Harnessing CRISPR–Cas9 immunity for genetic engineering. Curr Opin Microbiol 19:114–119
Chen K, Gao C (2014) Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep 33:575–583
Chylinski K, Makarova KS, Charpentier E, Koonin EV (2014) Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res 42:6091–6105
Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, Tibebu R, Davison S, Ray EE, Daulhac A, Coffman A, Yabandith A, Retterath A, Haun W, Baltes NJ, Mathis L, Voytas DF, Zhang F (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 14:169–176
Cong L, Zhang F (2015) Genome engineering using CRISPR–Cas9 system. Methods Mol Biol 1239:197–217
Cradick TJ, Fine EJ, Antico CJ, Bao G (2013) CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41:9584–9592
de Lange O, Binder A, Lahaye T (2014) From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J 78:753–771
Dow LE, Fisher J, O’Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, Lowe SW (2015) Inducible in vivo genome editing with CRISPR–Cas9. Nat Biotechnol 33:390–394
Droz-Georget Lathion S, Rochat A, Knott G, Recchia A, Martinet D, Benmohammed S, Grasset N, Zaffalon A, Besuchet Schmutz N, Savioz-Dayer E, Beckmann JS, Rougemont J, Mavilio F, Barrandon Y (2015) A single epidermal stem cell strategy for safe ex vivo gene therapy. EMBO Mol Med 7:380–393
Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217:90–97
Duan YB, Li J, Qin RY, Xu RF, Li H, Yang YC, Ma H, Li L, Wei PC, Yang JB (2016) Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol Biol 90:49–62
Duda K, Lonowski LA, Kofoed-Nielsen M, Ibarra A, Delay CM, Kang Q, Yang Z, Pruett-Miller SM, Bennett EP, Wandall HH, Davis GD, Hansen SH, Frodin M (2014) High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res 42:e84
Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33:5978–5990
Ebina H, Misawa N, Kanemura Y, Koyanagi Y (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 3:2510
Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56:41–47
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci Rep 5:12217
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Forner J, Pfeiffer A, Langenecker T, Manavella PA, Lohmann JU (2015) Germline-transmitted genome editing in Arabidopsis thaliana using TAL-effector-nucleases. PLoS ONE 10:e0121056
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110
Grosche C, Funk HT, Maier UG, Zauner S (2012) The chloroplast genome of Pellia endiviifolia: gene content, RNA-editing pattern, and the origin of chloroplast editing. Genome Biol Evol 4:1349–1357
Gupta RM, Musunuru K (2014) Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR–Cas9. J Clin Invest 124:4154–4161
Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA (2013) Targeted chromosomal deletions and inversions in zebrafish. Genome Res 23:1008–1017
Gurr GM, You M (2015) Conservation biological control of pests in the molecular era: new opportunities to address old constraints. Front Plant Sci 6:1255
Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, Du Y, Tate PH, Huang X, Zhang W (2014) Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol 11:829–835
Hansen K, Coussens MJ, Sago J, Subramanian S, Gjoka M, Briner D (2012) Genome editing with CompoZr custom zinc finger nucleases (ZFNs). J Vis Exp 64:e3304
Hayashi S, Wakasa Y, Ozawa K, Takaiwa F (2016) Characterization of IRE1 ribonuclease-mediated mRNA decay in plants using transient expression analyses in rice protoplasts. New Phytol 210:1259–1268
He Z, Proudfoot C, Mileham AJ, McLaren DG, Whitelaw CB, Lillico SG (2015) Highly efficient targeted chromosome deletions using CRISPR/Cas9. Biotechnol Bioeng 112:1060–1064
Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A (2015) Optical control of CRISPR/Cas9 gene editing. J Am Chem Soc 137:5642–5645
Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B (2013) Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140:4982–4987
Hutter G, Bodor J, Ledger S, Boyd M, Millington M, Tsie M, Symonds G (2015) CCR5 targeted cell therapy for HIV and prevention of viral escape. Viruses 7:4186–4203
Ikeda T, Tanaka W, Mikami M, Endo M, Hirano HY (2016) Generation of artificial drooping leaf mutants by CRISPR–Cas9 technology in rice. Genes Genet Syst 90:231–235
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun 467:76–82
Jabalameli HR, Zahednasab H, Karimi-Moghaddam A, Jabalameli MR (2015) Zinc finger nuclease technology: advances and obstacles in modelling and treating genetic disorders. Gene 558:1–5
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jiang W, Yang B, Weeks DP (2014) Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 9:e99225
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. Elife 2:e00471
Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997
Jinkerson RE, Jonikas MC (2015) Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J 82:393–412
Johnson RA, Gurevich V, Levy AA (2013) A rapid assay to quantify the cleavage efficiency of custom-designed nucleases in planta. Plant Mol Biol 82:207–221
Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20:856–874
Karcher D, Kahlau S, Bock R (2008) Faithful editing of a tomato-specific mRNA editing site in transgenic tobacco chloroplasts. RNA 14:217–224
Kim H, Kim JS (2014) A guide to genome engineering with programmable nucleases. Nat Rev Genet 15:321–334
Kumar V, Jain M (2015) The CRISPR–Cas system for plant genome editing: advances and opportunities. J Exp Bot 66:47–57
Lee J, Chung JH, Kim HM, Kim DW, Kim H (2016) Designed nucleases for targeted genome editing. Plant Biotechnol J 14:448–462
Li XJ, Zhang YF, Hou M, Sun F, Shen Y, Xiu ZH, Wang X, Chen ZL, Sun SS, Small I, Tan BC (2014) Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J Cell Mol Biol 79:797–809
Li JF, Zhang D, Sheen J (2015a) Targeted plant genome editing via the CRISPR/Cas9 technology. Methods Mol Biol 1284:239–255
Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015b) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169:960–970
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41:63–68
Liao JC, Hsieh WY, Tseng CC, Hsieh MH (2016) Dysfunctional chloroplasts up-regulate the expression of mitochondrial genes in Arabidopsis seedlings. Photosynth Res 127:151–159
Lin CP, Ko CY, Kuo CI, Liu MS, Schafleitner R, Chen LF (2015) Transcriptional slippage and RNA editing increase the diversity of transcripts in chloroplasts: insight from deep sequencing of Vigna radiata genome and transcriptome. PLoS ONE 10:e0129396
Lowder LG, Zhang D, Baltes NJ, Paul JW 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985
Luo Y, Liu C, Cerbini T, San H, Lin Y, Chen G, Rao MS, Zou J (2014) Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Transl Med 3:821–835
Ma L, Zhu F, Li Z, Zhang J, Li X, Dong J, Wang T (2015) TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds. PLoS ONE 10:e0143877
Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA 108:2623–2628
Martin-Ortigosa S, Peterson DJ, Valenstein JS, Lin VS, Trewyn BG, Lyznik LA, Wang K (2014) Mesoporous silica nanoparticle-mediated intracellular Cre protein delivery for maize genome editing via loxP site excision. Plant Physiol 164:537–547
Marton I, Zuker A, Shklarman E, Zeevi V, Tovkach A, Roffe S, Ovadis M, Tzfira T, Vainstein A (2010) Nontransgenic genome modification in plant cells. Plant Physiol 154:1079–1087
Marton I, Honig A, Omid A, De Costa N, Marhevka E, Cohen B, Zuker A, Vainstein A (2013) From Agrobacterium to viral vectors: genome modification of plant cells by rare cutting restriction enzymes. Int J Dev Biol 57:639–650
Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6:243
Mikami M, Toki S, Endo M (2015a) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88:561–572
Mikami M, Toki S, Endo M (2015b) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34:1807–1815
Mussolino C, Mlambo T, Cathomen T (2015) Proven and novel strategies for efficient editing of the human genome. Curr Opin Pharmacol 24:105–112
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123
Nejat N, Rookes J, Mantri NL, Cahill DM (2016) Plant–pathogen interactions: toward development of next-generation disease-resistant plants. Crit Rev Biotechnol 37:229
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM (2014) TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Nat 6:19–40
Nicolia A, Proux-Wera E, Ahman I, Onkokesung N, Andersson M, Andreasson E, Zhu LH (2015) Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J Biotechnol 204:17–24
Osakabe Y, Osakabe K (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol 56:389–400
Ousterout DG, Kabadi AM, Thakore PI, Perez-Pinera P, Brown MT, Majoros WH, Reddy TE, Gersbach CA (2015) Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol Ther 23:523–532
Peng R, Lin G, Li J (2015) Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J 283:1218–1231
Petersen B, Niemann H (2015a) Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN). Chromosome Res 23:7–15
Petersen B, Niemann H (2015b) Molecular scissors and their application in genetically modified farm animals. Transgenic Res 24:381–396
Petolino JF (2015) Genome editing in plants via designed zinc finger nucleases. In Vitro Cell Dev Biol Plant 51:1–8
Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57:629–637
Qin Y, Gao WQ (2016) Concise review: patient-derived stem cell research for monogenic disorders. Stem Cells 34:44–54
Redel BK, Prather RS (2015) Meganucleases revolutionize the production of genetically engineered pigs for the study of human diseases. Toxicol Pathol 44:428–433
Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji JY, Li JB, Ni JQ (2014) Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep 9:1151–1162
Reyon D, Khayter C, Regan MR, Joung JK, Sander JD (2012) Engineering designer transcription activator-like effector nucleases (TALENs) by REAL or REAL-Fast assembly. Curr Protoc Mol Biol. https://doi.org/10.1002/0471142727.mb1215s100
Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, Federici F, Sinha N, Deal RB, Bailey-Serres J, Brady SM (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166:455–469
Sauer NJ, Mozoruk J, Miller RB, Warburg ZJ, Walker KA, Beetham PR, Schopke CR, Gocal GF (2016) Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol J 14:496–502
Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, Muranaka T, Saito K, Umemoto N (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26:3763–3774
Schaeffer SM, Nakata PA (2015) CRISPR/Cas9-mediated genome editing and gene replacement in plants: transitioning from lab to field. Plant Sci 240:130–142
Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80:1139–1150
Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13:653–658
Scott JN, Kupinski AP, Boyes J (2014) Targeted genome regulation and modification using transcription activator-like effectors. FEBS J 281:4583–4597
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410
Shan Q, Zhang Y, Chen K, Zhang K, Gao C (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J 13:791–800
Sprink T, Metje J, Hartung F (2015) Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr Opin Biotechnol 32:47–53
Steentoft C, Bennett EP, Schjoldager KT, Vakhrushev SY, Wandall HH, Clausen H (2014) Precision genome editing: a small revolution for glycobiology. Glycobiology 24:663–680
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84:1295–1305
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62–67
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y (2015) Targeted mutagenesis in soybean using the CRISPR–Cas9 system. Sci Rep 5:10342
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L (2016) Engineering herbicide resistant rice plants through CRISPR/Cas9-mediated homologous recombination of the acetolactate synthase. Mol Plant 9:628
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169:931–945
Tang F, Yang S, Liu J, Zhu H (2016) Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol 170:26–32
Tingting L, Di F, Lingyu R, Yuanzhong J, Rui L, Keming L (2015) Highly efficient CRISPR/Cas9-mediated targeted mutagenesis of multiple genes in Populus. Yi Chuan 37:1044–1052
Tovkach A, Zeevi V, Tzfira T (2009) A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J 57:747–757
Tsai CJ, Xue LJ (2015) CRISPRing into the woods. GM Crops Food 6:1–10
Tzfira T, Weinthal D, Marton I, Zeevi V, Zuker A, Vainstein A (2012) Genome modifications in plant cells by custom-made restriction enzymes. Plant Biotechnol J 10:373–389
Ul Ain Q, Chung JY, Kim YH (2015) Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J Control Release 205:120–127
Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15:57–61
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3:2233–2238
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD (2010) Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11:636–646
Vaitilingom M, Stupar M, Grienenberger JM, Gualberto JM (1998) A gene coding for an RPS2 protein is present in the mitochondrial genome of several cereals, but not in dicotyledons. Mol Gen Genet 258:530–537
van Tol N, van der Zaal BJ (2014) Artificial transcription factor-mediated regulation of gene expression. Plant Sci 225:58–67
Vazquez-Vilar M, Bernabe-Orts JM, Fernandez-Del-Carmen A, Ziarsolo P, Blanca J, Granell A, Orzaez D (2016) A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 12:10
Waaijers S, Portegijs V, Kerver J, Lemmens BB, Tijsterman M, van den Heuvel S, Boxem M (2013) CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195:1187–1191
Wang M, Liu Y, Zhang C, Liu J, Liu X, Wang L, Wang W, Chen H, Wei C, Ye X, Li X, Tu J (2015) Gene editing by co-transformation of TALEN and chimeric RNA/DNA oligonucleotides on the rice OsEPSPS gene and the inheritance of mutations. PLoS ONE 10:e0122755
Weeks DP, Spalding MH, Yang B (2016) Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J 14:483–495
Weinthal D, Tovkach A, Zeevi V, Tzfira T (2010) Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci 15:308–321
Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ (2011) Targeted genome editing across species using ZFNs and TALENs. Science 333:307
Wright DA, Li T, Yang B, Spalding MH (2014) TALEN-mediated genome editing: prospects and perspectives. Biochem J 462:15–24
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR–Cas system. Mol Plant 6:1975–1983
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Xu R, Zhang S, Huang J, Zheng C (2013) Genome-wide comparative in silico analysis of the RNA helicase gene family in Zea mays and Glycine max: a comparison with Arabidopsis and Oryza sativa. PLoS ONE 8:e78982
Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5:11491
Yan W, Smith C, Cheng L (2013) Expanded activity of dimer nucleases by combining ZFN and TALEN for genome editing. Sci Rep 3:2376
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang B, Sun Q, Li H (2015a) Advances in genetic modification technologies. Sheng Wu Gong Cheng Xue Bao 31:1162–1174
Zhang G, Lin Y, Qi X, Li L, Wang Q, Ma Y (2015b) TALENs-assisted multiplex editing for accelerated genome evolution to improve yeast phenotypes. ACS Synth Biol 4:1101–1111
Zhang Z, Mao Y, Ha S, Liu W, Botella JR, Zhu JK (2015c) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep 35:1519
Zhang B, Yang X, Yang C, Li M, Guo Y (2016a) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in Petunia. Sci Rep 6:20315
Zhang H, Gou F, Zhang J, Liu W, Li Q, Mao Y, Botella JR, Zhu JK (2016b) TALEN-mediated targeted mutagenesis produces a large variety of heritable mutations in rice. Plant Biotechnol J 14:186–194
Zheng Z, Bao M, Wu F, Chen J, Deng X (2016) Predominance of single prophage carrying a CRISPR/cas system in “Candidatus Liberibacter asiaticus” strains in Southern China. PLoS ONE 11:e0146422
Zhou J, Peng Z, Long J, Sosso D, Liu B, Eom JS, Huang S, Liu S, Vera Cruz C, Frommer WB, White FF, Yang B (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J 82:632–643
Zlotorynski E (2015) Plant cell biology: CRISPR–Cas protection from plant viruses. Nat Rev Mol Cell Biol 16:642
Acknowledgements
We thank our colleagues in molecular genetics and genomics who helped us during the preparation of this manuscript. We express our great thanks to Drs. M. Lauressergues, A. Omidbakhshfard, M. Page, W. Thompson, and D. Whitley for their critical reading and valuable comments that improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Additional information
Project funding: None.
The online version is available at http://www.springerlink.com
Corresponding editor: Chai Ruihai.
Rights and permissions
About this article
Cite this article
Tang, W., Tang, A.Y. Genome engineering technologies for targeted genetic modification in plants. J. For. Res. 29, 875–887 (2018). https://doi.org/10.1007/s11676-017-0588-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-017-0588-z