Abstract
Recent studies have expanded the interests about microbial community and function following the rapid development of high-throughput sequencing techniques in the freshwater ecosystem. In this study, we aimed to attain a deep understanding of microbial community structure and potential nitrogen metabolism in Hulun Lake, a shallow hypereutrophic steppe lake in the Mongolian Plateau in China. The result demonstrated that cyanobacteria were the most dominant phylum. Network analysis showed both intra- and inter-phylum co-occurrence were pervasive, and there were modular structures in the microbial assemblages. The cluster dominated by proteobacteria was mainly negatively connected to the cluster dominated by both proteobacteria and actinobacteria. Cyanobacteria were tightly clustered together and positively connected to these two clusters. The major nitrogen metabolism pathways were glutamine synthetase–glutamate synthase and assimilatory nitrate reduction, indicating the nitrogen was mainly retained in the lake by microbial uptake. Cyanobacteria contributed 43.25% gene reads involved in the overall nitrogen metabolism but mainly contributed to assimilatory nitrate reduction and nitrogen fixation, aggravating the lake eutrophication. This study adds to our knowledge of microbial assemblages and nitrogen metabolism in the shallow hypereutrophic lake and provided an insight understanding for the purposes of lake ecosystem’s protection and efficient management in the Mongolian Plateau.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The major biogeochemical cycles on the earth have been dramatically altered by anthropogenic disturbances (Conley 1999; Falkowski et al. 2000). Large quantities of the nutrients have been delivered into aquatic ecosystems (Savage et al. 2010), stimulating primary production and accelerating eutrophication process (Cloern 2001). Excessive nutrient enrichment or eutrophication has been identified as the most important threat to aquatic ecosystems all over the world (Smith and Schindler 2009; Ansari et al. 2011; Woodward et al. 2012) and is widely associated with water quality degradation, biodiversity loss, and ecosystem function disruption (Rabalais 2002; Dudgeon et al. 2006).
Microbial community is the important fundamental composition of aquatic ecosystem processes and functions (Loreau 2001; Cotner and Biddanda 2002). As the major both producers and decomposers in the lake ecosystems, microbial community plays an important role in biogeochemical cycles, especially the carbon and nitrogen cycles (Cole et al. 2007; Cotner et al. 2010; Reverey et al. 2016). Nutrients in freshwater systems come from various sources which are major determinants of a bacterial community (Jardillier et al. 2005; Elser et al. 2007; Barlett and Leff 2010). However, the anthropogenic nutrient enrichment usually caused excessive cyanobacteria growth or severe cyanobacterial blooms (Xu et al. 2010; Qu et al. 2017), which has caused a critical concern for the environment management authorities. Considered one of the major reasons of aquatic ecosystem degradation (Paerl and Huisman 2009; Michalak et al. 2013), meanwhile, the cyanobacterial bloom also posed high threats to drinking water supply, fisheries, and food web alteration (Paerl et al. 2011; Zanchett and Oliveira-Filho 2013; Gobler et al. 2016). Understanding microbial community composition and functional potentials is thus of great interest of ecological processes and mechanisms, meanwhile, it also provides an important insight into the restoration and management of lakes (Schiewer 1998; Wu et al. 2007; Conty and Bécares 2013). Specifically, many cyanobacterial taxa are well known for their ability to fix atmospheric nitrogen, which is a potentially important route to maintain the demands of nitrogen (N) cycle during the period of excessive growth (Latysheva et al. 2012; Cottingham et al. 2015). The role of nutrient supply driving cyanobacteria in lakes has been extensively studied (Paerl et al. 2011; Wilhelm et al. 2011; Ma et al. 2015) and, nevertheless, is still obscure. Recently, the high-throughput sequencing provides a possible way to deeply understand the specific nitrogen cycling pathways based on the abundance of functional genes (Ren et al. 2017; Qu et al. 2017; Price et al. 2018). However, understanding of bacterial co-occurrence network and the nitrogen metabolism in the cyanobacterial bloom lake remains limited, especially in the hypereutrophic stepper shallow lakes.
Hulun Lake is the large freshwater shallow lake located in the steppe of northeast China. Due to climate change and anthropogenic activities, Hulun Lake is gradually changing from mesotrophic condition, eutrophic condition to hypereutrophic condition. The cyanobacterial bloom in summer had been reported (Zhai et al. 2013; Liang et al. 2016). As the largest freshwater lake in the Inner Mongolian Plateau, the National Wetland Nature Reserve and a Wetland of International Importance (Ramsar 2002) study of eutrophication in Hulun Lake is pressing. In this study, using high-throughput Illumina sequencing of 16S rRNA, we aimed to address these questions: (1) what is the microbial structure when dominated by cyanobacteria? (2) how do the cyanobacteria co-occur with other bacteria? and (3) what is the gene abundance for specific nitrogen cycling pathways? By understanding the microbial community structures and the potential nitrogen metabolism pathways in the lake with cyanobacteria bloom, this study can provide an insight into the eutrophication process in lakes.
Methods
Study area
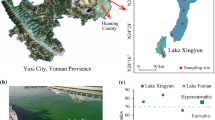
Hulun Lake (48° 33′–49° 20′ N, 116° 58′–117° 48′ E) is located in Hulun Buir of the Inner Mongolian Steppe in northeast China (Fig. 1). As the fifth largest lake and the fourth largest freshwater lake in China, Hulun Lake has a surface area of 2043 km2, an average perimeter of 447 km, and an average depth of 5.7 m in the wet season. Hulun Lake is located in the semi-arid high latitude westerly region belonging to the temperate continental climate region, with the mean annual air temperature, precipitation, and evaporation of − 0.2 °C, 290 mm, and 1600 mm, respectively (Xie et al. 2015; Lu et al. 2016). In the short spring and autumn period, the strong wind blow usually provides high effects on the sediment deposition. In the winter season, the lake surface is covered by ice and experienced an anaerobic environment about 170–180 days. Hulun Lake is mainly fed by precipitation and more than 80 tributaries (Li et al. 2013; Cai et al. 2016). The two major tributaries are the Wuerxun River and Kherlen River (Fig. 1). The Kherlen River originates from the Khentii Mountains in Mongolian People’s Republic with a length of 1090 km in Mongolia and 206 km in China. The Wuerxun River flows from Beier Lake, the sister lake of Hunlun Lake located in the China-Mongolia border region. The watershed of Hulun Lake belonged to the typical steppe grassland type and mainly covered by Stipa krylovii Roshev, Caragana stenophylla Pojark, and Artemisia frigida Willd (Liu et al. 2011). During the past decade, the rapid land use change, steppe degradation in the watershed, and the water contamination of its tributaries have caused the severe nutrient enrichment in Hulun Lake (Zhai et al. 2011; Wang et al. 2012), the cyanobacteria bloom was reported recently (Zhai et al. 2013; Liang et al. 2016).
Field sampling and analysis
Fieldwork was conducted and totally 15 sites were sampled in July 2016. The conductivity, dissolved oxygen, and water temperature were measured by using a handheld multiparameter water quality monitor (YSI Model 85) in situ. The pH was measured by using handheld YSI Pro 10. Microbial community samples were collected from the surface water samples at a depth of 0.5 m. A total of 600 ml water were filtered with Whatman Nylon membrane (pore size 0.2 μm) and immediately stored and transported in liquid nitrogen. In the lab, the microbial samples were stored at − 80 °C refrigerator shortly until DNA extraction. Meanwhile, surface water samples were collected and transported immediately to the laboratory at the portable incubator 4 °C filled with ice cubes at each site. Total nitrogen (TN), total phosphorus (TP), soluble reactive phosphorus (SRP), ammonium (NH4+), nitrate (NO3−), and dissolved organic carbon (DOC) were analyzed to provide the nutrient status of Hulun Lake (Table 1).
DNA extraction, PCR, and sequencing
The microbial community was measured and analyzed through 16S rRNA genes. Following manufacturer protocols of the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA, USA), genomic DNA of microbe was extracted. The 16S rRNA genes covering V3 to V4 regions were amplified using primers 806R and 338F (Invitrogen, Vienna, Austria). The polymerase chain reaction (PCR) was performed according to the standard procedures of Applied Biosystems 2720 Thermal Cycler (ABI, USA). By using a 1× TAE buffer, amplified DNA samples were verified by 1.0% agarose gel electrophoresis and purified using the Gel Extraction Kit (Qiagen, Hilden, Germany). The final sequencing process mainly based on a MiSeq sequencing platform (Illumina, USA). By using the software of QIIME (Caporaso et al. 2010), sequencing data were cleaned. Then, operational taxonomic units (OTUs) were clustered based on the complete linkage algorithm at an identity level of 97% sequence.
Statistical analysis
The relative abundances of the OTUs were calculated at each sampling site. Spearman correlation was used to calculate the pairwise correlation between OTUs. For the purpose of focusing on the most commonly occurring OTUs and decreasing the effects of rare OTUs, only those with relative abundance higher than 0.01% were analyzed by applying the Hmisc package in R program (version 3.3.2). Based on the statistical analysis, the significant (P < 0.001) and robust correlations (Spearman’s correlation coefficient of |R| > 0.75) remained. This filter cutoff values allow us to focus on the strongest correlations. The network visualization was made using Cytoscape (version 3.4.0). Each edge represents a robust and significant correlation and each node represents an OTU. A set of integrative metrics were calculated and compared to describe the network topology. For example, the average number of neighbors explains the complex pairwise connections. And the average path length describes node distribution. The subnetwork of cyanobacteria was extracted from the overall network. This subnetwork provided more detailed information about those nodes and edges which directly interacted with the nodes of Cyanobacteria. The modular structure of the networks was also analyzed and visualized using the clusterMaker in Cytoscape. To get insight into the nitrogen metabolism pathway, we used PICRUSt to identify the functional genes encoding the necessary enzymes associated with the specific nitrogen-cycling pathway based on Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Results
Bacterial community composition and structure
In our study, 3787 bacterial OTUs with 190,671 sequences were obtained. Totally, 39 bacterial phyla were detected in Hulun Lake samples. The most dominant phylum was Cyanobacteria (relative abundance of 39.1%) followed by Proteobacteria (20.2%), Bacteroidetes (12.2%), and Actinobacteria (12.2%) (Fig. 2). The top three orders were Nostocales (relative abundance of 13.0%), Synechococcales (11.1%), and Stramenopiles (8.6%), which were affiliated with Cyanobacteria (Fig. 3). In Hulun Lake, TN concentration was 1.453 ± 0.059 mg/L and TP concentration was 0.152 ± 0.013 mg/L (Table 1). Compared with the trophic classification boundary (Dodds and Whiles 2010), Hulun Lake was in a hypertrophic status enriched by both nitrogen and phosphorus.
The average relative abundance (bar) of the microbial community and the standard division (whisker) were shown at the phylum level. The dominant phyla with an average relative abundance ≥ 1% were showed, and the unclassified, unidentified, and the ones with an average relative abundance < 1% were showed in the group “Others”
Bacteria co-occurrence network
In the network analysis, the overall interaction patterns were explored in this hypereutrophic steppe lake (Fig. 4a). In addition, to get insights into co-occurrence interactions between Cyanobacteria and other bacteria, we extracted the cyanobacterial subnetwork (the network contained the nodes of Cyanobacteria and the nodes which directly interacted with the nodes of Cyanobacteria, Fig. 4b) from the overall network. The overall network contained 784 nodes and 6761 edges (Fig. 4a), and the cyanobacterial subnetwork contained 363 OTUs (145 OTUs belonging to Cyanobacteria) and 2156 edges (Fig. 4b). Moreover, the results showed that the positive correlations (co-occurrence interactions between OTUs) were dominant, containing 639 OTUs and 6574 edges (Fig. 4a). The negative correlations (co-exclusion interactions between OTUs), however, only contain 91 OTUs and 187 edges (Fig. 4a). The result of community cluster using clusterMaker showed community clusters of the microbial assemblages in Hulun Lake (Fig. 4c). It showed that the Cyanobacteria were tightly clustered together in Cluster-2 (Fig. 4c). Cluster-1 was dominated by Proteobacteria and Cluster-3 was dominated by both Actinobacteria and Proteobacteria (Fig. 4c). Cluster-1 and Cluster-3 were connected mainly by negative correlations, while Cluster-2 was connected to Cluster-1 and Cluster-3 mainly by positive correlations (Fig. 4a, c).
The edge-weighted interactions network based on the Spearman correlation coefficient between OTUs. The blue line and black line indicate positive (co-occurrence) and negative association (co-exclusion), respectively. The node size indicates the relative abundance. (a) the overall network; (b) the cyanobacterial subnetwork; (c) the result of community cluster using ClusterMaker app
A total of six topological parameters, describing the OTUs interactions, compactness, diversity, and modularity, were calculated and compared between the real networks and randomly generated networks (Table 2). The higher values of real network diameter, network centralization, network heterogeneity, characteristic path length, and clustering coefficient were confirmed in comparing with the random networks (Table 2). The results confirmed that both the overall network and cyanobacterial subnetwork showed a more clustered topology. Moreover, the modularity values were 0.935 and 0.924 at the overall network and Cyanobacterial network, respectively. The modular network suggested when the modularity value is higher than 0.4 (Newman 2006). And the results exhibited relative high modularity in Hulun Lake (Table 2).
Potential nitrogen metabolism
In this study, we probe into nitrogen cycling from the functional genes in a hypereutrophic lake, such as nitrogen fixation, nitrate reduction, nitrification, denitrification, and ammonium assimilation. Overall, cyanobacteria contributed 43.25% reads involved in nitrogen metabolism. Reads associated with nitrogen fixation represented 5.54% of the sequences assigned to N metabolism (Fig. 5). Reads associated with assimilatory nitrate reduction and dissimilatory nitrate reduction represented 10.31% and 6.50% of the sequences assigned to N metabolism, respectively (Fig. 5). Cyanobacteria contributed 76.87% in gene sequences encoding enzymes that catalyze the assimilatory nitrate reduction. In our study, 3.82% associated reads contributed to the denitrification process from nitric oxide to nitrogen (Fig. 5). Cyanobacteria only contributed 2.13% in gene sequences encoding enzymes that catalyze denitrification. In our study, reads associated with GDH and GS-GOGAT represented 7.00% and 40.54% of the sequences assigned to N metabolism, respectively. Cyanobacteria contributed 27.03% and 36.42% in gene sequences encoding enzymes that catalyze GDH and GS-GOGAT pathways, respectively.
A simplified nitrogen cycling schematic based on KEGG nitrogen metabolism pathway. The interconnected biogeochemical pathways major included (1) nitrogen fixation from nitrogen to ammonia, (2) nitrification from ammonia to nitrate, (3) both dissimilatory and assimilatory nitrate reduction at the reverse direction from nitrate to ammonia, (4) denitrification from nitrite to nitrogen, (5) anammox from both nitrite and ammonia to nitrogen, and (5) nitrate reduction through the GDH and GS-GOGAT. The nitrogen forms ranged from the oxidation states in nitrate (+5) to reduction states in ammonium (− 3). Based on KEGG database, the percentages of gene reads associated with each stage of nitrogen cycles were predicated using PICRUSt
Discussion
Cyanobacteria dominated the microorganism composition and its possible nitrogen and phosphorus resources
In general, Proteobacteria, Bacteroidetes, and Actinobacteria always dominated the bacteria phyla in lake water columns (Zwart et al. 2002; Kirchman 2002). In Hulun Lake, however, Cyanobacteria became the most abundant phylum. This result was in line with the previous studies of microorganisms in Hulun Lake (Zhao et al. 2007; Chen et al. 2012; Liang et al. 2016). The high relative abundance of Cyanobacteria usually stimulated by the high nutrient concentrations in lakes and streams (Smith et al. 1999; Guildford and Hecky 2000), especially the high P concentration (Rejmankova and Komarkova 2000; Xu et al. 2010; Ho and Michalak 2015). Through fixing dissolved N2 gas and releasing it into the water column in the forms of biological available, Cyanobacteria expanded the nitrogen pools in lake ecosystems (Howarth et al. 1988; Cottingham et al. 2015). The phosphorus (P) usually is a limiting factor for the community development of phytoplankton (Schindler 1977); however, some cyanobacterial taxa can utilize the P in sediments and increase available pools of phosphorus in the freshwater lake (Schindler et al. 2008; Zhu et al. 2013). Moreover, the microorganism can scavenge P and tie it up in biomass, resulting in legacy P stocks, which can sustain a high level of primary productivity for decades (McMahon and Read 2013). Meanwhile, the frequently disturbed sediment and particles falling from the atmosphere may also provide more phosphorus resources and need to be considered.
Complex network with high modularity
In Hulun Lake, the co-occurrence network of microbial assemblages represented a relatively complex structure with high modularity. Network analysis has been used to reveal relationships among individual and provides a useful tool to understand the structure, modularity, and interaction in the highly complex microbial communities (Newman 2006; Fuhrman 2009; de Menezes et al. 2015). The network also provided a deep insight into microbial co-occurrence patterns, community stability, and resilience (Barberan et al., 2012; Faust and Raes 2012; Freedman and Zak 2015). The more complex of the co-occurrence patterns, the more sustainable to human disturbance. Numerous studies have demonstrated that diverse heterotrophic microbes occurred within cyanobacterial blooming lakes and rivers (Berg et al. 2009; Wilhelm et al. 2011; Cai et al. 2013; Bagatini et al. 2014). However, we still have a limited understanding of microbial co-occurrence, seasonal variation, and responses to global climate change in the hypereutrophic steppe lake. Modularity usually is one of the important characteristics to understate the tightly-connected nodes (or individuals in the aquatic ecosystems) in the large complex system (Newman 2006), providing further understanding of microbial interactions across the view of whole lake ecosystems and to enhance understanding of their co-occurrence and co-exclusion relationships (de Menezes et al. 2015; Qu et al. 2017; Ren et al. 2017). In the Hunlun Lake study, the widely positive correlations were identified across different phyla of the microbial community. The symbiotic relationship and competition relationship usually showed more intensive species interactions than with the rest of the species in the community (Tonkin et al. 2018). And highly interconnected species grouped into a module represented as one of the characteristics of the eutrophic and hypereutrophic freshwater lakes or reservoirs (Newman 2006; Conty and Bécares 2013; Freedman and Zak 2015; Lee and Biggs 2015).
Microorganisms play a key role in nitrogen metabolism
Microbial community plays a key role in nitrogen metabolism, which is a complex process that involves various nitrogen chemical forms (Ollivier et al. 2011). Usually, understanding the nitrogen metabolism processes is important and would be the first step in order to properly manage aquatic ecosystems and prevent eutrophic lakes (Paerl and Otten 2013; McMahon and Read 2013; Ma et al. 2015). In the lake with cyanobacterial bloom, it is also interesting to compare the contributions of cyanobacteria and other bacteria to nitrogen metabolism (Rabalais 2002; Lehman 2011; Paerl et al. 2011), Cyanobacteria, mostly Nostoceae, Rivulariaceae, Scytonemataceae, and Stigonemataceae, and the other few organisms those which could carry out the nitrogen fixation process (Burris and Roberts 1993; Bernhard 2012). In our study, Cyanobacteria contributed 49.32% in gene sequences encoding enzymes that catalyze nitrogen fixation. Low concentration of ammonium and N:P ratios can stimulate some N2-fixing bacteria (Meeks et al. 1983; Smith et al. 1999; Guildford and Hecky 2000). The assimilatory process is a microbial process that starts with the reduction of NO3− to NO2− and then to NH4+, which is a kind of inorganic nitrogen and can easily be bioavailable to the cell to incorporate nitrogen into biomolecules, then can be transformed by nitrification (Richardson and Watmough 1999). It has been widely reported that nutrient enrichment, such as the increasing the concentration of NO3− could stimulate the growth rate of microorganisms. The functional genes including necessary enzyme-coding sequences required for nitrate reduction would increase significantly (Kirchman 2002; Förster et al. 2003). The other pathway retaining the bioavailable nitrogen in the form of ammonium is the dissimilatory nitrate reduction to ammonia (DNRA) (Tiedje et al. 1982; Zumft 1997). However, DNRA is also a competing process to denitrification, which is the dominant pathway to the removal of nitrogen from freshwater ecosystems by microorganisms (Tiedje et al. 1982; Seitzinger 1988). In our study, the denitrification was relatively low and only about 3.82% of the reads associated to denitrification. And Cyanobacteria only contributed 2.13% in gene sequences encoding enzymes that catalyze denitrification (Fig. 5).
The ammonia/ammonium incorporation pathways, the glutamine synthetase–glutamate synthase (GS-GOGAT) pathway and the NADP-dependent glutamate dehydrogenase (GDH), were well known in microorganisms. (Tempest et al. 1970). In our study, Cyanobacteria contributed 36.42% and 27.03% in gene sequences encoding enzymes that catalyze the GS-GOGAT and GDH pathways, respectively. The pathways of GS-GOGAT (ammonium-glutamine-glutamate) and GDH (ammonium-glutamate) provide important nitrogen resources for the synthesis of N-containing compounds in microorganisms (Stadtman 2001). Although it is also well known that the GS-GOGAT pathway usually dominated the glutamate synthesis when the concentration of NH4+ is low (Helling 1998), and it may highly related with cyanotoxins releasing (Banerjee et al. 2017).
Conclusion
This study explored the microbial community structure and potential function in Hulun Lake, a shallow hypereutrophic steppe lake, the fifth largest lake and the fourth largest freshwater lake in China. The most dominant phylum was Cyanobacteria (28.9%) which most likely be stimulated by the high nutrient concentrations and a relative low N:P ratio. Based on microbial community and its functional data, a bacterial co-occurrence network and potential pathways of nitrogen cycling in the form of the simplified model were showed. The microbial assemblages in Hulun Lake exhibited a modular structure and co-occurred with other bacteria. The major pathways were GS-GOGAT and assimilatory nitrate reduction, which highlight the importance of nitrate reduction and glutamate synthesis. The lower abundance of gene reads associated with the denitrification pathway suggested that the nitrogen in the water column is mainly retained in the lake. Based on metagenomic comparisons of cyanobacteria and other bacteria, cyanobacteria contributed more to the abundance of functional genes than OTUs. Most of the gene reads involved in assimilatory nitrate reduction and a half in nitrogen fixation were contributed by cyanobacteria. Eutrophication process and excessive growth of Cyanobacteria have been treated as one of the greatest threats to inland water during the past several decades. This result shed light on community structure and nitrogen metabolism of microbial assemblages in a hypereutrophic lake.
References
Ansari AA, Gill SS, Lanza GR, Walter R (2011) Eutrophication: causes, consequences and control. Springer, London
Bagatini IL, Eiler A, Bertilsson S, Klaveness D, Tessarolli LP, Henriques Vieira AA (2014) Host-specificity and dynamics in bacterial communities associated with bloom-forming freshwater phytoplankton. PLoS One 9:e85950
Banerjee S, Subramanian A, Chattopadhyaya J, Sarkar RR (2017) Exploring the role of GS–GOGAT cycle in microcystin synthesis and regulation – a model based analysis. Mol BioSyst 13:2603–2614
Barlett MA, Leff LG (2010) The effects of N:P ratio and nitrogen form on four major freshwater bacterial taxa in biofilms. Can J Microbiol 56:32–43
Berg KA, Lyra C, Sivonen K, Paulin L, Suomalainen S, Tuomi P, Rapala J (2009) High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J 3:314–325
Bernhard A (2012) The nitrogen cycle: processes, players, and human impact. Natural education knowledge 3:25
Burris RH, Roberts GP (1993) Biological nitrogen fixation. Annu Rev Nutr 13:317–335
Cai HY, Yan ZS, Wang AJ, Krumholz LR, Jiang HL (2013) Analysis of the attached microbial community on mucilaginous Cyanobacterial aggregates in the eutrophic Lake Taihu reveals the importance of Planctomycetes. Microb Ecol 66:73–83
Cai ZS, Jin TY, Li CY, Ofterdinger U, Zhang S, Ding AZ, Li JC (2016) Is China’s fifth-largest inland lake to dry-up? Incorporated hydrological and satellite-based methods for forecasting Hulun lake water levels. Adv Water Resour 94:185–199
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chen X, Chuai X, Yang L, Zhao H (2012) Climatic warming and overgrazing induced the high concentration of organic matter in Lake Hulun, a large shallow eutrophic steppe lake in northern China. Sci Total Environ 431:332–338
Cloern JE (2001) Our evolving conceptual model of the coastal eutrophication problem. Mar Ecol Prog Ser 210:223–253
Cole JJ, Prairie YT, Caraco NF, McDowell WH, Tranvik LJ, Striegl RG, Duarte CM, Kortelainen P, Downing JA, Middelburg JJ, Melack J (2007) Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems 10:172–185
Conley DJ (1999) Biogeochemical nutrient cycles and nutrient management strategies. Hydrobiologia 410:87–96
Conty A, Bécares E (2013) Unimodal patterns of microbial communities with eutrophication in Mediterranean shallow lakes. Hydrobiologia 700:257–265
Cotner JB, Biddanda BA (2002) Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5:105–121
Cotner JB, Hall EK, Scott JT, Heldal M (2010) Freshwater bacteria are stoichiometrically flexible with a nutrient composition similar to seston. Front Microbiol 1:132
Cottingham KL, Ewing HA, Greer ML, Carey CC, Weathers KC (2015) Cyanobacteria as biological drivers of lake nitrogen and phosphorus cycling. Ecosphere 6:1–19
Dodds WK, Whiles MR (2010) Freshwater ecology: concepts and environmental applications of limnology, Second edn. Academic press
Dudgeon D, Angela AH, Gessner MO, Kawabata ZI, Knowler DJ, Lévêque C, Naiman RJ, Prieur-Richard AH, Soto D, Stiassny MLJ, Sullivan CA (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biol Rev 81:163
Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, Ngai JT, Seabloom EW, Shurin JB, Smith JE (2007) Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett 10:1135–1142
Falkowski P, Scholes RJ, Boyle E, Canadell J, Canfield D, Elser J, Gruber N, Hibbard K, Högberg P, Linder S, Mackenzie FT, Moore B III, Pedersen T, Rosenthal Y, Seitzinger S, Smetacek V, Steffen W (2000) The global carbon cycle: a test of our knowledge of earth as a system. Science 290:291–296
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nature 10:538
Förster J, Famili I, Fu P, Palsson B, Nielson J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13:244–253
Freedman ZB, Zak DR (2015) Atmospheric N deposition alters connectance, but not functional potential among saprotrophic bacterial communities. Mol Ecol 24:3170–3180
Fuhrman JA (2009) Microbial community structure and its functional implications. Nature 459:193–199
Gobler CJ, Burkholder JM, Davis TW, Harke MJ, Johengen T, Stow CA, Van de Waal DB (2016) The dual role of nitrogen supply in controlling the growth and toxicity of cyanobacterial blooms. Harmful Algae 54:87–97
Guildford SJ, Hecky RE (2000) Total nitrogen, total phosphorus, and nutrient limitation in lakes and oceans: is there a common relationship? Limnol Oceanogr 45:1213–1223
Helling RB (1998) Pathway choice in glutamate synthesis in Escherichia coli. J Bacteriol 180:4571–4575
Ho JC, Michalak AM (2015) Challenges in tracking harmful algal blooms: a synthesis of evidence from Lake Erie. J Great Lakes Res 41:317–325
Howarth RW, Marino R, Cole JJ (1988) Nitrogen fixation in fresh-water, estuarine, and marine ecosystems 2: biogeochemical controls. Limnol Oceanogr 33:688–701
Jardillier L, Boucher D, Personnic S, Jacquet S, Thenot A, Sargos D, Amblard C, Debroas D (2005) Relative importance of nutrients and mortality factors on prokaryotic community composition in two lakes of different trophic status: microcosm experiments. FEMS Microbiol Ecol 53:429–443
Kirchman DL (2002) Inorganic nutrient use by marine bacteria. In: Bitton G (ed) Encyclopedia of environmental microbiology. Wiely, New York
Latysheva N, Junker VL, Palmer WJ, Codd GA, Barker D (2012) The evolution of nitrogen fixation in cyanobacteria. Bioinformatics 28:603–606
Lee RM, Biggs TW (2015) Impacts of land use, climate variability, and management on thermal structure, anoxia, and transparency in hypereutrophic urban water supply reservoirs. Hydrobiologia 745:263–284
Lehman JT (2011) Nuisance cyanobacteria in an urbanized impoundment: interacting internal phosphorus loading, nitrogen metabolism, and polymixis. Hydrobiologia 661:277–287
Li C, Sun B, Jia K, Zhang S, Li W, Shi X, M C, Cordovil DS, Pereira LS (2013) Multi-band remote sensing based retrieval model and 3D analysis of water depth in Hulun Lake, China. Math Comput Model 58:771–781
Liang LE, Li CY, Shi XH, Zhao SN, Tian Y, Zhang LJ (2016) Analysis on the eutrophication trends and affecting factors in Lake Hulun, 2006-2015. J Lake Sci 28:1265–1273
Liu H, Guo WL, Quan WJ (2011) Climatic division of the types and yields of grassland in Inner Mongolia. J Appl Meteorol Sci 22:329–335
Loreau M (2001) Microbial diversity, producer-decomposer interactions and ecosystem processes: a theoretical model. Proc R Soc B Biol Sci 268:303–309
Lu C, Wang B, He J, Vogt RD, Zhou B, Guan R, Zuo L, Wang W, Xie Z, Wang J, Yan D (2016) Responses of organic phosphorus fractionation to environmental conditions and lake evolution. Environ Sci Technol 50:5007–5016
Ma JR, Qin BQ, Wu P, Zhou J, Niu C, Deng JM, Niu HL (2015) Controlling cyanobacterial blooms by managing nutrient ratio and limitation in a large hyper-eutrophic lake: Lake Taihu, China. J Environ Sci 27:80–86
McMahon KD, Read EK (2013) Microbial contributions to phosphorus cycling in eutrophic lakes and wastewater. Annu Rev Microbiol 67:199–219
Meeks JC, Wycoff KL, Chapman JS, Enderlin CS (1983) Regulation of expression of nitrate and dinitrogen assimilation by Anabaena species. Appl Environ Microbiol 45:1351–1359
de Menezes AB, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH (2015) Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ Microbiol 17:2677–2689
Michalak AM, Anderson EJ, Beletsky D, Boland S, Bosch NS, Bridgeman TB, Chaffin JD, Cho K, Confesor R, Daloglu I, DePinto JV, Evans MA, Fahnenstiel GL, He L, Ho JC, Jenkins L, Johengen TH, Kuo KC, LaPorte E, Liu X, McWilliams MR, Moore MR, Posselt DJ, Richards RP, Scavia D, Steiner AL, Verhamme E, Wright DM, Zagorski MA (2013) Record-setting algal bloom in Lake Erie caused by agricultural and meteorological trends consistent with expected future conditions. Proc Natl Acad Sci U S A 110:6448–6452
Newman ME (2006) Modularity and community structure in networks. Proc Natl Acad Sci U S A 103:8577–8582
Ollivier J, Towe S, Bannert A, Hai B, Kastl EM, Meyer A, Su MX, Kleineidam K, Schloter M (2011) Nitrogen turnover in soil and global change. FEMS Microbiol Ecol 78:3–16
Paerl HW, Huisman J (2009) Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Environ Microbiol Rep 1:27–37
Paerl HW, Otten TG (2013) Harmful cyanobacterial blooms: causes, consequences, and controls. Microb Ecol 65:995–1010
Paerl HW, Xu H, McCarthy MJ, Zhu GW, Qin BQ, Li YP, Gardner WS (2011) Controlling harmful cyanobacterial blooms in a hyper-eutrophic lake (lake Taihu, China): the need for a dual nutrient (n & p) management strategy. Water Res 45:1973–1983
Price MN, Zane GM, Kuehl JV, Melnyk RA, Wall JD, Deutschbauer AM, Arkin AP (2018) Filling gaps in bacterial amino acid biosynthesis pathways with high-throughput genetics. PLoS Genet 14:e1007147
Qu XD, Ren Z, Zhang HP, Zhang M, Zhang YH, Liu XB, Peng WQ (2017) Influences of anthropogenic land use on microbial community structure and functional potentials of stream benthic biofilms. Sci Rep 7:15117.
Rabalais NN (2002) Nitrogen in aquatic ecosystems. AMBIO: A Journal of the Human Environment 31:102–112
Ramsar (2002) Ramsar site no. 1146. People’s Republic of China names 14 new Ramsar sites. http://www.ramsar.org/
Rejmankova E, Komarkova J (2000) A function of cyanobacterial mats in phosphorus-limited tropical wetlands. Hydrobiologia 431:135–153
Ren Z, Gao HK, Elser JJ, Zhao QD (2017) Microbial functional genes elucidate environmental drivers of biofilm metabolism in glacier-fed streams. Sci Rep 7:12668
Reverey F, Grossart HP, Premke K, Gunnar L (2016) Carbon and nutrient cycling in kettle hole sediments depending on hydrological dynamics: a review. Hydrobiologia 775:1–20
Richardson DJ, Watmough NJ (1999) Inorganic nitrogen metabolism in bacteria. Curr Opin Chem Biol 3:207–219
Savage C, Leavitt PR, Elmgren R (2010) Effects of land use, urbanization, and climate variability on coastal eutrophication in the Baltic Sea. Limnol Oceanogr 55:1033–1046
Schiewer U (1998) 30 years’ eutrophication in shallow brackish waters - lessons to be learned. Hydrobiologia 363:73–79
Schindler DW (1977) Evolution of phosphorus limitation in lakes. Science 195:260–262
Schindler DW, Hecky RE, Findlay DL, Stainton MP, Parker BR, Paterson MJ, Beaty KG, Lyng M, Kasian SEM (2008) Eutrophication of lakes cannot be controlled by reducing nitrogen input: results of a 37-year whole-ecosystem experiment. Proc Natl Acad Sci U S A 105:11254–11258
Seitzinger SP (1988) Denitrification in freshwater and coastal marine ecosystems: ecological and geochemical significance. Limnol Oceanogr 33:702–724
Smith VH, Schindler DW (2009) Eutrophication science: where do we go from here? Trends Ecol Evol 24:201–207
Smith VH, Tilman GD, Nekola JC (1999) Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environ Pollut 100:179–196
Stadtman ER (2001) The story of glutamine synthetase regulation. J Biol Chem 276:44357–44364
Tempest DW, Meers JL, Brown CM (1970) Synthesis of glutamate in Aerobacter aerogenes by a hitherto unknown route. Biochem J 117:405–407
Tiedje JM, Sexstone AJ, Myrold DD, Robinson JA (1982) Denitrification: ecological niches, competition and survival. Antonie Van Leeuwenhoek 48:569–583
Tonkin JD, Heino J, Altermatt F (2018) Metacommunities in river networks: the importance of network structure and connectivity on patterns and processes. Freshw Biol 63:1–5
Wang G, Song J, Xue B, Xu X, Otsuki K (2012) Land use and land cover change of Hulun Lake nature reserve in Inner Mongolia, China: a modeling analysis. J Fac Agric Kyushu Univ 57:219–225
Wilhelm SW, Farnsley SE, LeCleir GR, Layton AC, Satchwell MF, DeBruyn JM, Boyer GL, Zhu G, Paerl HW (2011) The relationships between nutrients, cyanobacterial toxins and the microbial community in Taihu (Lake Tai), China. Harmful Algae 10:207–215
Woodward G, Gessner MO, Giller PS, Gulis V, Hladyz S, Lecerf A, Malmqvist B, McKie BG, Tiegs SD, Cariss H, Dobson M, Elosegi A, Ferreira V, Graça MAS, Fleituch T, Lacoursière JO, Nistorescu M, Pozo J, Risnoveanu G, Schindler M, Vadineanu A, Vought LBM, Chauvet E (2012) Continental-scale effects of nutrient pollution on stream ecosystem functioning. Science 336:1438–1440
Wu QL, Chen YW, Xu KD, Liu ZW, Hahn MW (2007) Intra-habitat heterogeneity of microbial food web structure under the regime of eutrophication and sediment resuspension in the large subtropical shallow Lake Taihu, China. Hydrobiologia 581:241–254
Xie Z, He J, Lu C, Zhang R, Zhou B, Mao H, Song W, Zhao W, Hou D, Wang J, Li Y (2015) Organic carbon fractions and estimation of organic carbon storage in the lake sediments in Inner Mongolia Plateau, China. Environ Earth Sci 73:2169–2178
Xu H, Paerl HW, Qin BQ, Zhu GW, Guang G (2010) Nitrogen and phosphorus inputs control phytoplankton growth in eutrophic Lake Taihu, China. Limnol Oceanogr 55:420–432
Zanchett G, Oliveira-Filho EC (2013) Cyanobacteria and cyanotoxins: from impacts on aquatic ecosystems and human health to anticarcinogenic effects. Toxins 5:1896–1917
Zhai D, Xiao J, Zhou L, Wen R, Chang Z, Wang X, Jin X, Pang Q, Itoh S (2011) Holocene East Asian monsoon variation inferred from species assemblage and shell chemistry of the ostracodes from Hulun Lake, Inner Mongolia. Quat Res 75:512–522
Zhai D, Xiao J, Fan J, Zhou L, Wen R, Pang Q (2013) Spatial heterogeneity of the population age structure of the ostracode Limnocythere inopinata in Hulun Lake, Inner Mongolia and its implications. Hydrobiologia 716:29–46
Zhao HY, Li CC, Zhao HH, Tian HC, Song QW, Kou ZQ (2007) The climate change and its effect on the water environment in the Hulun Lake Wetland. J Glaciol Geocryol 29:795–801
Zhu MY, Zhu GW, Li W, Zhang YL, Zhao LL, Gu Z (2013) Estimation of the algal-available phosphorus pool in sediments of a large, shallow eutrophic lake (Taihu, China) using profiled SMT fractional analysis. Environ Pollut 173:216–223
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616
Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han SK (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155
Acknowledgments
We are grateful to the anonymous reviewers for the comments, to Mr. Bai and Mr. Xin for their great assistance in the field, to Dr. Lu Tan for the water chemistry analyses.
Funding
This study was financially supported by the National Natural Science Foundation of China (No. 41671048 and 51439007). This study was also supported by the Project of the State Key Laboratory of Simulation and Regulation of Water Cycle in River Basin (SKL2018CG02), National Key R & D Program of China (2017YFC0404506), and the IWHR Research and Development Support Program (WE0145B532017).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Robert Duran
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qu, X., Zhang, M., Yang, Y. et al. Taxonomic structure and potential nitrogen metabolism of microbial assemblage in a large hypereutrophic steppe lake. Environ Sci Pollut Res 26, 21151–21160 (2019). https://doi.org/10.1007/s11356-019-05411-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-05411-8