Abstract
Retroviruses comprise an ancient and varied group of viruses with the unique ability to integrate DNA from an RNA transcript into the genome, a subset of which are able to integrate in humans. The timing of these integrations during human history has dictated whether these viruses have remained exogenous and given rise to various human diseases or have become inseparable from the host genome (endogenous retroviruses). Given the ability of retroviruses to integrate into the host and subsequently co-opt host cellular process for viral propagation, retroviruses have been shown to be closely associated with several cellular processes including exosome formation. Exosomes are 30-150 nm unilamellar extracellular vesicles that originate from intraluminal vesicles (ILVs) that form in the endosomal compartment. Exosomes have been shown to be important in intercellular communication and immune cell function. Almost every cell type studied has been shown to produce these types of vesicles, with the cell type dictating the contents, which include proteins, mRNA, and miRNAs. Importantly, recent evidence has shown that infection by viruses, including retroviruses, alter the contents and subsequent function of produced exosomes. In this review, we will discuss the important retroviruses associated with human health and disease. Furthermore, we will delve into the impact of exosome formation and manipulation by integrated retroviruses on human health, survival, and human retroviral disease pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retroviruses first emerged into the general public in the 1980s during the beginnings of the AIDS epidemic. This disease is caused by Human Immunodeficiency Virus (HIV), which is a type of virus that requires reverse transcription of RNA to DNA and integration into the host DNA for successful infection. Indeed, while HIV is the best known and most recent retrovirus to enter the human population, retroviruses comprise a unique and diverse family of enveloped single-stranded RNA viruses that evidence suggests has a long evolutionary history dating back almost 450 million years ago in vertebrates (Aiewsakun and Katzourakis 2017; Hughes and Coffin 2005). Given the ancient history of retroviruses and their ability to integrate in the host genome, there have been instances when integrations have occurred in the germline, giving rise to permanent viral elements in the human genome. These elements, termed endogenous retroviruses, comprise almost 8% of the human genome (Griffiths 2001).
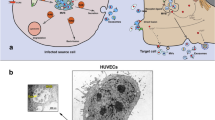
The close association between retroviruses and humans has allowed both endogenous and exogenous retroviruses to enter many human processes, both natural and pathogenic such that these viruses are thought to be involved in pregnancy, autoimmunity, cancer, and inflammatory disorders to name a few (Cloyd 1996; Ryan 2004). Indeed, retroviruses as a whole are large determinants of human morbidity and mortality. They have also become associated with intrinsic cellular processes such as genetic transfer (transposons) and intercellular communication (Izquierdo-Useros et al. 2011; Kitamura et al. 2003; Mittelbrunn and Sanchez-Madrid 2012). In particular, cells form extracellular vesicles including exosomes which are 30-150 nm in size to deliver messages as mRNA, miRNA, and proteins to local and distal sites as a form intercellular communication. This review will focus on the retroviruses associated with human health and mortality, as well as their association with exosomes and how the current understanding of this association impacts retroviral pathology.
Retroviruses
Retrovirues are RNA viruses that can reverse transcribe into the host-genome via a DNA intermediate, which can later serve as the template for viral mRNA and proteins. The family of retroviridae is split into two genera: orthoretrovirinae and spumaretrovirinae. All viruses known to cause to disease in humans and other animals are within the orthoretrovinae subfamilies. This subfamily is further classified into genera alpharetrovirus, betaretrovirus, delta retrovirus, epsilonretrovirus, gammaretrovirus, and lentivirus (Retrovidae 2012). Alpharetrovirus are not known to infect humans and include avian leukovirus and rous sarcoma virus. Betaretrovirus are also not known to infect humans but can infect some animals. This subfamily includes Langur virus and squirrel monkey retrovirus. Deltaretrovirus includes the important human pathogen Human T cell Lymphotropic virus Type 1 (HTLV-1) as well as HTLV-2, HTLV-3 and Bovine Leukemia Virus (BLV). Both episolonretrovirus and gammaretrovirus are not associated with human infection and include Walleye dermal sarcoma virus and feline leukemia virus respectively. Initial research had classified Xenotropic murine leukemia virus-related virus (XMRV) as a human pathogen within the subfamily of gammaretrovirus (Denner 2010). XMRV is now understood to be a result of a recombinant event and laboratory contaminant (Arias and Fan 2014). The subfamily of lentivirus is a large determinant of human disease, specifically due to Human Immunodeficiency virus (HIV), however it also includes HIV-2, feline immunodefiency virus (FIV) and simian immunodefiency virus (SIV). Alpha, beta, gamma and epsilonretroviruses are classified as simple while delta, lenti and spuma are classified as complex.
Endogenous retroviruses (ERV) comprise approximately 50 groups and are distributed amongst gamma-like, beta-like, epsilon-like and spuma-like groups (Escalera-Zamudio and Greenwood 2016). Well characterized human ERV (HERV) such as HERV-W and HERV-K (HML2) are classified in gamma and betaretrovirus respectively (Table 1).
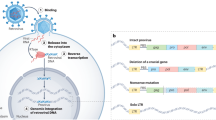
All retroviruses have a general composition of 2 single-stranded genomic RNA (gRNA) strands within a capsid surrounded by an envelope. Most particles measure approximately 100 nm in diameter and carry group-specific antigen (gag: capsid, nucleocapsid, matrix) as well as retrotranscriptase (RT), integrase (IN), and the viral protease (PR) (Kannian and Green 2010). While each individual retrovirus has specific cellular tropisms, they infect by attaching to the cell membrane via adhesion to a cellular receptor to viral env, followed by membrane fusion and viral entry (Kannian and Green 2010). Upon entry, the virus reverse transcribes the gRNA to dsDNA and integrates into the host genome as a provirus within the nucleus. There are multiple integration sites between viruses and certain viruses may favor integration into specific regions or have a non-redundant integration pattern when evaluating multiple infections (Desfarges and Ciuffi 2010; Serrao and Engelman 2016). Since retroviruses can potentially integrate anywhere in the human genome, they are de facto protooncogenic due to mutagenesis that occurs with insertions into genes or promoters (Downey et al. 2015). Once integrated into the host genome, viral gene transcription can be initiated from the long terminal repeats (LTR) at either end of the provirus. Viral gene transcription is tightly controlled, both by the virus and the host.
Viral gene expression and organization differs between simple and complex retroviruses. The RNA transcript encodes gag, pro, pol, and env but complex retroviruses also encode additional regulatory genes through alternate gene splicing. As an example, HTLV-1 also encodes tax, a pleiotropic regulatory gene. Reverse transcriptase and integrase are products of the pol coding region while the protease is a product of the pro coding region. Some of these genes, such as gag, pol and env are involved in viral particle assembly as well as integration into the host genome. Due to the small size of viral genome, several host genes are also involved in these processes. Indeed, retroviruses like other viruses are dependent on host processes for further propagation. Prior thinking restricted this to cooption of host transcription factors and DNA and RNA transcription machinery. However, in recent years viruses have also been noted to take advantage of another cellular process for viral propagation and/or escaping immune surveillance through the packaging of macromolecules into extracellular vesicles for distribution outside the cell.
Extracellular vesicles (EVs) are vesicles released by cells into the extracellular space. They comprise both microvesicles (MVs) which originate from the plasma membrane and range from 100 nm–1000 nm, apoptotic bodies which are the result of dying cells blebbing and compartmentalizing (1μm-5μm), and exosomes which range from 30 nm-100 or 150 nm (Helwa et al. 2017; Zeringer et al. 2015). Exosomes are intraluminal vesicles (ILVs) that formed in the endosomal compartment and are released into the extracellular space after fusion of multivesicular bodies (MVBs) with the plasma membrane instead of fusion with lysosomes for degradation (Bissig and Gruenberg 2014; Robbins and Morelli 2014). In a process called “back fusion”, ILVs deliver plasma membrane invaginations (through clathrin-mediated and clathrin-independent endocytosis) to the endosomal network, making them (and therefore exosomes) capable of carrying both intracellular and extracellular products (Alenquer and Amorim 2015; Nour and Modis 2014).
The exact intracellular signals that direct ILVs to the plasma membrane for release are still under investigation. Endosomal sorting complexes required for transport (ESCRT) machinery has been studied in its role for directing ubiquitin-labeled proteins into endosomes for delivery into MVBs, and, as such, ESCRT proteins, like Alix and TSG101, are enriched in exosomes (Matsuo et al. 2004). Lipid raft-associated proteins such as transferrin and caveolins, and other proteins involved in membrane trafficking, like the tetraspanins CD9, CD63, and CD81, which have been shown to bind to ESCRT machinery, are also enriched in exosomes (Meckes and Meckes Jr and Raab-Traub 2011; Sampey et al. 2014).
However, ESCRT is not the only method by which exosome formation can occur as there are also ESCRT independent methods by which proteins, lipids, and RNA can enter the endosomal pathway. For example, oligodendrocytes direct exosome formation via the ceramide pathway (Trajkovic et al. 2008), while other cell types rely on oligomerization of tetraspanin complexes (Perez-Hernandez et al. 2013; van Niel et al. 2011). Furthermore, while knockdown of some ESCRT components may abrogate exosome production, it does not completely knock it out (Stuffers et al. 2009; Tamai et al. 2010). Indeed, Rab GTPases, a known family of conserved proteins that regulate vesicular trafficking and membrane fusion events, are also involved in exosome formation as denoted by their high abundance in isolated exosomes (Schorey et al. 2015; Tamai et al. 2010). Several are implicated in the release of exosomes, including Rab11, Rab27, Rab5, Rab35, and Rab7, depending on cell type (Alenquer and Amorim 2015; Schorey et al. 2015). Rab 27a, in particular, regulates the fusion of MVBs at the plasma membrane to release ILVs (Ostrowski et al. 2010; Schorey et al. 2015). Knockdown of Rab 27a inhibits exosome secretion from tumor cell lines (Ostrowski et al. 2010; Robbins and Morelli 2014). There are several other Rabs that have also proven essential through a diminution in exosome levels after their knockdown, including Rab 2B, Rab9A, Rab5A, and Rab27b (Robbins and Morelli 2014). Being GTPases, the activation of each Rab is dependent on an influx of calcium, as is the case for Rab 11 in the K562 cell line, which may involve SNARE complexes (Colombo et al. 2014; Fader et al. 2009; Savina et al. 2005). Altogether, there are clearly several players within the cell that contribute to the endosomal compartment and, ultimately, to the release of exosomes, further emphasizing the importance of this pathway in normal biology.
The contents of exosomes vary based on the cell of origin but can include miRNAs, proteins, mRNAs, enzymes, lipids, and carbohydrates. Almost every cell investigated has been found to be capable of producing exosomes. The cargo of exosomes released by regulatory T cells will necessarily differ from the exosomes released by dendritic cells or activated T cells. Indeed, while immunosuppressive exosomes have largely been attributed to the Tregs, other cells can produce exosomes that help reduce inflammation like mesenchymal stem cells (MSCs) (Baglio et al. 2012; Narayanan et al. 2013). Since exosomes are a reflection of the content and function of the cells from which they arise, they have become another tool to assess the state of the host. Indeed, there is an ever increasing number of studies utilizing exosomes as biomarkers in cancer, autoimmune disease and neurodegenerative disorders (Lin et al. 2015; Perez-Hernandez and Cortes 2015; Properzi et al. 2013).
Given their important role in intracellular communication, methods of exosome isolation have been heavily investigated. Their microscopic size and potential overlap with other microscopic organisms has necessitated careful isolation methods. Traditionally, serial steps of ultracentrifugation have been favored for the removal of these vesicles from fluid preparations. However, this method is time consuming, volume limiting, wasteful, and can often result in exosomes that are altered/ damaged (Lamparski et al. 2002; Zeringer et al. 2015). As such other technologies have been employed that utilize size, charge, and chemical means for isolation of exosomes. For example, there are certain products, like ExoQuick®, which allow for polymer-based exosomal precipitation while other technologies allow for bead-based immunonologic separation of exosomes based on tetraspanin expression (Rider et al. 2016). Still other technologies, such as qEV® columns, separate exosomes based on chromatography and filtration (Lezin et al. 2005). Each method has its advantages and disadvantages in the ability to specifically isolate exosomes separately from other microvesicles (Helwa et al. 2017). Another method utilizing synthetic hydrogel nanoparticles called Nanotrap particles ® has demonstrated specific isolation of exosomes. The method of isolation requires size and charge exclusion as well as binding to sugars on tetraspanins (unpublished), which may explain the increased specificity of the technology.
Exosomes in Viral Infection
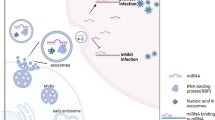
Considering that the function of released exosomes is dependent on the cargo and receptors on the exosomes, it follows that any alteration in the cell contents of the exosome-producing cell would alter the contents of released exosomes. Indeed, infected cells have been shown to vastly alter exosomal contents. Hepatitis C Virus (HCV) infection represents a well-documented case of viral coopting of exosomal communication for the delivery of viral components (Anderson et al. 2014). It is well known that the HCV viral genome can remain in ILVs and be secreted within exosomes, where they can operate as infectious particles (Longatti et al. 2015; Ramakrishnaiah et al. 2013). By using a transwell assay, Longatti et al. showed that they were able to infect Hu7 cells after exposure to these shed exosomes without the need for direct cell–cell contact. Further, this infection was inhibited by blocking exosomal release with a sphingomyelinase inhibitor. Other pathogens are closely associated with the Rab GTPases and other components of the ESCRT pathway which allows for incorporation of viral components into developing exosomes. For example, HIV Gag has been shown to interact with tetraspanins, especially CD63 and CD81, to aid in virion egress (Madison and Okeoma 2015).
Trafficking of Antigens into Exosomes
Understanding how viral components incorporate into exosomes requires knowledge of how macromolecules can be generally directed to this pathway. Further elaborating on the ESCRT pathway, the endosomal sorting complexes required for transport (ESCRT) is actually composed of four sets of machinery referred to as ESCRT 0, I, ESCRT II, and ESCRT III (Urbanelli et al. 2013). ESCRT 0 recognizes ubiquinated proteins and recruits them for endosomal sorting through interaction of its ubiquitin-binding Hrs FYVE domain with phosphatidyl inositol 3-phosphate (PI3P) (Henne et al. 2011). ESCRT 1 is then recruited through tumor susceptibility gene 101(TSG101) interaction with the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) PSAP of ESCRT 0 (Schmidt and Teis 2012). ESCRT 1 then recruits ESCRT II proteins which then recruit ESCRT III, which is stabilized by the recruitment of Alix (ALG-2 interacting protein X) (Villarroya-Beltri et al. 2014). For the complex to dissociate from the plasma membrane requires energy in the form of ATP. This is provided by the ATPase VsP4 (Henne et al. 2011).
While ubiquitination is a tool for protein sorting into ESCRT-dependent exosomes, there are other mechanisms as well. Syndecans, which are a main source of heparin sulfate in the cell membrane, bind to syntenin which can interact with CD63 and Alix (Villarroya-Beltri et al. 2014). Syndecans possess lateral heparin sulfate polysacharide chains which can be cleaved into shorter chains by heparanse activity in the endosomes (Roucourt et al. 2015). Shorter heparain sulfate chains condense and cluster leading to syndecan oligermerization, which appears to allow for syntenin binding in a cargo-dependent manner (Stoorvogel 2015). This ultimately allows for sorting into endosomes. Indeed, silencing of either syntenin or syndecans reduced exosome production (Baietti et al. 2012). Therefore, any protein that can bind to syndecans can also sort to exosomes. This suggest that ubiquitination, which is usually the first step in the ESCRT pathway, is not strictly necessary for sorting of cellular proteins into exosome.
In addition to ESCRT dependent pathways, exosomes are also known to form through ceramide oligomerization. Ceramide can cause spontaneous bending and coalescence of microdomains in endosomal membranes (Trajkovic et al. 2008). Furthermore, T cell CD63+ exosome shuttling to APCs involved ceramide synthesis (Mittelbrunn et al. 2011). Tetraspanin clustering in tetraspanin-enriched domains (TEMs) also appears to be involved in exosome formation and protein sorting. In particular, there are a number of viral proteins known to preferentially sort to exosomes through interactions with tetraspanins. In particular, EBV LMP1 binds to CD63 and sorts to exosomes (Verweij et al. 2011). Separately, CD81 plays an important role in exosome composition through interactions of its cytoplasmic domain (Andreu and Yanez-Mo 2014). Another cited mechanism for protein sorting into exosomes is through higher order oligomerization (oligomerization of oligomers) of cytoplasmic proteins in a process similar to pole formation in polarized cells which is termed immediate mode exosome biogenesis (Fang et al. 2007). However, this mechanism was shown to be important in protein sorting to shedding vesicles from the plasma membrane, although the authors noted that it is often difficult to differentiate these from endosomally derived vesicles (Shen et al. 2011). Additionally, it has been shown that acylation serves as another method of protein tagging for export in microvesicles (Fang et al. 2007).
RNA localization to exosomes involves additional mechanisms. It has been observed that miRNAs carry specific EXOmotifs that allow for preferential packaging into exosomes, as enrichment of specific miRNAs in exosomes has been observed in human studies (Villarroya-Beltri et al. 2013). These motifs facilitate binding to heterogeneous ribonucleoprotein A2B1 (hnRNPA2b1), which is preferentially sumoylated in exosomes (Villarroya-Beltri et al. 2013). It is an RNA binding and transport protein known to be particularly important in RNA trafficking in neurons (Han et al. 2010). Additionally, Annexin-2 may also play a role in RNA sorting into exosomes (Hagiwara et al. 2015) (Filipenko et al. 2004). Sorting of mRNAs into exosomes seems to involve binding to the mRNA 3’UTR and the short motif CTGCC (Villarroya-Beltri et al. 2014). Interestingly, mRNA sorting also appeared to be dependent on certain miRNAs (Zhang et al. 2015).
As has been illustrated, the mechanisms involved in the formation of exosomes and the steps leading to macromolecule incorporation prior to their release is still under investigation. An overview of retroviruses and specifically important human pathogens in the retrovirus family have been given to exemplify their importance in human pathology. Furthermore, examples of how viral antigens can be incorporated at each step of exosome formation has been briefly illustrated. For the remainder of this review, how retroviruses specifically become associated with exosomes and how this process aids or alters retrovirus-associated diseases will be discussed through the examples of HIV, HTLV-1 and endogenous retroviruses.
HIV
As mentioned HIV, a lentivirus, is the most recent retrovirus to enter the human population. Most agree that infection in humans likely occurred in the early twentieth century through exposure to SIV from chimps and eventual mutation to an efficient human pathogen (Sharp and Hahn 2011). While AIDS was not recognized as a disease until the 1980s, HIV retains many of the mechanisms for infection and egress that are seen with SIV infection. This includes close association with exosome formation machinery for virion particle release. Indeed, SIV has also been shown to incorporate viral antigens into produced exosomes (Yelamanchili et al. 2015). In particular, both viruses require interactions with cellular factors like TSG101 and Alix for viral budding, both of which are involved in exosome biogenesis (Fisher et al. 2007; Jesus da Costa et al. 2009). Additionally, HIV-1 tends to favor budding from tetraspanin-enriched microdomains (TEM) containing CD9, CD63, CD81, and CD82 (Krementsov et al. 2009; Thali 2009). It has even been reported that tetraspanins can be present in the envelope of budding HIV-1 (Thali 2011; Tremblay et al. 1998), although there are conflicting reports as to the significance of this finding. Some research supports increased particle release with incorporation of tetraspanins while other reports were equivocal in the change to virion budding with tetraspanins (Krementsov et al. 2009; Thali 2009). Most agree that tetraspanins present on the virion membrane did have a negative effect on subsequent Env-induced virus-cell fusion (Gordon-Alonso et al. 2006).
Importantly, HIV virion assembly and egress closely resembles exosome biogenesis (Pelchen-Matthews et al. 2003), which may explain why HIV has been noted to take advantage of several steps in exosome biogenesis. Gould et al. (Gould et al. 2003) hypothesize that with the similarities between HIV assembly and egress and exosome biogenesis, HIV has evolved to co-opt the exosome system and infect cells through packaging of the viral genome in the “Trojan horse” hypothesis (Gould et al. 2003). It is a theory supported by observations that HIV virions are released with exosomes and have enhanced infectivity in the presence of these vesicles (Wiley and Gummuluru 2006). However, this mechanism occurred via uptake by dendritic cells (DCs), which subsequently transferred endocytosed HIV to closely associated uninfected T cells (Piguet and Steinman 2007). Specifically, HIV is endocytosed via DC sign into the endocytic pathway and trafficked back to the cell surface in intact DCs for presentation to T cells. While direct packaging of HIV genomic RNA into exosomes has been observed in the U937 cell line, it was not infectious (Columba Cabezas and Federico 2013) and this observation has not been made in vivo. Furthermore, evidence supports HIV virion budding from the plasma membrane (Sundquist and Krausslich 2012), which would more closely resemble the release of microvesicles than exosomes (Raposo and Stoorvogel 2013).
Nevertheless, besides close association with components of the exosome biogenesis machinery, HIV has also been noted to preferentially incorporate mRNA, protein and miRNA into released exosomes. For example, HIV has been shown to traffic transactivating response element (TAR) RNA into exosomes (Narayanan et al. 2013; Sampey et al. 2016). TAR RNA is a pre-miRNA necessary for activation of the viral promoter and viral replication through binding to HIV Tat protein and interaction with the LTR promoter (Das et al. 2011; Swaminathan et al. 2014). The uptake of exosomal TAR in recipient cells can downregulate apoptosis and is postulated to have a role in supporting HIV infection (Narayanan et al. 2013). Importantly, TAR RNA was still able to be detected in exosomes isolated from the serum of HIV-positive patients on highly active antiretroviral therapy, indicating that even with antiretroviral therapy, short transcripts remain present in these exosomes (Jaworski et al. 2014). Indeed, the same group later went on to find that exosomal TAR RNA could stimulate proinflammatory cytokines in recipient cells through activation of the nuclear factor kappa b (NFκB) pathway (Sampey et al. 2016).
In a separate study, the HIV Nef protein was found in released exosomes (Campbell et al. 2008). Later uptake of these Nef+ exosomes led to increased susceptibility of naïve T cells to HIV infection (Arenaccio et al. 2014; Campbell et al. 2008). Indeed, another report found that exosomes from HIV-infected cells could reactivate HIV in latently infected cells (Arenaccio et al. 2015). This is similar to an observation in SIV where plasma-derived exosomes could reactivate resting CD4+ T cells (Hong, Schouest, and Xu. Scientific Reports Nov 2017). Exosomal Nef has also been shown to increase T-cell apoptosis in vitro, which may contribute to the CD4+ T-cell depletion in AIDS pathogenesis (Lenassi et al. 2010). Interestingly, Nef expression in CD4+ T cells was also noted to decrease CD4 and MHC I export to released exosomes (de Carvalho et al. 2014). The authors postulated that this decreases the ability of CD4+ T cells to inhibit HIV infection in uninfected cells by using exosomes as decoys to soak up HIV virions. This would further explain how Nef+ exosomes enhance HIV infectivity. More recently, Luo disputed the incorporation of Nef into exosomes at all (Luo et al. 2015), despite several reports finding motifs within Nef that are necessary for exosomal incorporation (Ali et al. 2010; Campbell et al. 2012). Nef may traffic to exosomes through association with lipid-raft domains. Moreover, transfection of cell lines with Nef increased exosome production (Madison and Okeoma 2015).
In addition to HIV TAR RNA, gRNA, and proteins, HIV-deried miRNAs can also be detected in exosomes isolated from cell line cultures and HIV positive individuals. In particular, HIV-1 vmiRTAR, vmiRT88, vmiRT99 can be found in exosomes isolated from the blood of HIV positive individuals (Bernard et al. 2014). The authors showed that the presence of vmi88 and vmi99 in released exosomes could stimulate the release of TNFα from macrophages through TLR8 signaling while separately vmiRTAR in exosomes reduced Bim and Cdk9 expression in target cells leading to their reduced apoptosis (Madison and Okeoma 2015).
It is clear that HIV has evolved mechanisms to alter the cellular microenvironment to its advantage through exosomal cellular communication. Compounded with data indicating the presence of exosomes containing HIV proteins in the serum of HIV patients undergoing highly active antiretroviral therapy (HAART), it becomes apparent that incorporation of viral components into released exosomes may play a role in HIV infection and viral persistence (Jaworski et al. 2014). Indeed, the ability to detect viral antigens in exosomes even in the absence of viral detection has yielded increased interest in exosomal detection in HIV associated neurologic disorders (HAND), as well as in other disorders in which immune responses to a pathogen are suspected but where the pathogen is difficult to detect.
HTLV
Like HIV, there are several other viruses that also target viral RNAs and proteins for exosomal export. HTLV-1, another human retrovirus and the cause of adult T-cell leukemia and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), has also been shown to incorporate viral proteins into shed exosomes (Anderson et al. 2016). Unlike HIV, HTLV-1 has been in the human population for millennia (Li et al. 1999; Novak 1999; Verdonck et al. 2007), likely explaining the low incidence of human disease associated with infection (less than 5% for both ATLL and HAM/TSP). Jaworski and colleagues found that HTLV-1-infected cell lines shed exosomes containing Tax (Jaworski et al. 2014), a pleiotropic transactivating protein implicated in the immune dysregulation associated with infection (Currer et al. 2012; Romanelli et al. 2013). Tax appears to be targeted for exosome entry by ubiquitination (Jaworski et al. 2014; Shembade and Harhaj 2010),which was noted earlier to be an important method for protein trafficking using ESCRT machinery. Indeed, prior studies have shown Tax colocalization with organelles undergoing exocytosis (Alefantis et al. 2005; Alefantis et al. 2005). Exosomes shed from HTLV-1+ cell lines were found to also contain viral mRNA and miRNAs such as tax and hbz (Jaworski et al. 2014). Additionally, exosomes shed from HTLV-1+ infected cell lines showed a different cell miRNA profile, as well as a unique set of host proteins and lipids, compared to those shed by uninfected cell lines. In work from our laboratory in collaboration with Dr. Fatah Kashanchi, it was further demonstrated that HTLV-1 Tax could be found in exosomes isolated from the cerebrospinal fluid of some patients with HAM/TSP, while exosomes from uninfected controls were negative (in submission). Similarly, cultured, unstimulated peripheral blood mononuclear cells (PBMCs) from patients with HAM/ TSP were shown to shed exosomes that contained Tax protein, as well as tax mRNA. HBZ protein could also be detected in PBMC-derived exosomes although no hbz mRNA could be found in these exosome samples. While both tax and hbz mRNA are expressed by HAM/TSP patient PBMCs in culture, their expression diverges in scale, time, and localization (Li et al. 2009; Rende et al. 2011). It was thought that the absence of hbz mRNA in isolated exosomes likely results from hbz mRNA preferential localization to the nucleus, whereas tax mRNA can be found equally in both the nucleus and cytoplasm. Additionally, HBZ protein preferentially localizes to the cytoplasm in HAM/TSP patients (Baratella et al. 2017).
These findings may have functional consequences as HTLV-1 is a cell-associated virus and shedding of viral antigens may contribute to the inflammatory immune response particularly observed in patients with HTLV-I associated neurologic disease (HAM/TSP). Indeed, it has previously been shown that extracellular Tax can have damaging consequences for neurons (Alefantis et al. 2005; Cowan et al. 1997), although neither study specifically implicated exosomes. Once Tax protein or tax mRNA enter recipient cells, it can stimulate the production of proinflammatory cytokines, like interleukin (IL)-6 and tumor necrosis factor-α (Dhib-Jalbut et al. 1994). Ongoing work in our laboratory has found further evidence for functional consequences to HTLV-1 antigen exosomal incorporation. We noted that target cells exposed to PMBC-derived exosomes were susceptible to lysis by HTLV-1-specific cytotoxic T cells. Specifically, HTLV-1 Tax+ exosomes sensitized targets for lysis while ND PBMC-derived exosomes were unable to elicit this response (in submission). In addition, Tax+ exosomes isolated from the HTLV-I infected C8166 cell line were noted to increase the survival of IL-2 dependent cytotoxic T-cell line CTLL-2 and uninfected PBMCs in culture (Jaworski et al. 2014). Moreover, the majority of HAM/TSP PBMC exosomes produced in culture appeared to be produced by CD4+CD25+ T cells (in submission). HAM/TSP PBMC-derived exosomes were also shown to reduce the CD4+CD25+ T cell population when exposed to uninfected PBMCs (Anderson, unpublished observations). The observation was particularly intriguing given the known dysfunction of regulatory T cells (Tregs) in HAM/TSP patients (Anderson et al. 2014; Grant et al. 2008; Yamano et al. 2005). This supported the theory that HTLV-1 antigen trafficking to exosomes may support inflammatory responses in HAM/TSP through diminished Treg function, since uninfected Treg exosomes are typically suppressive through incorporation of anti-inflammatory miRNA (Okoye et al. 2014). More work must be done to understand if targeting of HTLV-1 Tax or other antigens changes the host miRNA profile of produced Treg exosomes or otherwise changes the function of released exosomes.
Of interest in the pathogenesis of HAM/TSP is the lack of documented infection of resident neuronal cells. While astrocytes and microglia can be infected in vitro, they have not been shown to be infected in vivo (Lepoutre et al. 2009). Moreover, the loss of oligodendrocytes that occurs with disease progression cannot be explained by active infection. One hypothesis is that the proinflamatory environment contributes to their eventual breakdown, while others have proposed direct targeting of oligodendrocytes through a mechanism of molecular mimicry of HTLV-I Tax with a neuronal protein (Irish et al. 2009; Kubota et al. 2000; Levin et al. 2002). Exosomal Tax may therefore ultimately explain both possibilities by contributing to the inflammatory cytokine production through packaging of these cytokines, as well as induction in recipient cells. Additionally, exosomal Tax uptake in uninfected cells could explain targeting by HTLV-1-specific T cells in the presence of uninfected resident neuronal cells. Clearly, further research must be undertaken to further elucidate the role of exosomes in the disease progression of HAM/TSP and potential targeting of therapeutics.
Endogenous Retroviruses
Endogenous retroviruses represent exogenous retroviruses that integrated into the germline and have since been maintained in human hosts. As mentioned they represent close to 8% of the human genome and are a type of transposable element (Griffiths 2001). Each has a similar structure to a typical retrovirus with a 5′ and 3′ LTR flanking the gag, pro, pol, and env genes. However, many of these proviruses have acquired fatal mutations such as stop codons that prevent the translation of functional proteins or the production of infectious retroviral particles (Downey et al. 2015); as such they are thought to be relatively silent. However, a growing pool of evidence is showing that their expression can be increased in conditions such as autoimmunity, cancer and pregnancy, making them a potential large cause of human morbidity and mortality (Douville and Nath 2014; Gonzalez-Cao et al. 2016; Nexo et al. 2016; Ryan 2004). Indeed, the HERV-K group, which is closely related to betaretroviruses, is characterized by the most complete proviruses and has been the best studied in association with clinical disease (Barbulescu et al. 1999). HERV-K consists of 11 subgroups, of which HML-2 is the most important due to its potential oncogenic activity (Downey et al. 2015; Subramanian et al. 2011).
Though endogenous retroviruses are generally silenced (Schulz et al. 2006), in the environment of hypomethylation that can occur with cancer, there can be instances of increased gene expression (Ehrlich 2009). Demethylation of HERV-K, and HERV-W has been reported in several different types of cancer (Downey et al. 2015; Gimenez et al. 2010; Stengel et al. 2010). Once demethylated, inflammatory factors can then allow for activation of the provirus and subsequent protein expression. Therefore, these endogenous retrovirus proteins and mRNAs can potentially be trafficked into exosomes, similar to what has been observed for HIV and HTLV-1. Indeed, exosome analysis of tumor samples showed increased endogenous retrovirus mRNA content compared to epidermoid carcinoma tumor cell exosomes (Balaj et al. 2011). Specifically, in analyzing exosomes from patients with frequent c-MYC amplification such as those with glioblastoma multiforme (GBM), medulloblastoma, atypical teratoid rhabdoid tumor (AT/RT), and malignant melanoma, it was found that these exosomes were enriched for mRNA from HERV-K, HERV-C, and HERV-W (Balaj et al. 2011). Moreover, HERV-K RNA could be transferred to human umbilical vein endothelial cells (HUVECs) in vitro via medulloblastoma exosomes suggesting that endogenous retrovirus expression can potentially spread through cells. It is as yet unclear if HERV product detection in exosomes is a biomarker of cancer or a potential cause of disease (Downey et al. 2015). Further investigation is necessary in order to understand the dynamics of this relationship between endogenous retrovirus expression, exosome production, and oncogenesis.
As mentioned, endogenous retroviruses have also been implicated in the field of autoimmunity. In mice, MuLV Gag and Env proteins were detected in exosomes produced by mesenchymal stem cells (MSCs) in Type 1 diabetes NOD mice (Dai et al. 2017). Furthermore, autoantibodies against the gp70 subunit of MuLV Env could be detected in addition to the presence of T cells specific to the MuLV Gag protein (Bashratyan et al. 2017). These T cells were later shown to contribute to the loss of islet cells in the mouse model of T1D. This demonstrates in an animal model that endogenous retroviral antigens can potentially precipitate autoimmune responses through immune targeting, similar to the sensitization for cell lysis observed after exposure to exosomes carrying HTLV-1 Tax (Anderson et al. 2016).
Although there still remains controversy, several families of endogenous retrovirus have been associated with multiple sclerosis (MS) (Morandi et al. 2017), a debilitating progressive neuroinflammatory disorder thought to be caused by damaging immune responses to self. This includes HERV-W and HERV-K (Ryan 2011). In particular, MS-associated retrovirus (MSRV) of the family HERV-W had been demonstrated to have increased env and pol mRNA present in PBMC, serum and CSF samples as compared to healthy controls (Garson et al. 1998; Mameli et al. 2007; Morandi et al. 2015; Nowak et al. 2003). Even more suggestive was a reported increased expression of MSRV Env in MS lesions (Perron et al. 2012). However, there are conflicting reports in the literatures as to expression levels of HERV-W in MS patients and what role it may ultimately play in disease pathogenesis (Downey et al. 2015).
Besides being potential pathologic determinants, the longevity of the presence of endogenous retrovirus sequences in the human genome suggests a potential beneficial or even necessary role in human function. Syncytin-1, a protein expressed by HERV-W is important in trophoblast fusion in the formation of the placenta (Mi et al. 2000; Potgens et al. 2004). Syncytin-1 is mostly limited to cytotrophoblast, synctiotrophoblast, and extravillous trophoblasts, mediating fusion between cytotrophoblasts and synctiotrophoblasts (Holder et al. 2012; Noorali et al. 2009). Synctin-2 is separately encoded by ERVFRD-1 (Malassine et al. 2008). Both synctins are retroviral like envelope proteins and both can be detected in exosomes produced by the placenta (Lokossou et al. 2014). Synctin-1 and Synctin-2 were found to be incorporated at the surface of placental exosomes (Lokossou et al. 2014), potentially aiding in binding upon release or maintenance of the synctitial trophoblast layer that allows for proper separation of fetal and maternal circulation. The authors suggest that the presence of these proteins may allow for a type of exosomal tropism so that released exosomes deliver products to specific locations (Vargas et al. 2014).
Conclusion
The detection of retroviral antigens in exosomes has implications in many diseases. As has been illustrated in the examples discussed in this review, viral antigens in exosomes could potentially aid in viral persistence and propagation such as occurs in HIV. These antigens could also serve to alter immune response or even enhance inflammatory responses, as was discussed in the examples of HAM/TSP. Interestingly, HTLV-1 is a primarily cell-associated virus, such that cell-free viral particles are often difficult to detect in patient serologic samples (Bangham and Matsuoka 2017). Therefore, the virus typically exists as a provirus, much like endogenous retroviruses. The most obvious difference between the state of the HTLV-1 provirus and endogenous proviruses is that the HTLV-1 provirus typically has not been mutated (Mansky 2000). However as was discussed, some HERVs like HML-2 have intact genomes and can undergo demethylation given the correct conditions in the host, as can occur in cancer or other conditions associated with global hypomethylation. This allows for the gene transcription and translation of endogenous retroviral proteins, although the production of intact infectious viral particles has not been reported (Bannert and Kurth 2004). In these scenarios, endogenous retrovirus antigens such as Env proteins have been found in serum exosomes. Intriguingly, exosomes carrying endogenous retrovirus antigens induced anti-self responses in B and T cells in an autoimmune disease model in mice (Dai et al. 2017), supporting the theory that endogenous retroviruses are associated with pathological responses in autoimmune disease (Nexo et al. 2016).
Given these examples of detection of retroviral antigens (proteins, mRNA, etc) in exosomes despite the absence of free-viral particles, exosome isolation and characterization of the contents becomes of import in inflammatory disorders, cancer, and neurologic diseases. Indeed, exosomes represent a potential detection tool for the myriad of meningitides and encephalitides for which an inciting pathogen is suspected. The etiology for over 50% of meningitis and encephalitis cases is currently unknown (Jarrin et al. 2016; Tunkel et al. 2008). Furthermore, there are several neurologic diseases such as MS, amyotrophic lateral sclerosis (ALS), and Parkinson’s disease (PD) which have an inflammatory component and have been hypothesized to have a viral trigger (Douville and Nath 2014; Zhou et al. 2013). ALS has even been thought to be associated with increased expression of HERV-K proteins (Alfahad and Nath 2013; Douville et al. 2011). Additionally, recent data has demonstrated that exosomes containing HIV antigens are still present in the serum of patients under HAART for whom there is no detectable virus (Jaworski et al. 2014), indicating that tracking of exosomal cargo could monitor potential viral reservoirs. Collectively, the observations presented in this report on exosomes containing retroviral antigens in the CSF of patients with HTLV-1 associated neurologic disease, HAND, and diseases associated with increased endogenous retroviral expression will be of relevance to many systemic and neurologic diseases in which viruses are thought to play a role.
References
Aiewsakun P, Katzourakis A (2017) Marine origin of retroviruses in the early Palaeozoic era. Nat Commun 8:13954. https://doi.org/10.1038/ncomms13954
Alefantis T, Jain P, Ahuja J, Mostoller K, Wigdahl B (2005) HTLV-1 tax nucleocytoplasmic shuttling, interaction with the secretory pathway, extracellular signaling, and implications for neurologic disease. J Biomed Sci 12:961–974. https://doi.org/10.1007/s11373-005-9026-x
Alefantis T, Mostoller K, Jain P, Harhaj E, Grant C, Wigdahl B (2005) Secretion of the human T cell leukemia virus type I transactivator protein tax. J Biol Chem 280:17353–17362. https://doi.org/10.1074/jbc.M409851200
Alenquer M, Amorim MJ (2015) Exosome biogenesis, regulation, and function in viral infection. Viruses 7:5066–5083. https://doi.org/10.3390/v7092862
Alfahad T, Nath A (2013) Retroviruses and amyotrophic lateral sclerosis. Antivir Res 99:180–187. https://doi.org/10.1016/j.antiviral.2013.05.006
Ali SA, Huang MB, Campbell PE, Roth WW, Campbell T, Khan M, Newman G, Villinger F, Powell MD, Bond VC (2010) Genetic characterization of HIV type 1 Nef-induced vesicle secretion. AIDS Res Hum Retrovir 26:173–192. https://doi.org/10.1089/aid.2009.0068
Anderson MR, Enose-Akahata Y, Massoud R, Ngouth N, Tanaka Y, Oh U, Jacobson S (2014) Epigenetic modification of the FoxP3 TSDR in HAM/TSP decreases the functional suppression of Tregs. J NeuroImmune Pharmacol 9:522–532. https://doi.org/10.1007/s11481-014-9547-z
Anderson MR, Kashanchi F, Jacobson S (2016) Exosomes in viral disease. Neurotherapeutics 13:535–546. https://doi.org/10.1007/s13311-016-0450-6
Andreu Z, Yanez-Mo M (2014) Tetraspanins in extracellular vesicle formation and function. Front Immunol 5:442. https://doi.org/10.3389/fimmu.2014.00442
Arenaccio C, Anticoli S, Manfredi F, Chiozzini C, Olivetta E, Federico M (2015) Latent HIV-1 is activated by exosomes from cells infected with either replication-competent or defective HIV-1. Retrovirology 12:87. https://doi.org/10.1186/s12977-015-0216-y
Arenaccio C, Chiozzini C, Columba-Cabezas S, Manfredi F, Affabris E, Baur A, Federico M (2014) Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. J Virol 88:11529–11539. https://doi.org/10.1128/JVI.01712-14
Arias M, Fan H (2014) The saga of XMRV: a virus that infects human cells but is not a human virus Emerg Microbes Infect 3:e doi:https://doi.org/10.1038/emi.2014.25
Baglio SR, Pegtel DM, Baldini N (2012) Mesenchymal stem cell secreted vesicles provide novel opportunities in (stem) cell-free therapy. Front Physiol 3:359. https://doi.org/10.3389/fphys.2012.00359
Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G (2012) Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14:677–685. https://doi.org/10.1038/ncb2502
Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J (2011) Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2:180. https://doi.org/10.1038/ncomms1180
Bangham CRM, Matsuoka M (2017) Human T-cell leukaemia virus type 1: parasitism and pathogenesis. Philos Trans R Soc Lond Ser B Biol Sci 372:20160272. https://doi.org/10.1098/rstb.2016.0272
Bannert N, Kurth R (2004) Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A 101(Suppl 2):14572–14579. https://doi.org/10.1073/pnas.0404838101
Baratella M, Forlani G, Raval GU, Tedeschi A, Gout O, Gessain A, Tosi G, Accolla RS (2017) Cytoplasmic localization of HTLV-1 HBZ protein: a biomarker of HTLV-1-associated myelopathy/tropical spastic Paraparesis (HAM/TSP). PLoS Negl Trop Dis 11:e0005285. https://doi.org/10.1371/journal.pntd.0005285
Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J (1999) Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol 9:861–868
Bashratyan R, Regn D, Rahman MJ, Marquardt K, Fink E, Hu WY, Elder JH, Binley J, Sherman LA, Dai YD (2017) Type 1 diabetes pathogenesis is modulated by spontaneous autoimmune responses to endogenous retrovirus antigens in NOD mice. Eur J Immunol 47:575–584. https://doi.org/10.1002/eji.201646755
Bernard MA, Zhao H, Yue SC, Anandaiah A, Koziel H, Tachado SD (2014) Novel HIV-1 miRNAs stimulate TNFalpha release in human macrophages via TLR8 signaling pathway. PLoS One 9:e106006. https://doi.org/10.1371/journal.pone.0106006
Bissig C, Gruenberg J (2014) ALIX and the multivesicular endosome: ALIX in wonderland. Trends Cell Biol 24:19–25. https://doi.org/10.1016/j.tcb.2013.10.009
Campbell PE, Isayev O, Ali SA, Roth WW, Huang MB, Powell MD, Leszczynski J, Bond VC (2012) Validation of a novel secretion modification region (SMR) of HIV-1 Nef using cohort sequence analysis and molecular modeling. J Mol Model 18:4603–4613. https://doi.org/10.1007/s00894-012-1452-x
Campbell TD, Khan M, Huang MB, Bond VC, Powell MD (2008) HIV-1 Nef protein is secreted into vesicles that can fuse with target cells and virions. Ethnicity & disease 18:S2–14-19
Cloyd MW (1996) Human Retroviruses. In: th, Baron S (eds) Medical Microbiology. Galveston (TX),
Colombo M, Raposo G, Thery C (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. https://doi.org/10.1146/annurev-cellbio-101512-122326
Columba Cabezas S, Federico M (2013) Sequences within RNA coding for HIV-1 gag p17 are efficiently targeted to exosomes. Cell Microbiol 15:412–429. https://doi.org/10.1111/cmi.12046
Cowan EP, Alexander RK, Daniel S, Kashanchi F, Brady JN (1997) Induction of tumor necrosis factor alpha in human neuronal cells by extracellular human T-cell lymphotropic virus type 1 tax. J Virol 71:6982–6989
Currer R, van Duyne R, Jaworski E, Guendel I, Sampey G, Das R, Narayanan A, Kashanchi F (2012) HTLV tax: a fascinating multifunctional co-regulator of viral and cellular pathways. Front Microbiol 3:406. https://doi.org/10.3389/fmicb.2012.00406
Dai YD, Sheng H, Dias P, Jubayer Rahman M, Bashratyan R, Regn D, Marquardt K (2017) Autoimmune responses to exosomes and candidate antigens contribute to type 1 diabetes in non-obese diabetic mice. Curr Diab Rep 17:130. https://doi.org/10.1007/s11892-017-0962-4
Das AT, Harwig A, Berkhout B (2011) The HIV-1 tat protein has a versatile role in activating viral transcription. J Virol 85:9506–9516. https://doi.org/10.1128/JVI.00650-11
de Carvalho JV, de Castro RO, da Silva EZM, Silveira PP, da Silva-Januário ME, Arruda E, Jamur MC, Oliver C, Aguiar RS, daSilva LLP (2014) Nef neutralizes the ability of exosomes from CD4+ T cells to act as decoys during HIV-1 infection. PLoS One 9:e113691. https://doi.org/10.1371/journal.pone.0113691
Denner J (2010) Detection of a gammaretrovirus, XMRV, in the human population: open questions and implications for xenotransplantation. Retrovirology 7:16. https://doi.org/10.1186/1742-4690-7-16
Desfarges S, Ciuffi A (2010) Retroviral integration site selection. Viruses 2:111–130. https://doi.org/10.3390/v2010111
Dhib-Jalbut S, Hoffman PM, Yamabe T, Sun D, Xia J, Eisenberg H, Bergey G, Ruscetti FW (1994) Extracellular human T-cell lymphotropic virus type I tax protein induces cytokine production in adult human microglial cells. Ann Neurol 36:787–790. https://doi.org/10.1002/ana.410360516
Douville R, Liu J, Rothstein J, Nath A (2011) Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol 69:141–151. https://doi.org/10.1002/ana.22149
Douville RN, Nath A (2014) Human endogenous retroviruses and the nervous system. Handb Clin Neurol 123:465–485. https://doi.org/10.1016/B978-0-444-53488-0.00022-5
Downey RF, Sullivan FJ, Wang-Johanning F, Ambs S, Giles FJ, Glynn SA (2015) Human endogenous retrovirus K and cancer: innocent bystander or tumorigenic accomplice? Int J Cancer 137:1249–1257. https://doi.org/10.1002/ijc.29003
Ehrlich M (2009) DNA hypomethylation in cancer cells. Epigenomics 1:239–259. https://doi.org/10.2217/epi.09.33
Escalera-Zamudio M, Greenwood AD (2016) On the classification and evolution of endogenous retrovirus: human endogenous retroviruses may not be 'human' after all. APMIS 124:44–51. https://doi.org/10.1111/apm.12489
Fader CM, Sanchez DG, Mestre MB, Colombo MI (2009) TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 1793:1901–1916. https://doi.org/10.1016/j.bbamcr.2009.09.011
Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ (2007) Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol 5:e158. https://doi.org/10.1371/journal.pbio.0050158
Filipenko NR, MacLeod TJ, Yoon CS, Waisman DM (2004) Annexin A2 is a novel RNA-binding protein. J Biol Chem 279:8723–8731. https://doi.org/10.1074/jbc.M311951200
Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP (2007) Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841–852. https://doi.org/10.1016/j.cell.2007.01.035
Garson JA, Tuke PW, Giraud P, Paranhos-Baccala G, Perron H (1998) Detection of virion-associated MSRV-RNA in serum of patients with multiple sclerosis. Lancet 351:33
Gimenez J, Montgiraud C, Pichon JP, Bonnaud B, Arsac M, Ruel K, Bouton O, Mallet F (2010) Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res 38:2229–2246. https://doi.org/10.1093/nar/gkp1214
Gonzalez-Cao M, Iduma P, Karachaliou N, Santarpia M, Blanco J, Rosell R (2016) Human endogenous retroviruses and cancer. Cancer Biol Med 13:483–488. https://doi.org/10.20892/j.issn.2095-3941.2016.0080
Gordon-Alonso M, Yanez-Mo M, Barreiro O, Alvarez S, Munoz-Fernandez MA, Valenzuela-Fernandez A, Sanchez-Madrid F (2006) Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol 177:5129–5137
Gould SJ, Booth AM, Hildreth JE (2003) The Trojan exosome hypothesis. Proc Natl Acad Sci U S A 100:10592–10597. https://doi.org/10.1073/pnas.1831413100
Grant C, Oh U, Yao K, Yamano Y, Jacobson S (2008) Dysregulation of TGF-beta signaling and regulatory and effector T-cell function in virus-induced neuroinflammatory disease. Blood 111:5601–5609. https://doi.org/10.1182/blood-2007-11-123430
Griffiths DJ (2001) Endogenous retroviruses in the human genome sequence Genome Biol 2:REVIEWS1017
Hagiwara K, Katsuda T, Gailhouste L, Kosaka N, Ochiya T (2015) Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett 589:4071–4078. https://doi.org/10.1016/j.febslet.2015.11.036
Han SP, Friend LR, Carson JH, Korza G, Barbarese E, Maggipinto M, Hatfield JT, Rothnagel JA, Smith R (2010) Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms. Traffic 11:886–898. https://doi.org/10.1111/j.1600-0854.2010.01072.x
Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, Hamrick MW, Liu Y (2017) A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One 12:e0170628. https://doi.org/10.1371/journal.pone.0170628
Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21:77–91. https://doi.org/10.1016/j.devcel.2011.05.015
Holder BS, Tower CL, Abrahams VM, Aplin JD (2012) Syncytin 1 in the human placenta. Placenta 33:460–466. https://doi.org/10.1016/j.placenta.2012.02.012
Hughes JF, Coffin JM (2005) Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171:1183–1194. https://doi.org/10.1534/genetics.105.043976
Irish BP, Khan ZK, Jain P, Nonnemacher MR, Pirrone V, Rahman S, Rajagopalan N, Suchitra JB, Mostoller K, Wigdahl B (2009) Molecular mechanisms of neurodegenerative diseases induced by human retroviruses: a review. Am J Infect Dis 5:231–258
Izquierdo-Useros N, Puertas MC, Borras FE, Blanco J, Martinez-Picado J (2011) Exosomes and retroviruses: the chicken or the egg? Cell Microbiol 13:10–17. https://doi.org/10.1111/j.1462-5822.2010.01542.x
Jarrin I, Sellier P, Lopes A, Morgand M, Makovec T, Delcey V, Champion K, Simoneau G, Green A, Mouly S, Bergmann JF, Lloret-Linares C (2016) Etiologies and Management of Aseptic Meningitis in patients admitted to an internal medicine department. Medicine (Baltimore) 95:e2372. https://doi.org/10.1097/MD.0000000000002372
Jaworski E, Narayanan A, van Duyne R, Shabbeer-Meyering S, Iordanskiy S, Saifuddin M, Das R, Afonso PV, Sampey GC, Chung M, Popratiloff A, Shrestha B, Sehgal M, Jain P, Vertes A, Mahieux R, Kashanchi F (2014) Human T-lymphotropic virus type 1-infected cells secrete exosomes that contain tax protein. J Biol Chem 289:22284–22305. https://doi.org/10.1074/jbc.M114.549659
Jaworski E, Saifuddin M, Sampey G, Shafagati N, van Duyne R, Iordanskiy S, Kehn-Hall K, Liotta L, Petricoin E, Young M, Lepene B, Kashanchi F (2014) The use of Nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS One 9:e96778. https://doi.org/10.1371/journal.pone.0096778
Jesus da Costa L, Lopes dos Santos A, Mandic R, Shaw K, Santana de Aguiar R, Tanuri A, Luciw PA, Peterlin BM (2009) Interactions between SIVNef, SIVGagPol and Alix correlate with viral replication and progression to AIDS in rhesus macaques. Virology 394:47–56. https://doi.org/10.1016/j.virol.2009.08.024
Kannian P, Green PL (2010) Human T Lymphotropic virus type 1 (HTLV-1): molecular biology and oncogenesis. Viruses 2:2037–2077. https://doi.org/10.3390/v2092037
Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H (2003) Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol 31:1007–1014
Krementsov DN, Weng J, Lambele M, Roy NH, Thali M (2009) Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology 6:64. https://doi.org/10.1186/1742-4690-6-64
Kubota R, Nagai M, Kawanishi T, Osame M, Jacobson S (2000) Increased HTLV type 1 tax specific CD8+ cells in HTLV type 1-asociated myelopathy/tropical spastic paraparesis: correlation with HTLV type 1 proviral load. AIDS Res Hum Retrovir 16:1705–1709. https://doi.org/10.1089/08892220050193182
Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB (2002) Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods 270:211–226
Lenassi M, Cagney G, Liao M, Vaupotič T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitaš A, Peterlin BM (2010) HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11:110–122. https://doi.org/10.1111/j.1600-0854.2009.01006.x
Lepoutre V, Jain P, Quann K, Wigdahl B, Khan ZK (2009) Role of resident CNS cell populations in HTLV-1-associated neuroinflammatory disease. Front Biosci 14:1152–1168
Levin MC, Lee SM, Morcos Y, Brady J, Stuart J (2002) Cross-reactivity between immunodominant human T lymphotropic virus type I tax and neurons: implications for molecular mimicry. J Infect Dis 186:1514–1517. https://doi.org/10.1086/344734
Lezin A, Olindo S, Olière S, Varrin-Doyer M, Marlin R, Cabre P, Smadja D, Cesaire R (2005) Human T lymphotropic virus type I (HTLV-I) proviral load in cerebrospinal fluid: a new criterion for the diagnosis of HTLV-I-associated myelopathy/tropical spastic paraparesis? J Infect Dis 191:1830–1834. https://doi.org/10.1086/429962
Li HC, Fujiyoshi T, Lou H, Yashiki S, Sonoda S, Cartier L, Nunez L, Munoz I, Horai S, Tajima K (1999) The presence of ancient human T-cell lymphotropic virus type I provirus DNA in an Andean mummy. Nat Med 5:1428–1432. https://doi.org/10.1038/71006
Li M, Kesic M, Yin H, Yu L, Green PL (2009) Kinetic analysis of human T-cell leukemia virus type 1 gene expression in cell culture and infected animals. J Virol 83:3788–3797. https://doi.org/10.1128/JVI.02315-08
Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, Xu YM, Huang LF, Wang XZ (2015) Exosomes: novel biomarkers for clinical diagnosis. Sci World J 2015:657086. https://doi.org/10.1155/2015/657086
Lokossou AG, Toudic C, Barbeau B (2014) Implication of human endogenous retrovirus envelope proteins in placental functions. Viruses 6:4609–4627. https://doi.org/10.3390/v6114609
Longatti A, Boyd B, Chisari FV (2015) Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol 89:2956–2961. https://doi.org/10.1128/JVI.02721-14
Luo X, Fan Y, Park IW, He JJ (2015) Exosomes are unlikely involved in intercellular Nef transfer. PLoS One 10:e0124436. https://doi.org/10.1371/journal.pone.0124436
Madison MN, Okeoma CM (2015) Exosomes: implications in HIV-1 pathogenesis. Viruses 7:4093–4118. https://doi.org/10.3390/v7072810
Malassine A, Frendo JL, Blaise S, Handschuh K, Gerbaud P, Tsatsaris V, Heidmann T, Evain-Brion D (2008) Human endogenous retrovirus-FRD envelope protein (syncytin 2) expression in normal and trisomy 21-affected placenta. Retrovirology 5:6. https://doi.org/10.1186/1742-4690-5-6
Mameli G, Astone V, Arru G, Marconi S, Lovato L, Serra C, Sotgiu S, Bonetti B, Dolei A (2007) Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not human herpesvirus 6. Journal Gen Virol 88:264–274. https://doi.org/10.1099/vir.0.81890-0
Mansky LM (2000) In vivo analysis of human T-cell leukemia virus type 1 reverse transcription accuracy. J Virol 74:9525–9531
Matsuo H, Chevallier J, Mayran N, le Blanc I, Ferguson C, Fauré J, Blanc NS, Matile S, Dubochet J, Sadoul R, Parton RG, Vilbois F, Gruenberg J (2004) Role of LBPA and Alix in multivesicular liposome formation and endosome. Organization. Science 303:531–534. https://doi.org/10.1126/science.1092425
Meckes DG Jr, Raab-Traub N (2011) Microvesicles and viral infection. J Virol 85:12844–12854. https://doi.org/10.1128/JVI.05853-11
Mi S, Lee X, Li XP, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, McCoy JM (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. https://doi.org/10.1038/35001608
Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2:282. https://doi.org/10.1038/ncomms1285
Mittelbrunn M, Sanchez-Madrid F (2012) Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 13:328–335. https://doi.org/10.1038/nrm3335
Morandi E, Tanasescu R, Tarlinton RE, Constantinescu CS, Zhang W, Tench C, Gran B (2017) The association between human endogenous retroviruses and multiple sclerosis: a systematic review and meta-analysis. PLoS One 12:e0172415. https://doi.org/10.1371/journal.pone.0172415
Morandi E, Tarlinton RE, Gran B (2015) Multiple sclerosis between genetics and infections: human endogenous retroviruses in monocytes and macrophages. Front Immunol 6:647. https://doi.org/10.3389/fimmu.2015.00647
Narayanan A, Iordanskiy S, Das R, van Duyne R, Santos S, Jaworski E, Guendel I, Sampey G, Dalby E, Iglesias-Ussel M, Popratiloff A, Hakami R, Kehn-Hall K, Young M, Subra C, Gilbert C, Bailey C, Romerio F, Kashanchi F (2013) Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J Biol Chem 288:20014–20033. https://doi.org/10.1074/jbc.M112.438895
Nexo BA, Jensen SB, Hansen B, Laska MJ (2016) Endogenous retroviruses are associated with autoimmune diseases Ugeskr Laeger 178
Nexo BA et al (2016) Are human endogenous retroviruses triggers of autoimmune diseases? Unveiling associations of three diseases and viral loci. Immunol Res 64:55–63. https://doi.org/10.1007/s12026-015-8671-z
Noorali S, Rotar IC, Lewis C, Pestaner JP, Pace DG, Sison A, Bagasra O (2009) Role of HERV-W syncytin-1 in placentation and maintenance of human pregnancy. Appl Immunohistochem Mol Morphol 17:319–328. https://doi.org/10.1097/PAI.0b013e31819640f9
Nour AM, Modis Y (2014) Endosomal vesicles as vehicles for viral genomes. Trends Cell Biol 24:449–454. https://doi.org/10.1016/j.tcb.2014.03.006
Novak K (1999) Ancient HTLV-1. Nat Med 5:1357. https://doi.org/10.1038/70923
Nowak J, Januszkiewicz D, Pernak M, Liweń I, Zawada M, Rembowska J, Nowicka K, Lewandowski K, Hertmanowska H, Wender M (2003) Multiple sclerosis-associated virus-related pol sequences found both in multiple sclerosis and healthy donors are more frequently expressed in multiple sclerosis patients. J Neurovirol 9:112–117. https://doi.org/10.1080/13550280390173355
Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, Seabra MC, Wilson MS (2014) MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41:89–103. https://doi.org/10.1016/j.immuni.2014.05.019
Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12:19–30; sup pp 11–13 doi:https://doi.org/10.1038/ncb2000
Pelchen-Matthews A, Kramer B, Marsh M (2003) Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol 162:443–455. https://doi.org/10.1083/jcb.200304008
Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, Vázquez J, Yáñez-Mó M (2013) The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem 288:11649–11661. https://doi.org/10.1074/jbc.M112.445304
Perez-Hernandez J, Cortes R (2015) Extracellular Vesicles as Biomarkers of Systemic Lupus Erythematosus. Dis Markers 2015:613536. https://doi.org/10.1155/2015/613536
Perron H, Germi R, Bernard C, Garcia-Montojo M, Deluen C, Farinelli L, Faucard R, Veas F, Stefas I, Fabriek BO, van-Horssen J, van-der-Valk P, Gerdil C, Mancuso R, Saresella M, Clerici M, Marcel S, Creange A, Cavaretta R, Caputo D, Arru G, Morand P, Lang AB, Sotgiu S, Ruprecht K, Rieckmann P, Villoslada P, Chofflon M, Boucraut J, Pelletier J, Hartung HP (2012) Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult Scler 18:1721–1736. https://doi.org/10.1177/1352458512441381
Piguet V, Steinman RM (2007) The interaction of HIV with dendritic cells: outcomes and pathways. Trends Immunol 28:503–510. https://doi.org/10.1016/j.it.2007.07.010
Potgens AJ, Drewlo S, Kokozidou M, Kaufmann P (2004) Syncytin: the major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum Reprod Update 10:487–496. https://doi.org/10.1093/humupd/dmh039
Properzi F, Logozzi M, Fais S (2013) Exosomes: the future of biomarkers in medicine. Biomark Med 7:769–778. https://doi.org/10.2217/bmm.13.63
Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JAA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJW (2013) Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. https://doi.org/10.1073/pnas.1221899110
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. https://doi.org/10.1083/jcb.201211138
Rende F, Cavallari I, Corradin A, Silic-Benussi M, Toulza F, Toffolo GM, Tanaka Y, Jacobson S, Taylor GP, D'Agostino DM, Bangham CRM, Ciminale V (2011) Kinetics and intracellular compartmentalization of HTLV-1 gene expression: nuclear retention of HBZ mRNAs. Blood 117:4855–4859. https://doi.org/10.1182/blood-2010-11-316463
Retrovidae (2012). In: King A, Lefkowitz E, Adams MJ, Carstens EB (ed) Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses edn. Elsevier, pp 477–495
Rider MA, Hurwitz SN, Meckes DG Jr (2016) ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep 6:23978. https://doi.org/10.1038/srep23978
Robbins PD, Morelli AE (2014) Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 14:195–208. https://doi.org/10.1038/nri3622
Romanelli MG, Diani E, Bergamo E, Casoli C, Ciminale V, Bex F, Bertazzoni U (2013) Highlights on distinctive structural and functional properties of HTLV tax proteins. Front Microbiol 4:271. https://doi.org/10.3389/fmicb.2013.00271
Roucourt B, Meeussen S, Bao J, Zimmermann P, David G (2015) Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res 25:412–428. https://doi.org/10.1038/cr.2015.29
Ryan FP (2004) Human endogenous retroviruses in health and disease: a symbiotic perspective. J R Soc Med 97:560–565. https://doi.org/10.1258/jrsm.97.12.560
Ryan FP (2011) Human endogenous retroviruses in multiple sclerosis: potential for novel neuro-pharmacological research. Curr Neuropharmacol 9:360–369. https://doi.org/10.2174/157015911795596568
Sampey GC, Meyering SS, Asad Zadeh M, Saifuddin M, Hakami RM, Kashanchi F (2014) Exosomes and their role in CNS viral infections. J Neurovirol 20:199–208. https://doi.org/10.1007/s13365-014-0238-6
Sampey GC, Saifuddin M, Schwab A, Barclay R, Punya S, Chung MC, Hakami RM, Asad Zadeh M, Lepene B, Klase ZA, el-Hage N, Young M, Iordanskiy S, Kashanchi F (2016) Exosomes from HIV-1-infected cells stimulate production of pro-inflammatory cytokines through trans-activating response (TAR) RNA. J Biol Chem 291:1251–1266. https://doi.org/10.1074/jbc.M115.662171
Savina A, Fader CM, Damiani MT, Colombo MI (2005) Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 6:131–143. https://doi.org/10.1111/j.1600-0854.2004.00257.x
Schmidt O, Teis D (2012) The ESCRT machinery. Curr Biol 22:R116–R120. https://doi.org/10.1016/j.cub.2012.01.028
Schorey JS, Cheng Y, Singh PP, Smith VL (2015) Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep 16:24–43. https://doi.org/10.15252/embr.201439363
Schulz WA, Steinhoff C, Florl AR (2006) Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol 310:211–250
Serrao E, Engelman AN (2016) Sites of retroviral DNA integration: from basic research to clinical applications. Crit Rev Biochem Mol Biol 51:26–42. https://doi.org/10.3109/10409238.2015.1102859
Sharp PM, Hahn BH (2011) Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. https://doi.org/10.1101/cshperspect.a006841
Shembade N, Harhaj EW (2010) Role of post-translational modifications of HTLV-1 tax in NF-kappaB activation. World J Biol Chem 1:13–20. https://doi.org/10.4331/wjbc.v1.i1.13
Shen B, Wu N, Yang JM, Gould SJ (2011) Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem 286:14383–14395. https://doi.org/10.1074/jbc.M110.208660
Stengel S, Fiebig U, Kurth R, Denner J (2010) Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer 49:401–411. https://doi.org/10.1002/gcc.20751
Stoorvogel W (2015) Resolving sorting mechanisms into exosomes. Cell Res 25:531–532. https://doi.org/10.1038/cr.2015.39
Stuffers S, Sem Wegner C, Stenmark H, Brech A (2009) Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10:925–937. https://doi.org/10.1111/j.1600-0854.2009.00920.x
Subramanian RP, Wildschutte JH, Russo C, Coffin JM (2011) Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. https://doi.org/10.1186/1742-4690-8-90
Sundquist WI, Krausslich HG (2012) HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. https://doi.org/10.1101/cshperspect.a006924
Swaminathan G, Navas-Martin S, Martin-Garcia J (2014) MicroRNAs and HIV-1 infection: antiviral activities and beyond. J Mol Biol 426:1178–1197. https://doi.org/10.1016/j.jmb.2013.12.017
Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K (2010) Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun 399:384–390. https://doi.org/10.1016/j.bbrc.2010.07.083
Thali M (2009) The roles of tetraspanins in HIV-1 replication. Curr Top Microbiol Immunol 339:85–102. https://doi.org/10.1007/978-3-642-02175-6_5
Thali M (2011) Tetraspanin functions during HIV-1 and influenza virus replication. Biochem Soc Trans 39:529–531. https://doi.org/10.1042/BST0390529
Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. https://doi.org/10.1126/science.1153124
Tremblay MJ, Fortin JF, Cantin R (1998) The acquisition of host-encoded proteins by nascent HIV-1. Immunol Today 19:346–351
Tunkel AR, Glaser CA, Bloch KC, Sejvar JJ, Marra CM, Roos KL, Hartman BJ, Kaplan SL, Scheld WM, Whitley RJ, Infectious Diseases Society of America (2008) The management of encephalitis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis : An Official Publication of the Infectious Diseases Society of America 47:303–327. https://doi.org/10.1086/589747
Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, Polchi A, Tancini B, Emiliani C (2013) Signaling pathways in exosomes biogenesis, secretion and fate. Genes (Basel) 4:152–170. https://doi.org/10.3390/genes4020152
van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G (2011) The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 21:708–721. https://doi.org/10.1016/j.devcel.2011.08.019
Vargas A, Zhou S, Ethier-Chiasson M, Flipo D, Lafond J, Gilbert C, Barbeau B (2014) Syncytin proteins incorporated in placenta exosomes are important for cell uptake and show variation in abundance in serum exosomes from patients with preeclampsia. FASEB J 28:3703–3719. https://doi.org/10.1096/fj.13-239053
Verdonck K, Gonzalez E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E (2007) Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect Dis 7:266–281. https://doi.org/10.1016/S1473-3099(07)70081-6
Verweij FJ, van Eijndhoven MAJ, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM (2011) LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-kappaB activation. EMBO J 30:2115–2129. https://doi.org/10.1038/emboj.2011.123
Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M (2014) Sorting it out: regulation of exosome loading. Semin Cancer Biol 28:3–13. https://doi.org/10.1016/j.semcancer.2014.04.009
Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4:2980. https://doi.org/10.1038/ncomms3980
Wiley RD, Gummuluru S (2006) Immature dendritic cell-derived exosomes can mediate HIV-1 trans infection. Proc Natl Acad Sci U S A 103:738–743. https://doi.org/10.1073/pnas.0507995103
Yamano Y, Takenouchi N, Li HC, Tomaru U, Yao K, Grant CW, Maric DA, Jacobson S (2005) Virus-induced dysfunction of CD4+CD25+ T cells in patients with HTLV-I-associated neuroimmunological disease. J Clin Invest 115:1361–1368. https://doi.org/10.1172/JCI23913
Yelamanchili SV, Lamberty BG, Rennard DA, Morsey BM, Hochfelder CG, Meays BM, Levy E, Fox HS (2015) MiR-21 in extracellular vesicles leads to neurotoxicity via TLR7 signaling in SIV neurological disease. PLoS Pathog 11:e1005032. https://doi.org/10.1371/journal.ppat.1005032
Zeringer E, Barta T, Li M, Vlassov AV (2015) Strategies for isolation of exosomes. Cold Spring Harb Protoc 2015:319–323. https://doi.org/10.1101/pdb.top074476
Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S (2015) Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13:17–24. https://doi.org/10.1016/j.gpb.2015.02.001
Zhou L, Miranda-Saksena M, Saksena NK (2013) Viruses and neurodegeneration. Virol J 10:172. https://doi.org/10.1186/1743-422X-10-172
Funding
Studies involving HTLV-1 were funded internally by NINDS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Anderson M declares that she has no conflicts of interest. Kashanchi F declares that he has no conflicts of interest. Jacobson S declares that he has no conflicts of interest.
Human and Animal Rights and Informed Consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consents were obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Anderson, M., Kashanchi, F. & Jacobson, S. Role of Exosomes in Human Retroviral Mediated Disorders. J Neuroimmune Pharmacol 13, 279–291 (2018). https://doi.org/10.1007/s11481-018-9784-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11481-018-9784-7