Abstract
Drinking water treatment techniques are used globally in the context of water security and public health, yet they are not applicable to antibiotic resistance gene (ARG) contamination. Using high-throughput quantitative PCR, we analyzed the prevalence and diversity of ARGs and mobile genetic elements (MGEs) in water supplies. A total of 224 ARGs and MGEs were detected in all sampling sites. Absolute abundance and detected number of ARGs decreased significantly (P < 0.05) in sand filter water after drinking water treatment and increased thereafter at point-of-use (household tap water). Changes in the composition and diversity of the bacterial community were observed in water samples at different steps. A significant correlation (P < 0.001) between microbial communities and ARG profiles was observed, and variance in ARG profiles could be primarily attributed to community composition (11.9%), and interaction between community composition, environmental factors and MGEs (30.7%). A network analysis was performed, and the results showed eight bacterial phyla were significantly correlated with nine different classes of ARGs, suggesting the potential bacterial host for ARGs. This study suggested that although the absolute abundance of ARGs decreased after treatment of drinking water treatment plants (DWTPs), the rebounded of ARGs in the water distribution system should not be neglected.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The contamination of the aquatic environment with antibiotic resistance genes (ARGs) is considered deleterious to human health (Witte 1998; Chee-Sanford et al. 2009; Marshall and Levy 2011). Although the presence of ARGs in the environment is known (Wellington et al. 2013), anthropogenic activities and recalcitrant antibiotic residues promote the distribution of ARGs. Selection pressure on antibiotics results in the distribution of ARGs and propagation of antibiotic-resistant bacteria (ARB), which further contaminate the natural environment (Pruden et al. 2006; Zhu et al. 2013). The overuse of antibiotics has resulted in the detection of ARGs in water sources and the other surface waters (Bai et al. 2015) which have potential to be enriched at drinking water treatment plants (DWTPs) (Guo et al. 2014; Sanganyado and Gwenzi 2019).
DWTPs improve drinking water quality and reduce the incidence of waterborne diseases (Li et al. 2016). However, conventional DWTPs are not designed to eliminate ARGs and ARB (Xu et al. 2016). Several studies have suggested the ARGs and ARB are prevalent in effluent from conventional DWTPs, even after disinfection using techniques such as chlorination (Jia et al. 2015; Liu et al. 2018). The ARGs are persistent in water source and cannot be completely eliminated in the conventional drinking water processes (Ouyang et al. 2015; Xu et al. 2016), posing a potential risk to human health.
Water distribution infrastructure also provides an opportunity for the spread of ARGs and propagation of ARB (Garner et al. 2018; Tan et al. 2019). For example, biofilms are an ideal environment for ARGs and ARB survival (Zhang et al. 2019). Although water pipelines provide a harsh environment lacking in nutrients, biofilms with high densities of microorganisms are common (Yang et al. 2011). It has been reported that the abundance and diversity of the resistome observed in water distribution systems are higher than in water treatment plant effluent, potentially due to biofilm growth (Xi et al. 2009). Additionally, chlorine residues on water pipes may be responsible for the enrichment of ARGs (Bai et al. 2015). Previous studies have also highlighted that heavy metals such as copper in water accelerate the spread of ARGs and ARB, while promoting horizontal gene transfer (HGT) (Baker-Austin et al. 2006; Seiler and Berendonk 2012). On the basis of previous studies, heavy metals used in the construction of distribution pipeline are also considered to have the same effect after metal corrosion (Baker-Austin et al. 2006; Khan et al. 2019).
Nevertheless, information on ARGs associated with the drinking water source, conventional DWTPs and residential areas is relatively sparse. Although antibiotics and ARGs are frequently detected in DWTPs and their distribution systems may be important reservoirs of ARGs (Benotti et al. 2008; Xi et al. 2009), the dynamics and underlying mechanisms of ARGs throughout the water supply from the source to the point-of-use have not been well studied. More importantly, the number and diversity of ARGs detected in previous studies are not sufficient to provide a comprehensive understanding of ARG dynamics. In the current study, we investigated the ARG dynamics from the drinking water source to DWTPs. On the other hand, we compared the differences in ARGs between DWTP effluent and the tap water. The absolute abundance of ARGs is fully quantified by linking the water source to the tap water. Thanks to the high-throughput quantitative PCR (HT-qPCR), we targeted 295 individual ARGs, which represent the major classes of ARGs. Combining the Illumina sequencing, in this study we aimed to (1) characterize ARG profiles along the whole water supply, (2) evaluate any shift in water microbiota at different points of the water supply, (3) characterize the interactions, if any, between the among antibiotic resistome, environmental factors and bacterial community.
Material and methods
Sampling sites, sample collection and DNA extraction
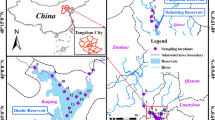
The sampling sites for this study were located in Zhangzhou City, Fujian Province, in the southeast of China. The drinking water source was located on the Jiulongjiang River, whereas both the DWTP and residential area were located within the city (Fig. 1).
River water (RW), influent water of coagulation-flocculation-sedimentation tank (IW), effluent water from the coagulation-flocculation-sedimentation (SW), sand filter water (SFW), finished water (FW), household tap water (HTW) were collected from the drinking water source (River Water, RW), DWTP and residential area, respectively (Fig. 1). Sterile plastic bottles were used for water sample collection. To minimize sampling variation, three replicate water samples (5 L per sample) were collected simultaneously at each sampling site (18 water samples in total) in July 2017. Samples were immediately transported to the laboratory in ice coolers within 6 h. Water samples were filtered using 0.22 μm nitrocellulose ester membranes (Toyo Roshi Co., Ltd., Japan), and the membrane filters were placed in tinfoil bags at −20 °C. The membranes were subsequently cut into small pieces using sterilized scissors. A FastDNA® SPIN Kit for Soil (MP Biomedicals, USA) was used for DNA extraction following the manufacturer’s protocol. The concentration and purity of DNA were measured by microspectrophotometry (NanoDrop ND-1000, Thermo Scientific, USA). DNA (80 μL) was stored at −20 °C prior to use.
The determination of physicochemical parameters and heavy metals
Water samples were acidified prior to heavy metal determination (Vu et al. 2017; Wijesiri et al. 2019). The water samples (100 mL per sample) were filtered through glass filter paper (pore size 0.45 μm) (Haiyan China). Metal determination was done by Inductively Coupled Plasma-Mass Spectrometer or ICP-MS (Agilent 7500cx, Agilent Technologies, USA). A Shimadzu TOC-V cph analyzer instrument (Wijesiri et al. 2019) was used to determine total nitrogen (TN) and total organic carbon (TOC). Turbidity of water samples was determined by a turbidimeter (HACH 2100AN, USA).
ARG and MGE quantification using high-throughput quantitative PCR
The Wafergen SmartChip Real-time PCR system (WaferGen Bio-systems, Inc., USA) was used for high-throughput qPCR analysis. A total of 296 of primer sets (Genscript, China) (Table S1) and PCR conditions were as previously described (Chen et al. 2017). A threshold cycle (Ct) of 31 was used as the detection limit for this study. Three technical replicates were conducted in all sampling sites. Results only adopted if three replicates were positive after HT-qPCR. Relative abundance of ARGs was calculated as previously (Zhu et al. 2013), and the absolute abundance of ARGs was normalized to 16S rRNA gene copy number (Ouyang et al. 2015). The mechanisms of antibiotic resistance genes were classified according to the previous study (Chen et al. 2017).
Illumina sequencing, data processing and analysis
The V4–V5 region of the 16s rRNA gene was used to characterize the composition and structure of bacterial community. PCR reactions followed (Zhou et al. 2019). Purification of PCR products used a TIANGEN DNA purification kit (TIANGEN Biotech, China), and products were brought to equal concentration prior to sequencing (Illumina Hiseq 2500 platform; Novogene, Beijing, China). Data processing was performed by Quantitative Insights Into Microbial Ecology (QIIME) (Caporaso et al. 2010). Open-reference operational taxonomic units (OTU) were set at 97% of similarity level by UCLUST clustering (Edgar 2010). Taxonomic characterization of OTUs used the Greengenes (version 13.8) database (McDonald et al. 2012).
Statistical analysis
Excel 2016 was used for calculating means and standard deviations. Principal coordinate analysis (PCoA), redundancy analysis (RDA), variation partitioning analysis (VPA), Heatmap, Procrustes and Mantel test were analyzed in R Studio with the following packages: vegan (version 2.2.0) (Dixon 2003), labdsv (version 1.8.0) (https://CRAN.R-project.org/package=labdsv) and pheatmap (version 0.7.7) (https://CRAN.R-project.org/package=pheatmap). Prior to the network analysis, pair wise Spearman rank correlations between detected ARGs were calculated. Only when the correlation coefficient (ρ) was > 0.8 and P < 0.01 was network construction generated (Li et al. 2015). Network visualization was provided by Cytoscape (version 3.3.0) software (Smoot et al. 2011). SPSS 21 was conducted for statistical assessments. Only when P <0.05 is considered significant in all statistical test.
Results
The profiles of antibiotic resistance genes
A total of 214 ARGs and 10 MGEs were detected in all water samples. Nine classes of ARGs were identified as aminoglycoside, beta-lactams, chloramphenicol, Macrolide-Lincosamide-Streptogramin B (MLSB), multidrug, sulfonamide, tetracycline, vancomycin and others. The detected number of ARGs and MGEs range from 38 to 173 (Fig. 2a). Those ARGs detected were classified into 4 major mechanisms: antibiotic deactivation (40.9%), cellular protection (15.2%), efflux pumps (35.5%) and others/unknown (8.3%) (Fig. 2b). A PCoA of ARGs based on the Bray-Curtis distance separated sample sites into three groups (Fig. 2c). The group which consisted of RW, IW and SW separated from the group that consisted of SFW, FW, along the first coordinate (P < 0.05, PERMANOVA). The composition of ARGs from the residential area (HTW) was significantly different to all sampling sites (P < 0.05, PERMANOVA) in both PC1 and PC2. The total absolute abundance of ARGs ranged from 2.1 × 105 to 1.9 × 1010 copies L−1. The absolute abundance of ARGs was highest at RW (P < 0.05), while samples from SFW had the lowest abundance compared to other sample points (P < 0.05). The total absolute abundance of ARGs in each sampling point was 1.9 × 1010 (RW), 5.1 × 109 (IW), 3.7 × 109 (SW), 2.1 × 105 (SFW), 5.3 × 105 (FW) and 1.8 × 107 (HTW) respectively. The absolute abundance of aminoglycoside, multidrug, tetracycline was significantly greater than beta-lactams, chloramphenicol, MLSB, others, sulfonamide and vancomycin in samples of RW respectively (P < 0.05) (Fig. 2d). Three chloramphenicol resistance genes (cmlA1-01, cmlA1-02 and cmx (A)) were detected in the study. The absolute abundance of chloramphenicol resistance genes was highest in SW and lowest in SFW. In HTW, the absolute abundance of cmlA1-01, cmlA1-02 and cmx (A) was significantly higher than FW and SFW respectively. The absolute abundance of 9 classes of ARGs and MGEs from household tap water (HTW) was significantly greater (P < 0.05) than SFW and FW. Overall, the absolute abundance of ARGs was significantly reduced (P < 0.05) throughout the water treatment and supply process (Fig. 3).
(a): Detected numbers of ARGs and MGEs. (b): The major antibiotic-resistant mechanisms. (c): Principal Coordinate Analysis (PCoA) of ARGs based on Bray-Curtis distance. (d): Absolute abundance of ARGs and MGEs at different sampling sites. A total of 9 classes of ARGs were classified: aminoglycoside, beta-lactams, chloramphenicol, macrolide-lincosamide-streptog-ramin B (MLSB), multidrug, sulfonamide, tetracycline, vancomycin and others. RW, IW, SW, SFW, FW and HTW represent river water, influent water of coagulation-flocculation-sedimentation tank, effluent water of coagulation-flocculation-sedimentation, send filter water, finished water and household tap water respectively. Data is calculated and presented as mean (n = 3) value
The profiles of bacterial community
A total of 1,639,725 high-quality sequences were detected in water samples from all sites, which ranged from 26,561 to 146,444. At the OTU level, SFW samples had the greatest bacterial diversity, followed by RW, IW, SW and HTW (Fig. 4a). Samples from FW had the lowest bacterial diversity, which was further confirmed by PD whole tree, Shannon index and chao1 analyses (Fig. S1). Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Cyanobacteria, Acidobacteria and Unassigned were the dominant phyla, which accounted for > 85% of recovered from water samples (Fig. 4b). A PCoA analysis of bacterial OTUs indicated a significant shift of bacterial communities during DWTP (P < 0.05, PERMANOVA) (Fig. 4c). DWTP effluent was separated from influent along PC1, which accounted for 40.32% of the variation (P < 0.05, PERMANOVA), while PC2 accounted for 19.61% of the variation.
Co-occurrence patterns between ARGs and bacterial phyla
Correlations between ARGs and bacterial taxa were evaluated by the Mantel test and Procrustes analysis. ARGs were strongly correlated (P < 0.0001, r = 0.818, Mantel test; M2 = 0.2809, P < 0.0001, r = 0.848, Procrustes analysis) with bacterial taxa at all sampling sites (Fig. 5a). Furthermore, a network analysis consisted of 55 nodes and 123 edges (Fig. 5b) and showed that Bacteriodetes had the most correlations with different ARGs including resistance genes for all 9 classes of antibiotic and MGEs.
(a): Procrustes analysis showing a significant correlation between antibiotic resistance genes (ARGs) and microbial community structure (ARG/OTU distribution patterns) based on Bray-Curtis dissimilarity metrics (sum of squares M2 = 0.2809, r = 0.848, P < 0.0001, 9999 permutations). Red line connecting the two data sets of groups. (b): Network analysis between resistomes and bacterial taxa in water samples. Node size was defined by the number of correlations, while edge was defined by the correlation coefficient. Blue and black edges represent the positive and negative correlations, respectively
Relationship among environmental factors, bacterial communities, MGEs and ARGs
The concentration of As in household tap water (HTW) was significantly higher than DWTP effluent (FW) (P < 0.05, Table S2). Most ARG classes significantly (P < 0.05, Table S2) correlated with Integron, Transposon and total absolute abundance of MGEs respectively (Table S3).
Redundancy analysis (RDA, Fig. 6a) identified that contributors to ARG profiles along the whole water supply were different. TOCs, MGEs and Cyanobacteria were major contributors in river water (RW); Cu and Actinobacteria contributed mainly in the first steps of drinking water treatment, while Acidobacteria, Firmicutes and Proteobacteria were key contributors during the second half of drinking water treatment (SFW, FW) and tap water. Environmental factors, bacterial community and MGEs jointly accounted for 92.7% of the total variation of ARGs (Fig. 6b). Bacterial community contributed 11.9% to the shift in ARGs, which was higher than environmental factors (1.3%) and MGEs (2.8%).
(a): Redundancy analysis (RDA) of the quantitative correlations between major bacterial phyla (> 1% in any sample) (Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Cyanobacteria, Acidobacteria), mobile genetic elements (MGEs) and antibiotic resistance genes (ARGs). (b): Variation partitioning analysis (VPA) differentiating effects of bacterial community, environmental factors and MGEs on changes of the antibiotic resistome
Discussion
Impact of drinking water treatment on dynamics of ARGs
In the current study, the ARG profile was assessed by HT-qPCR, describing the full dynamics and changes in ARG at different stages from drinking water sources, through DWTP to residential areas. The absolute abundance of ARGs in water samples was calculated and provided reference for further risk assessment. A total of 224 ARGs and MGEs out of 295 targeted genes were detected in all water samples. The detected number of ARGs was greatest in raw water (RW) and the first half of the water treatment process (IW and SW) (P <0.05). Post sand filtration, water samples (SFW) had the lowest detected number and absolute abundance of ARGs, which suggested a positive impact of the first half of DWTP. However, numbers and absolute abundance of ARGs increased after chlorination disinfection, suggesting that current disinfection methods may have a limited effect in reducing ARGs. It has been reported that the dosage rate of chlorine was critical in disinfection of ARGs and antibiotic-resistant bacteria (ARB), as low doses may facilitate the permeability of bacterial membranes, promoting gene transfer between bacteria (Zhang et al. 2020a). Furthermore, the enlarged number of ARB under a low dose of chlorine may trigger secondary pollution of ARGs after treatment (Huang et al. 2011). Similar to this result, the detected numbers and absolute abundance of aminoglycosides, beta-lactam and tetracycline resistance genes and MGEs were significantly higher (P <0.05) in this study after chlorination than in the sand filtered (SFW) effluent, which may be attributed to the change of bacterial community in the water samples. And it further suggested that the effluent water after the sand filtration step was contaminated. Therefore, a more effective and cost-friendly method is needed for removal of ARGs in DWTP effluent.
The final step of the drinking water supply, household tap water, links water supply infrastructure to human residences and thus is critical to human health. However, the total detected number and absolute abundance of ARGs, even the “last-resort” vancomycin, detected in HTW were significantly higher than in DWTP effluent, suggesting that ARGs may be transmitted through drinking water distribution systems (Chen et al. 2020). The significant increase of chloramphenicol resistance genes in HTW suggests that chlorine residues in pipes may contribute to the enrichment of ARGs in tap water (Shi et al. 2013). One alternative, similar to that found associated with heavy metal concentration, is the effect of nocturnal water stagnation in pipes (Zietz et al. 2003). Co-selection of heavy metal was considered to enrich ARGs in water samples (Seiler and Berendonk 2012). For example, arsenic and copper pollution can accelerate the enrichment of ARGs (Zhang et al. 2018; Zhang et al. 2020b). Biofilms are known to have considerable biomass in drinking water distribution systems (Waak et al. 2018) and may facilitate ARG enrichment through horizontal gene transfer due to the high cell density and close proximity in pipe systems (Flemming et al. 2002; Król et al. 2013). Also, water retention time in pipes may facilitate the potential risk of ARG distribution (Lautenschlager et al. 2010). Besides, in combination with the possible contributors mentioned above, the water residence time in the pipe system may facilitate the enrichment and distribution of ARGs (Lautenschlager et al. 2010). The enrichment of ARGs in HTW, suggesting the secondary pollution of ARGs in the drinking water distribution system, should not be neglected (Huang et al. 2011). The assessment of the potential impact of ARGs to human health is urgent and critical in the near future.
The correlation among environmental factors, microbiome and resistome
In the current study, the ARGs were significantly correlated with environmental factors and microbial communities respectively, suggesting that environmental variables may be responsible for the transmission of ARGs in the water samples. TOC and TN were positively correlated with ARGs in RW, indicating both of the factors were one of the important contributors in distribution of ARGs in that sampling site. The nutrient levels in water decreased after the treatment of DWTPs, and therefore, the contributors changed. The environmental variables such as carbon, nitrogen and other essential nutrients not only facilitate the growth of bacteria or ARB but also promote horizontal transfer of ARGs (Yuan et al. 2018; Zhang et al. 2020c). However, based on the VPA analysis, the contribution of environmental factors solely to ARGs was relatively low compared to the joint contribution of both environmental variables and bacteria community, suggesting the environmental factors may affect the ARGs indirectly through bacterial communities.
The contribution of phyla to ARGs changed with different sampling sites, likely due to differences in the composition of the bacterial community. The shift of bacterial community explained 11.9% of the variation of ARGs, higher than environmental variables and MGEs. These results were consistent with previous studies, which suggested the bacterial phylogeny was the major contributor to profile of ARGs (Forsberg et al. 2014; Zhou et al. 2019). Furthermore, the network analysis indicated co-occurrence between bacterial phyla and ARGs, suggesting that some bacteria are the potential host of ARGs. Notably, the dominant phyla in the current study may serve as the hosts for multiple ARGs. For instance, the aminoglycoside resistance gene (aphA1.aka.kan), MLSB resistance genes (ermx and mphA_02) and tetracycline resistance genes (tetQ and tetM_01) were strongly and significantly correlated with Bacteroidetes, which deserved further study to investigate potential risks.
MGEs are critical to horizontal gene transfer of ARGs between disparate groups of microorganisms (Gaze et al. 2011). In the present study, the major classes of ARGs were significantly associated with total MGEs, integrons and transposons, respectively, and suggest that MGEs may play an important role in shaping the characteristics of ARGs in water samples (Marshall and Levy 2011). Similar to environmental factors, the contribution of MGEs to ARGs was low. However, a combination of factors (bacterial community, environment and MGEs) rather than a single factor may drive the major changes in ARGs in water samples. In this study, factors that may affect ARGs in tap water, such as chlorine, were not monitored, which is one of the limitations of our work. Further evaluation is needed in the future to better assess the risk of ARG to human health.
Conclusion
In this study, the ARG profiles were evaluated by HT-qPCR and a total of 224 ARGs and MGEs out of 295 targeted resistance genes were identified from water sources, DWTP and household tap water samples. Although drinking water treatment from source to finished water reduced ARG number and absolute abundance, an increase was noted in tap water samples. Environmental factors, MGEs and bacterial community were the main drivers of ARG profiles. New treatment technologies, such as the treatment of powdered activated carbon, are needed to eliminate ARGs in drinking water. This study presents a dynamic distribution profile of ARGs from raw water source to tap water thus providing a baseline reference for subsequent risk assessment.
Data availability
All relevant data are within the manuscript and available from the corresponding author upon request
References
Bai XH, Ma XL, Xu FM, Li J, Zhang H, Xiao X (2015) The drinking water treatment process as a potential source of affecting the bacterial antibiotic resistance. Sci Total Environ 533:24–31. https://doi.org/10.1016/j.scitotenv.2015.06.082
Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. https://doi.org/10.1016/j.tim.2006.02.006
Benotti MJ, Trenholm RA, Vanderford BJ, Holady JC, Stanford BD, Snyder SA (2008) Pharmaceuticals and endocrine disrupting compounds in US drinking water. Environ Sci Technol 43:597–603
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303https://www.nature.com/articles/nmeth.f.303#supplementary-information
Chee-Sanford JC, Mackie RI, Koike S, Krapac IG, Lin YF, Yannarell AC, Maxwell S, Aminov RI (2009) Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J Environ Qual 38:1086–1108
Chen QL, Li H, Zhou XY, Zhao Y, Su JQ, Zhang X, Huang FY (2017) An underappreciated hotspot of antibiotic resistance: the groundwater near the municipal solid waste landfill. Sci Total Environ 609:966–973. https://doi.org/10.1016/j.scitotenv.2017.07.164
Chen J, Li W, Zhang J, Qi W, Li Y, Chen S, Zhou W (2020) Prevalence of antibiotic resistance genes in drinking water and biofilms: the correlation with the microbial community and opportunistic pathogens. Chemosphere 259:127483. https://doi.org/10.1016/j.chemosphere.2020.127483
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. https://doi.org/10.1111/j.1654-1103.2003.tb02228.x
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Flemming HC, Percival SL, Walker JT (2002) Contamination potential of biofilms in water distribution systems. In: Malzer HJ, Gimbel R, Schippers JC (eds) Innovations in conventional and advanced water treatment processes, vol 2. Water Science and Technology: Water Supply, vol 1. Iwa Publishing, London, pp 271–280
Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G (2014) Bacterial phylogeny structures soil resistomes across habitats. Nature 509:612–616. https://doi.org/10.1038/nature13377
Garner E, Chen C, Xia K, Bowers J, Engelthaler DM, McLain J, Edwards MA, Pruden A (2018) Metagenomic characterization of antibiotic resistance genes in full-scale reclaimed water distribution systems and corresponding potable systems. Environ Sci Technol 52:6113–6125. https://doi.org/10.1021/acs.est.7b05419
Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, Brown H, Davis S, Kay P, Boxall ABA, Wellington EMH (2011) Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. Isme J 5:1253–1261. https://doi.org/10.1038/ismej.2011.15
Guo XP, Li J, Yang F, Yang J, Yin DQ (2014) Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci Total Environ 493:626–631. https://doi.org/10.1016/j.scitotenv.2014.06.035
Huang JJ, Hu HY, Tang F, Li Y, Lu SQ, Lu Y (2011) Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res 45:2775–2781. https://doi.org/10.1016/j.watres.2011.02.026
Jia SY, Shi P, Hu Q, Li B, Zhang T, Zhang XX (2015) Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ Sci Technol 49:12271–12279. https://doi.org/10.1021/acs.est.5b03521
Khan S, Beattie TK, Knapp CW (2019) Rapid selection of antimicrobial-resistant bacteria in complex water systems by chlorine and pipe materials. Environ Chem Lett 17:1367–1373. https://doi.org/10.1007/s10311-019-00867-z
Król JE, Wojtowicz AJ, Rogers LM, Heuer H, Smalla K, Krone SM, Top EM (2013) Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid 70:110–119. https://doi.org/10.1016/j.plasmid.2013.03.003
Lautenschlager K, Boon N, Wang YY, Egli T, Hammes F (2010) Overnight stagnation of drinking water in household taps induces microbial growth and changes in community composition. Water Res 44:4868–4877. https://doi.org/10.1016/j.watres.2010.07.032
Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM, Zhang T (2015) Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9:2490–2502. https://doi.org/10.1038/ismej.2015.59. https://www.nature.com/articles/ismej201559#supplementary-information
Li W, Wang F, Zhang J, Qiao Y, Xu C, Liu Y, Qian L, Li W, Dong B (2016) Community shift of biofilms developed in a full-scale drinking water distribution system switching from different water sources. Sci Total Environ 544:499–506. https://doi.org/10.1016/j.scitotenv.2015.11.121
Liu SS, Qu HM, Yang D, Hu H, Liu WL, Qiu ZG, Hou AM, Guo J, Li JW, Shen ZQ, Jin M (2018) Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res 136:131–136. https://doi.org/10.1016/j.watres.2018.02.036
Marshall BM, Levy SB (2011) Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 24:718–733
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J 6:610–618. https://doi.org/10.1038/ismej.2011.139
Ouyang WY, Huang FY, Zhao Y, Li H, Su JQ (2015) Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl Microbiol Biotechnol 99:5697–5707. https://doi.org/10.1007/s00253-015-6416-5
Pruden A, Pei RT, Storteboom H, Carlson KH (2006) Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ Sci Technol 40:7445–7450. https://doi.org/10.1021/es0604131
Sanganyado E, Gwenzi W (2019) Antibiotic resistance in drinking water systems: occurrence, removal, and human health risks. Sci Total Environ 669:785–797. https://doi.org/10.1016/j.scitotenv.2019.03.162
Seiler C, Berendonk TU (2012) Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:10. https://doi.org/10.3389/fmicb.2012.00399
Shi P, Jia S, Zhang XX, Zhang T, Cheng S, Li A (2013) Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res 47:111–120. https://doi.org/10.1016/j.watres.2012.09.046
Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432. https://doi.org/10.1093/bioinformatics/btq675
Tan QW, Li WY, Zhang JP, Zhou W, Chen JP, Li Y, Ma J (2019) Presence, dissemination and removal of antibiotic resistant bacteria and antibiotic resistance genes in urban drinking water system: a review. Front Environ Sci Eng 13:15. https://doi.org/10.1007/s11783-019-1120-9
Vu CT, Lin C, Shern CC, Yeh G, Le VG, Tran HT (2017) Contamination, ecological risk and source apportionment of heavy metals in sediments and water of a contaminated river in Taiwan. Ecol Indicatorsecol Indic 82:32–42. https://doi.org/10.1016/j.ecolind.2017.06.008
Waak MB, LaPara TM, Hallé C, Hozalski RM (2018) Occurrence of Legionella spp. in water-main biofilms from two drinking water distribution systems. Environ Sci Technol 52:7630–7639. https://doi.org/10.1021/acs.est.8b01170
Wellington EMH, Boxall ABA, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM, Williams AP (2013) The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis 13:155–165. https://doi.org/10.1016/s1473-3099(12)70317-1
Wijesiri B, Liu A, Deilami K, He B, Hong N, Yang B, Zhao X, Ayoko G, Goonetilleke A (2019) Nutrients and metals interactions between water and sediment phases: an urban river case study. Environ Pollut 251:354–362. https://doi.org/10.1016/j.envpol.2019.05.018
Witte W (1998) Medical consequences of antibiotic use in agriculture. Science 279:996–997
Xi C, Zhang Y, Marrs CF, Ye W, Simon C, Foxman B, Nriagu J (2009) Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol 75:5714–5718
Xu L, Ouyang W, Qian Y, Su C, Su J, Chen H (2016) High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ Pollut 213:119–126. https://doi.org/10.1016/j.envpol.2016.02.013
Yang L, Hu YF, Liu Y, Zhang JD, Ulstrup J, Molin S (2011) Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol 13:1705–1717. https://doi.org/10.1111/j.1462-2920.2011.02503.x
Yuan QB, Zhai YF, Mao BY, Hu N (2018) Antibiotic resistance genes and intI1 prevalence in a swine wastewater treatment plant and correlation with metal resistance, bacterial community and waste. Water Parameters Ecotox Environ Saf 161:251–259. https://doi.org/10.1016/j.ecoenv.2018.05.049
Zhang ML, Chen LH, Ye CS, Yu X (2018) Co-selection of antibiotic resistance via copper shock loading on bacteria from a drinking water bio-filter. Environ Pollut 233:132–141. https://doi.org/10.1016/j.envpol.2017.09.084
Zhang ML, Wang L, Xu M, Zhou H, Wang S, Wang YJ, Bai M, Zhang C (2019) Selective antibiotic resistance genes in multiphase samples during biofilm growth in a simulated drinking water distribution system: occurrence, correlation and low-pressure ultraviolet removal. Sci Total Environ 649:146–155. https://doi.org/10.1016/j.scitotenv.2018.08.297
Zhang GS, Li WY, Chen S, Zhou W, Chen JP (2020a) Problems of conventional disinfection and new sterilization methods for antibiotic resistance control. Chemosphere 254:12. https://doi.org/10.1016/j.chemosphere.2020.126831
Zhang ML, Wan K, Zeng J, Lin WF, Ye CS, Yu X (2020b) Co-selection and stability of bacterial antibiotic resistance by arsenic pollution accidents in source water. Environ Int 135:11. https://doi.org/10.1016/j.envint.2019.105351
Zhang Y, Wang J, Lu J, Wu J (2020c) Antibiotic resistance genes might serve as new indicators for wastewater contamination of coastal waters: spatial distribution and source apportionment of antibiotic resistance genes in a coastal bay. Ecol Indicatorsecol Indic 114:106299. https://doi.org/10.1016/j.ecolind.2020.106299
Zhou S-Y-D, Zhu D, Giles M, Yang X-R, Daniell T, Neilson R, Zhu Y-G (2019) Phyllosphere of staple crops under pig manure fertilization, a reservoir of antibiotic resistance genes. Environ Pollut 252:227–235. https://doi.org/10.1016/j.envpol.2019.05.098
Zhu Y-G, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM (2013) Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci 110:3435–3440. https://doi.org/10.1073/pnas.1222743110
Zietz BP, de Vergara JD, Dunkelberg H (2003) Copper concentrations in tap water and possible effects on infant's health—results of a study in Lower Saxony, Germany. Environ Res 92:129–138. https://doi.org/10.1016/s0013-9351(03)00037-9
Funding
This work was funded by the National Natural Science Foundation of China (41807460, 41977210).
Author information
Authors and Affiliations
Contributions
F-YH and Q-LC designed the experimental protocol and carried out the experiments. XZ and S-Y-DZ assisted with the experiments. F-YH, Q-LC and S-Y-DZ wrote the manuscript. RN, J-QS revised the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 306 kb)
Rights and permissions
About this article
Cite this article
Huang, FY., Chen, QL., Zhang, X. et al. Dynamics of antibiotic resistance and its association with bacterial community in a drinking water treatment plant and the residential area. Environ Sci Pollut Res 28, 55690–55699 (2021). https://doi.org/10.1007/s11356-021-14896-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-14896-1