Abstract
To investigate the impact of an exotic Frankia nodulated tree (Alnus trabeculosa) on soil nitrogen content, soil microbial composition, and the abundance of N turnover-related functional microorganism community, we compared the community structure and abundance of key functional genes (nifH, bacterial/archaeal amoA, and nosZ) in the rhizosphere and nonrhizosphere of monoculture of Phragmites australis and A.trabeculosa–P.australis mixed communities by MiSeq Illumina sequencing and real-time PCR, respectively. The introduction of Frankia nodulated tree to recover degraded wetland was effective in the accumulation of soil organic carbon and nitrogen, which was the key factor to impact on the bacterial community composition revealed by canonical correspondence analysis. Acidobacteria and Proteobacteria were the dominant bacterial phylums while seven rare phyla appeared the most phylogenetically different among the investigated soil of two vegetations, including Chlorobi, Cyanobacteria, OD1, OP11, TM6, TM7, and GN02. The gene copy numbers of nifH were ranged from 2.28 × 108 to 2.96 × 109 copies g-1 dry soil in the wetland, and which were significantly higher in soil samples from P. australis than that from A.trabeculosa. While the abundance of nosZ in both rhizosphere and nonrhizosphere soils of A.trabeculosa–P.australis mixed communities was significantly lower compared with P.australis monoculture. The potential nitrification (PNA) (0.15–0.41 mg NOx-N kg−1 dry soil d−1) in the rhizosphere of A. trabeculosa was significantly higher than that of P. australis, and the soil denitrification enzyme activity (DEA) (0.42–0.90 nmol N2O-N g−1 dry soil h−1) was lower in the mixed community compared with monoculture of P. australis. The introduced planting of Frankia nodulated tree effectively accumulated soil organic carbon and nitrogen and reduce the relative abundance and activity of nitrogen-fixing bacteria and denitrification bacteria.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Tidal wetlands occupy the ecotone between terrestrial and aquatic coastal ecosystems, are characterized by the highest biodiversity and various important ecological functions (Bodelier and Dedsh, 2013). However, wetland degradation has been the major environmental problem in China in recent years (Zhang et al., 2010). Because of frequent and intensive disturbances, human-induced environmental changes often conflict with biodiversity conservation targets, which lead to changes in plant species composition. Therefore, in view of the roles of plants as ecosystem engineers, effective restoration of damaged and degraded wetlands through vegetation management is needed to restore wetlands to support both a diversity of species and ecosystem services (Zedler, 2000).
A fundamental question in establishing mixed-species restoration plantings is to choose appropriate plant species with consideration of native species. Phragmites australis, regarded to be one of the most widely distributed angiosperms, is a widely spread native species in China. Due to high cellulose levels, P. australis is harvested annually for papermaking with a productivity of up to 18–28 tons of dry weight per acre per year (Sathitsuksanoh et al., 2009). Many studies have documented that P. australis is responsible for the majority of inorganic nitrogen removal through direct sequestration into plant tissues (Findlay et al., 2002), thereby depleting sediment nitrogen pools. In order to achieve a fast recovery by woody vegetation and to increase soil fertility in wetland ecosystems, many revegetation programs have been conducted by planting fast growing exotic trees such as Alnus trabeculosa (Boudiaf et al., 2013). Because this Frankia nodulated tree is supposed to increase soil nitrogen availability (Selmants et al., 2005), which was believed to be the limiting nutrient in coastal wetlands (Scott et al., 2007), it has often been recommended to enhance productivity and biodiversity of disturbed wetland. Therefore, these Frankia nodulated trees may have an important impact upon N dynamics in these systems.
Increased N levels under N-fixing trees may shift soil microbial community, which play a vital role in soil nutrition cycling (Chowdhury et al., 2009). They participate in important ecological processes such as nitrogen fixation, ammonification, nitrification, and denitrification. However, the diversity of functional microbial communities in wetland after introduction of woody vegetation is poorly understood, which could potentially control vegetation composition in wetlands (Lamers et al. 2012). In particular, we hypothesize that replacing native vegetation with these Frankia nodulated trees will change soil N-cycling microbial abundance and improve nitrogen sustainability of the wetland ecosystem. However, the influence of introducing planting of Frankia nodulated tree on the microorganisms involved in nitrogen turnover is not clear.
In this study, we investigated two important aspects of restoration planting with introduction of N-fixing tree: (1) N dynamic and deposit in wetland sediment with or without the introduction of an N-fixing tree. (2) the effect of N-fixing tree on the soil microbial composition, especially the abundance of functional microorganisms involved in key processes of the inorganic nitrogen cycle, and to link these results to abiotic and biotic properties of the soils of different vegetations.
Materials and methods
Study sites and soil sampling
Soil samples were collected in the Chongxi wetland (31° 42′–31° 44′ N, 121° 12′–121° 16′ E) located in the southwest of Chongming Island in the Yangtze Estuary near Shanghai, China. The wetland has an area of about 300 hm2 with a mean annual temperature of 16 °C and precipitation of 1100 mm. The wetland is a semi-diurnal tidal regime with water salinity of less than 1 ‰ (Gao and Liu, 2008). The tidal marsh is dominated by monospecific plant of P. australis, and which is harvested annually by local peasants for papermaking. The woody plant, A.trabeculosa, was introduced into the high tidal plants of the Chongxi wetland for forested wetland restoration in January 2006. It was planted in a patch shape in the P. australis community, with an interplant distance of 2 m. With the integrated indexes of survival ratio, height, crown width, and diameter at breast height, A.trabeculosa grew excellently and the most quickly after 2 years of transplantation.
Rhizosphere (R) and nonrhizosphere (N) soil samples of pure P. australis in community (PA-R, PA-N) and A.trabeculosa–P. australis mixed community (mPA-R, mPA-N, mAT-R, mAT-N) were collected at each site with three independent field replicates using a soil auger from 0- to 20-cm depth, each replicates being composed of five individual soil cores (taken in a distance of 1 m2), which were pooled and homogenized to reduce heterogeneity. To collect nonrhizosphere soil, plants and soil were transferred in polyethylene bags and vigorously shaken by hand for 10 min until roots non-adhering soil particles were completely removed. Rhizosphere soil was afterwards collected by shaking roots for 10 min (150 rpm) in sterile NaCl (0.9 %) solution to remove the adhering soil, and subsequently centrifuged (1700 g, 10 min) to concentrate soil particles in the pellet (Aboudrar et al., 2007). Aliquots of 5–10 g for DNA extraction were shock frozen in liquid nitrogen directly after sampling and stored at −80 °C, whereas the remaining soil was stored at 4 °C and analyzed in the following 2 weeks. The soils were air-dried and sieved (mesh size 2 mm) and soil physicochemical properties were determined as follows. Soil pH was measured with a soil/water ratio of 1:5. Soil organic matter content (SOM), total C (TC) and N (TN) were determined by TOC analyzer HT1300 (Multi N/C 3100, Analytik Jena AG, Germany). Determination of total phosphorus (TP), ammonium (NH4 +-N), and nitrate (NO3 −-N) concentrations was done by spectrophotometer. These physicochemical properties of the six soils are listed in Table 1.
Potential nitrification and denitrification enzyme activity assay
The potential nitrification (PNA) was measured according to Kurola et al. (2005) with minor modification. In brief, approximately 5 g fresh paddy soil without gravels and plant roots was added to 50 mL jars containing 20 mL of 1 mM phosphate-buffered saline (PBS) and 1 mM of (NH4)2SO4 at 25 °C in the dark for 24 h. Potassium chlorate (final concentration 10 mg l−1) was added to inhibit nitrite oxidation. For extracting NOx-N content, 5 mL of 2 M KCl was added at the end of incubation, and the supernatant of which after centrifugation was analyzed immediately using a continuous flow injection analyzer (FIA QC8500, Lachat America).
The soil denitrification enzyme activity (DEA) was performed as described by Barton et al., (2000) with some modification. 5 g fresh soil was weighed into a glass serum bottle (120 mL) and sealed with rubber septa and aluminum crimp cap. Each bottle was flushed with high-purity N2 gas, and amended with a 10 mL solution containing 1 mM glucose, 1 mM potassium nitrate and 0.39 mM chloramphenicol. Approximately 15 % (V = V) acetylene gas was added to inhibit N2O reduction. After incubated at 25 °C for 0.25 and 1.0 h, 5 mL of headspace was sampled and stored in a 3 mL evacuated vacutainer for N2O analysis.
DNA extraction and real-time PCR
DNA was extracted from ca. 0.5 g (fresh weight) of soil using a MoBio PowerSoil™ DNA extraction kit (MoBio laboratories; Carlsbad, CA, USA), following the manufacturer’s instructions. The concentration and quality of the extracted DNA was determined by spectroscopic analysis (Nano-drop, Wilmington, Delaware, USA) and agarose gelelectrophoresis. DNA was stored at −20 °C.
Copy numbers of the functional genes (amoA, nosZ) were determined by real-time PCR using an iCycleriQ 5 thermocycler (Bio-Rad, Calif, USA). The PCR primers were shown in Table S1. To optimize the real-time PCR reaction system, some of the DNA extracts were diluted 100-fold or 10-fold and used as template. As standards, serial plasmid dilutions of the respective functional genes were used, and the parameters of the standard curves were listed in Table S1, including slope, intercept, the linear coefficient correlation (R 2) and PCR efficiency.
The 25 μL reaction mixtures included 1 μl of template DNA, 12.5 μL of SYBR Premix Ex Taq (Takara BioInc, Shiga, Japan), 500 nM of the primers. All PCR runs started with an initial enzyme activation step performed at 95 °C for 5 min. The subsequent thermal profile was different for each gene as shown in Table S1, followed by a melting curve analysis was from 65 to 98 °C, 0.2 °C per read, 6-s hold. Fluorescence was read during each cycle at 83 °C.
Illumina sequencing and analysis
In total, the microbial communities of 18 soil samples (3 plants × 2 treatments with rhizosphere or nonrhizosphere × 3 plots) were analyzed by Illumina MiSeq sequencing. Microbial sequencing was performed using the MiSeq Illumina platform at Meiji Biotechnology Company (Shanghai, China), as previously described by Caporaso et al. (2010). Briefly, the V4 region of 16S bacterial ribosomal DNA (rDNA) was amplified using the custom degenerate primer pair 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCCTTTGAGTTT-3′) to generate an amplicon size of 392 bp. Sequences of <300 bp were removed from the resulting data, with 1193,282 unique sequences ultimately being obtained.
The total PCR reaction volume was 25 μL and contained 2.5 μL of 10× buffer, 25 mM MgCl2, 2.5 mM of each dNTP, 0.5 U of Taq DNA polymerase (rTaq, TaKaRa, Dalian, China), 3.0 pmol of each of the forward and reverse primers, and ~50 ng of DNA template. The PCR cycling conditions were as follows: 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 52–64 °C for 30 s and 72 °C for 40 s, with a 72 °C extension period for 7 min.
Sequence analysis was performed with both QIIME (Caporaso et al. 2010) and Mothur (Schloss et al. 2009). The sequences were also denoised using Amplicon Noise and checked for chimeras using Perseus (Quince et al. 2011). This output was clustered with Uclust (Edgar 2010) at the 97 % sequence identity level, which resulted in 8810 operational taxonomic units (OTUs). A representative sequence from each OTU was classified using the Ribosomal Database Project (RDP) classifier (Cole et al. 2007) and a training set extracted from the Silva108 database (Quast et al. 2012). All sequences have been submitted to Sequence Read Archive under the BioProject accession number SRP051305.
Statistical analyses
Real-time PCR data and soil chemical parameters were subjected to analysis of variance (ANOVA) with Duncan test performed at the significance level of p < 0.05 using the statistical software program SPSS (vesion18.0, Chicago, Illinois, USA). The Pearson correlation analysis was used to test the correlations between PNA, DEA, functional gene abundance, and different soil chemical properties, using the software SPSS. The effects of soil properties on the microbial community were analyzed by canonical correspondence analysis (CCA) using the community ecology package ‘vegan’ of R (Oksanen et al., 2010). Canonical correspondence analysis (CCA) ordination diagrams were generated using 1000 times permutation tests, and p values ≤0.05 were considered statistically significant. Bacterial beta diversity patterns across the rhizosphere and nonrhizosphere of vegetations were assessed using unweighted UniFrac distance matrix (Lozupone and Knight, 2005). The plots in this study were created using the Origin program (version 9.0).
Results
Physicochemical properties and microbial N transform activity
Soil properties including SOC, TP, and TN showed a significant difference between samples from the P. australis monoculture and A. trabeculosa–P. australis mixed communities (Table 1). Soil organic carbon contents in both rhizosphere and nonrhizosphere soil of A. trabeculosa were significantly higher than that in soil of P. australis, regardless in monoculture or in mixed communities. In addition, SOC content was higher in the rhizosphere compared with the nonrhizosphere of A. trabeculosa and P. australis. Similarly, total N content was increased after the introduction of A. trabeculosa. The nonrhizosphere of P. australis in the mixed community had much higher TN content (3.6 g kg−1) than that in the monoculture (1.16 g kg−1). Ammonium was the prevalent inorganic nitrogen in wetland. Both of ammonium and nitrate were significant increased in the rhizosphere and nonrhizosphere soil of mixed community compared with monoculture of P. australis. However, TP was decreased with the introduction of A. trabeculosa, and which was lowest in the nonrhizophere of A. trabeculosa (1.95 g kg−1). The N:P ratios in soils of A. trabeculosa and P. australis ranged from 0.28 to 1.97.
The potential nitrification (PNA) ranged from 0.15 to 0.41 mg NOx-N kg−1 dry soil d−1, with A. trabeculosa was significantly higher than P. australis in the rhizosphere, but the trend was quite opposite in the nonrhizosphere. The soil denitrification enzyme activity (DEA) was lower in the mixed community compared with monoculture of P. australis. As revealed by the Pearson correlation analysis, PNA had no significant relationship with soil properties, while DEA was significantly positively correlated with TP and negatively correlated with ratio of N:P (Table 2).
Quantification of functional genes
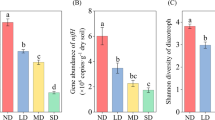
After gene copy numbers normalized to the amount of extracted DNA, four detected functional genes were various between six soils and ranged in the order of magnitude 107–109 copies g-1 dry soil (Fig. 1). Gene copy numbers of nifH were significantly higher than the other three genes, and ranged between 2.28 × 108 and 2.96 × 109 copies g-1 dry soil. For soil samples from P. australis, significantly higher copy numbers were measured than the soil from A.trabeculosa, with the exception of mPA-N, where values were comparable. The nifH gene copy numbers in the rhizosphere were significantly higher than that in the nonrhizosphere in A.trabeculosa–P. australis mixed communities, but there was no significant difference between the rhizosphere and nonrhizosphere in pure P. australis community.
The gene copy numbers for nifH, amoA of AOA and AOB, nosZ in rhizosphere and nonrhizosphere soil samples of Phragmites australis monoculture, and A. trabeculosa–P. australis mixed communities. Error bars indicate the standard deviation of three replicates (n = 4). Within the abundance values for each gene in the soils of different vegetations, the different letters indicate a significant difference at p < 0.05 (Duncan’s ANOVA)
The abundance of archaeal amoA gene ranged between 4.75 × 106and 8.93 × 107 copies g-1 dry soil, and was different between different plant types. Gene copy numbers of archaeal amoA were highest in the soil sample from PA-N, while the lowest in the soil sample of mAT-R. In addition, compared with archaeal amoA gene copy numbers in PA-N, the abundance of archaeal amoA gene were significantly decreased in mPA-N (1.64x107 copies g-1 dry soil) by a factor of 5.44, while there was no significant difference between PA-R and mPA-R. Bacterial amoA gene copy numbers were comparable between all six soils and ranged between 1.24 × 107 and 2.01 × 107 copies g-1 dry soil. In general, archaeal amoA gene copy numbers dominated over bacterial amoA genes by a factor of 1.62 to 7.27 except for mPA-N and mAT-N.
For soil samples from PA-R, a significantly higher copy number of nosZ gene (7.10 × 107 copies g-1 dry soil) was measured than with the other five soils. The nosZ gene copy numbers in rhizosphere were significantly higher than that in nonrhizosphere in both pure P. australis communities and A.trabeculosa–P. australis mixed communities. After introducing planting of A.trabeculosa, the abundance of nosZ in both rhizosphere and nonrhizosphere soils of A.trabeculosa–P. australis mixed communities (mPA-R and mPA-N) was significantly decreased (for mPA-R and mPA-N, 5.59 × 107 and 2.25 × 107 copies g-1 dry soil, respectively; for PA-R and PA-N, 7.10 × 107 and 5.32 × 107copies g-1 dry soil, respectively).
As revealed by the Pearson correlation analysis, the abundance of archaeal amoA was negative correlated with SOM, TN, nitrate and ratio of N:P, while abundance of bacteria amoA was positive correlated with these properties. In addition, the abundance of nosZ was negative correlated with soil nitrogen content (Table 2).
Microbial community structure
A total of 1,193,282 16S rRNA sequences were generated through MiSeq Illumina sequencing, with an average read length of 396 bp. A total of 8809 OTUs (defined at the 97 % sequence similarity level) were found in soil samples from the investigated vegetations. The bacterial composition derived from the two vegetations were dominated by Proteobacteria, which showed a relative abundance varying from 31.4 % at mPAN to 35.4 % at PAR. Within this phylum, Deltaproteobacteria (13.0–14.6 %) exhibited a relatively higher abundance than other sub-classes (Fig. 2a). At the phylum level, Acidobacteria contributed 15.2 % (PAN) to 20.1 % (mATR) of the total sequences. Compared with monoculture P. australis, the abundance of Acidobacteria in the soil of mixed community (mPA, mAT) was increased irrespective of the rhizosphere and nonrhizosphere. The total contributions of the seven rare phyla (<0.1 %), including Chlorobi, Cyanobacteria, OD1, OP11, TM6, TM7, and GN02 appeared the most phylogenetically different among the investigated soil of two vegetations (Fig. S1, Fig. 2b). The abundance of Chlorobi and OD1 in the soil of A.trabeculosa (mATR, mATN) was significantly higher than that in P. australis (PAR, PAN). In addition, the abundance of TM6 was increased in both mPAR, mPAN compared with PAR, PAN, respectively.
Besides the shifts in rare abundance phyla, the relative abundance of some major individual taxa was impacted by the vegetations (Table S2). In the class of Alphaproteobacteria, bacteria of the order Rhizobiales were significantly different between various plants and which was significantly more abundance in the rhizosphere (on average, 1.61–2.64 %) than that in the nonrhizosphere (on average, 1.19–2.10 %). The proportions of Thermodesulfovibrinaceae in the order Nitrospirales were lower in the A.trabeculosa–P. australis mixed community than that in P. australis. Lower proportions of other taxa were also observed in the A.trabeculosa–P. australis mixed community, including the Deltaproteobacteria genus Desulfobacterales, the Desulfuromonadales, NB1-j, and the Actinobacteria order Actinomycetales, family Micrococcaceae. OD-1 had much higher proportions in the A.trabeculosa–P. australis mixed community than in P. australis.
Correlations of the microbial community with physicochemical characteristics
Canonical correspondence analysis (CCA) was performed to explore the key environmental factors influencing community composition (Fig. 3b). Total N, ammonia, and SOC significantly influenced the microbial community derived from the rhizosphere and nonrhizosphere of A.trabeculosa and P. australis (p < 0.05). Microorganisms in the mixed vegetation community, with the exception of mPAR, were significantly positively correlated with the above environmental factors. Principal coordinate analysis with unweighted UniFrac distance matrix indicated that bacterial composition was more distinct across vegetations than across the rhizosphere and nonrhizosphere (Fig. 3a) (p < 0.05). Bacterial composition in mATR was quite different from that in the mixed vegetation communities. On the other hand, bacterial composition in mATN was intermixed and not different from other bacterial composition from the mixed vegetation.
Principal coordinate analysis (PCoA) plots of unweighted UniFrac distances (a) and canonical correspondence analysis (CCA) ordination plots (b) showing patterns of beta diversity in bacterial communities across vegetations and the relationship between the bacterial community composition and environmental factors, respectively. The optimal CCA models represented by the diagrams were produced with standardization explanatory variables by the weighting and scaling of scores focusing on inter-group correlations using F statistic significance tests. Correlations between the environmental variables and CCA axes are represented by the length and angle of the lines (environmental factor vectors)
Discussion
In the restoration of degraded fields, C input and an increase in soil N content are of great importance because they can enhance the capacity of the system to support a more complex community (Macedo et al. 2008). Frankia nodulated plants have been widely used in the recovering of tropical and sub-tropical forests (Siddique et al. 2008; Macedo et al. 2008). In this study, not only soil N but also SOM has been increased by Frankia nodulated A. trabeculosa, which was consistent with previous studies (Macedo et al. 2008; Wang et al., 2010). Results of SOM and TN in the present study also showed that A. trabeculosa was able to restore C- and N-cycling better than the native species of P. australis. Huang et al. (2014) have revealed that mixing N-fixing species into Eucalyptus urophylla plantations can enhance soil C sequestration by increasing TOC and microbial biomass carbon (MBC). Including N-fixing tree species in mixed-species restoration plantings may increase and accelerate the carbon sequestration potential of the ecosystem (Kaye et al., 2000), which attributed to higher growth rates of N-fixing trees and subsequent higher C inputs into the soil via litter and root exudates (e.g., Resh et al., 2002; Wang et al., 2010). In this respect, introducing planting of N-fixing species of A. trabeculosa could enhance soil C and N sequestration and retention, which improved the soil quality and could be potentially used for wetland restoration.
Our results showed that the introduction of A. trabeculosa exerts strong effects on soil microbial community composition, which was altered not only in the soil of A. trabeculosa but also in the soil of P. australis in the mixed community. The distinction between bacterial communities which was pronounced in different vegetations could be ascribed to the differences in edaphic factors, i.e., C and N contents were significantly different with the introduction of A. trabeculosa. This is consistent with previous studies showing that introduction of exotic plants exerted significant effects on soil properties, which in turn affect microbial abundance and community composition (Liu et al. 2010). Presumably, differences in quality and quantity of root exudates and in root morphology known to drive microbial communities and to form the basis for microbial activity in the rhizosphere might have contributed to the different effects observed (Mao et al., 2011). Typically, despite belonging to the same vegetation, bacterial composition in the rhizosphere and nonrhizosphere in the present study also showed quite a difference. This reinforces the notion of exotic Frankia nodulated tree impact on soil microbial communities and consistent with Remigi et al. (2008) who found that an exotic legume tree species induced strong modifications in soil microbial functionalities and the structure of the Arbuscular Mycorrhizal community.
Acidobacteria, Bacteroidetes, Chloroflexi, and Proteobacteria were the dominant bacterial groups in the investigated tidal wetland, especially for the Acidobacteria and Deltaproteobacteria contributed >10 % of overall sequences. These groups are abundant in most ecosystems and their dominance was also observed in other coastal tidal wetlands (Feng et al., 2009; Yu et al., 2012). While this study showed mixed community (mPA, mAT) have higher abundance of Acidobacteria than the monoculture P. australis irrespective of the rhizosphere and nonrhizosphere. The consistent abundance of recalcitrant carbon degrading Acidobacteria found in this study is congruent with a recent study (Lin et al., 2014), which showed that the dominant phylum of Acidobacteria was increased in a cedar plantation invaded by bamboo (Lin et al., 2014). The distribution and abundance of Acidobacteria in soil have been demonstrated to be influenced by the types and availability of organic resources, such as readily oxidizable carbon from root exudates and plant polymers from plant litter degradation (Eichorst et al., 2011). Similarly, Proteobacteria, appeared as the most dominant phyla in other coastal tidal wetlands (Yu et al., 2012), encompasses an enormous level of morphological, physiological, and metabolic diversity, altered with the introduction of A.trabeculosa. The relative abundance of the order Rhizobiales, which fix nitrogen and are symbiotic with plant roots, was significantly impacted by various plants and rhizosphere effects. Notably, the order Nitrospirales were significantly decreased with the introduction of A.trabeculosa. Some evidence suggests that Nitrospira spp. is specialized nitrite oxidizer. Therefore, increased N levels under N-fixing trees may shift the microbial community towards bacterial dominance, which may in turn influence the rate of decomposition of organic matter and increasing the rate of soil C sequestration (Sun et al., 2015).
Rare species occur at low frequency or in a low number in a sample, have important ecological roles as reservoirs of genetic and functional diversity (Coveley et al., 2015). Seven rare phyla (<0.1 %) in this study displayed obvious phylogenetic difference among the investigated soil of two vegetations. In particular, the abundance of Chlorobi and OD1 in soil of A.trabeculosa (mATR, mATN) was significantly higher than that in P. australis (PAR, PAN)(p < 0.5). It has been reported that members of the OD1 group are most likely to be anaerobes and may play roles in sulfur cycling and fermentation of refractory sedimentary carbon to produce acetate, ethanol, lactate, and hydrogen (Peura et al., 2012; Wrighton et al., 2012). Similarly, the Chlorobi, which are also commonly known as green sulfur bacteria, are all anoxygenic obligate photoautotrophs, which obtain electrons for anaerobic photosynthesis from hydrogen sulfide (Bryant et al., 2006). Due to tides transportation, the tidal wetland investigated in this study had relatively high amount of sulfate (Bu et al., 2015), which supplied as a substrate for sulfate reducers and subsequently driven the sulfur redox cycling in wetland. On the other hand, the introduction of A.trabeculosa significantly increased the carbon content in soil, which might stimulate the growth of OD1 and Chlorobi; Thus, we presumed that the introduction of A. trabeculosa significantly affected the rare biosphere involved in sulfur cycling.
Furthermore, this study aimed to identify the effect of N-fixing tree on the abundance of functional microorganisms involved in key processes of the inorganic nitrogen cycle. As revealed by CCA analysis, our results demonstrate significant correlation between soil N content (e.g., total N, ammonia, and nitrate) and microbial community. Accordingly, gene copy numbers and relative genotype richness of functional microbial communities involved in nitrogen transformation showed differences between the investigated soils before and after planting the exotic A.trabeculosa. For nifH gene investigated in this study, the gene copy numbers were significantly higher in soil samples from P. australis than that from A.trabeculosa. In general, legume roots exude various flavonoid and isoflavonoid molecules that are known to induce development of symbiotic interactions between the plant and nitrogen-fixing Alphaproteobacteria within root nodules (Squartini, 2003). However, it has to be taken into account that the used assay only covered the performance of free-living heterotrophic N fixers and did not consider symbiotic N fixation by Frankia, plant-associated N fixation or phototrophic N fixation. The observation that the abundance of free-living N fixers is low in the soil of A.trabeculosa might be due to high nitrogenase activity and the balance with symbiotic N fixation by Frankia, which occurs abundantly in large masses. Hence, it can be speculated that symbiotic and plant-associated N-fixing microbes mainly play roles for N input in the soil of A.trabeculosa. In addition, once C-rich nutrients were delivered like exudates in the rhizosphere of plants, N-fixation activity significantly increased (Duc et al. 2009); hence, the nifH gene copy numbers in the rhizosphere were significantly higher than that in the nonrhizosphere in A.trabeculosa–P. australis mixed communities. Thus, P. australis was putatively to utilize the N nutrition derived from N-fixing trees through root connection or organic forms of N from the litter layer (Hoogmoed et al., 2014). On the other hand, both rhizosphere and nonrhizosphere soil in the mix vegetation community with lower abundance of nosZ gene, showed lower denitrification activity (EDA). The AOA amoA gene abundance in the nonrhizosphere of mono culture of P. australis was significantly higher than that in the nonrhizosphere of mixed vegetation community, and this is consistent with their activity in PNA analysis, which indicated that AOA have more prominent role in ammonia oxidation than AOB in the wetland.
Overall, our study indicated evidence of the introduction of Frankia nodulated tree to recover degraded wetland was effective in accumulation of soil organic carbon and nitrogen, which further impact on the bacterial community composition. Acidobacteria and Proteobacteria are the dominant bacterial groups, while seven rare phyla appeared the most phylogenetically different among the investigated soil of two vegetations, including Chlorobi, Cyanobacteria, OD1, OP11, TM6, TM7, and GN02. The gene copy numbers of free-living N fixers (nifH) were significantly higher in soil samples from P. australis than that from A.trabeculosa, which implicated a balance between N source from atmosphere N fixing by symbiotic Frankia and free-living N fixers. Both decreasing in denitrification gene abundance (nosZ) and enzyme activity (DEA) in soils with the introduction of A.trabeculosa indicated suppressed soil denitrification in the wetland.
References
Aboudrar W, Schwartz C, Benizri C, Morel JL, Boularbah A (2007) Soil microbial diversity as affected by the rhizosphere of the hyperaccumulator Thlaspicae rulescens under natural conditions. Int J Phytoremediat 9:41–52
Barton L, Schipper LA, Smith CT, McLay CDA (2000) Denitrification enzyme activity is limited by soil aeration in a wastewater-irrigated forest soil. Biol Fertil Soils 32:385–389
Bodelier PLE, Dedysh SN (2013) Microbiology of wetlands. Front Microbiol 4:79. doi: 10.3389/fmicb.2013.00079
Boudiaf I, Baudoin E, Sanguin H, Beddiar A, Thioulouse J, Galiana A, Prin Y, Le Roux C, Lebrun M, Duponnois R (2013) The exotic legume tree species, Acacia mearnsii, alters microbial soil functionalities and the early development of a native tree species, Quercussuber, in North Africa. Soil Biol Biochem 65:172–179
Bryant DA, Frigaard NU (2006) Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14:488–496
Bu NS, Qu JF, Zhao H, Yan QW, Zhao B, Fan JL, Fang CM, Gang L (2015) Effects of semi-lunar tidal cycling on soil CO2 and CH4 emissions: a case study in the Yangtze River estuary, China. Wetl Ecol Manag 23(4):727–736. doi:10.1007/s11273-015-9415-5
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chowdhury SP, Schmid M, Hartmann A, Tripathi AK (2009) Diversity of 16S-rRNA and nifH genes derived from rhizosphere soil and roots of an endemic drought tolerant grass, Lasiurus sindicus. Eur J Soil Biol 45:114–122
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM (2007) The ribosomal database project (RDP-II): introducing my RDP space and quality controlled public data. Nucleic Acids Res 35:169–172
Coveley S, Elshahed MS, Youssef NH (2015) Response of the rare biosphere to environmental stressors in a highly diverse ecosystem (Zodletone spring, OK, USA). PeerJ 3:e1182
Duc L, Noll M, Meier B, Burgmann H, Zeyer J (2009) High diversity of diazotrophs in the forefield of a receding alpine glacier. Microbiol Ecol 57:179–190
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Eichorst SA, Kuske CR, Schmidt TM (2011) Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl Environ Microbiol 77:586–596
Feng BW, Li XR, Wang JH, Hu ZY, Meng H, Xiang LY, Quan ZX (2009) Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea. FEMS Microbiol Ecol 70:236–248
Findlay SEG, Dye S, Kuehn KA (2002) Microbial growth and nitrogen retention in litter of Phragmites australis compared to Typha angustifolia. Wetlands 22:616–625
Gao W, JJ L (2008) A restoration trial of bird habitat on the intertidal flats in the Yangtze estuary and its short-term effects. Acta Ecol (Chinese) 28:2080–2089
Hoogmoed M, Cunningham SC, Baker P, Beringer J, Cavagnaro TR (2014) N-fixing trees in restoration plantings: effects on nitrogen supply and soil microbial communities. Soil Biol Biochem 77:203–212
Huang XM, Liu SR, Wang H, Hu ZD, Li ZG, You YM (2014) Changes of soil microbial biomass carbon and communitycomposition through mixing nitrogen-fixing species with Eucalyptus urophylla in subtropical China. Soil Biol Biochem 73:42–48
Kaye JP, Resh SC, Kaye MW, Chimner RA (2000) Nutrient and carbon dynamics in a replacement series of eucalyptus and albizia trees. Ecology 81:3267–3273
Kurola J, Salkinoja-Salonen M, Aarnio T, Hultman J, Romantschuk M (2005) Activity, diversity and population size of ammonia-oxidising bacteria in oil-contaminated landfarming soil. FEMS Microbiol Let 250:33–38
Lamers LPM, Van Diggelen JMH, OpDenCamp HJM, Visser EJW, Lucassen ECHET, Vile MA (2012) Microbial transformations of nitrogen, sulfur, and iron dictate vegetation composition in wetlands: a review. Front Microbiol 3:156. doi:10.3389/fmicb.2012.00156
Lin YT, Tang SL, Pai CW, Whitman WB, Coleman DC, Chiu CY (2014) Changes in the soil bacterial communities in a cedar plantation invaded by Moso bamboo. Microb Ecol 67:421–429
Liu L, Duan ZH, Xu MK, Hu JC, Wang SL, Hu ZG, Zhang QR, Wang SJ (2010) Effect of monospecific and mixed Cunninghamia lanceolata plantations on microbial community and two functional genes involved in nitrogen cycling. Plant Soil 327:413–428
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb(AEM) 71(12):8828–8835
Macedo MO, Resende AS, Garcia PC (2008) Changes in soil C and N stocks and nutrient dynamics 13 years after recovery of degraded land using leguminous nitrogen-fixing trees. Forest Ecol Manag 255:1516–1524
Mao YJ, Yannarell AC, Mackie RI (2011) Changes in N-transforming archaea and bacteria in soil during the establishment of bioenergy crops. PLoS One 6(9):e24750
Oksanen J, Blanchet G, Kindt R, Legendre P, O’Hara RG, Simpson GL (2010) Vegan: community ecology package. R package version 1:17–11
Peura S, Eiler A, Bertilsson S, Nykänen H, Tiirola M, Jones RI (2012) Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J 6:1640–1652
Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinf 12:38
Remigi P, FayeA KA, Deruaz M, Thioulouse J, Cissoko M, Prin Y, Galiana A, Dreyfus B, Duponnois R (2008) The exotic legume tree species Acacia holosericea alters microbial soil functionalities and the structure of the arbuscularmycorrhizal community. Appl Environ Microbiol 74:1485–1493
Resh SC, Binkley D, Parrotta JA (2002) Greater soil carbon sequestration under nitrogen-fixing trees compared with eucalyptus species. Ecosystems 5:217–231
Sathitsuksanoh N, Zhu ZG, Templeton N, Rollin JA, Harvey SP, Zhang Y, Percival H (2009) Saccharification of a potential bioenergy crop, Phragmites australis (common reed), by lignocellulose fractionation followed by enzymatic hydrolysis at decreased cellulase loadings. Ind Eng Chem Res 48:6441–6447
Scott JT, Doyle RD, Back JA, Dworkin SI (2007) The role of N2 fixation in alleviating N limitation in wetland metaphyton: enzymatic, isotopic, and elemental evidence. Biogeochemistry 84:207–218
Selmants PC, Hart SC, Boyle SI, Stark JM (2005) Red alder (Alnusrubra) alters community-level soil microbial function in conifer forests of the Pacific Northwest, USA. Soil Biol Biochem 37:1860–1868
Siddique I, Engel V, Parrotta J (2008) Dominance of legume trees alters nutrient relations inmixed species forest restoration plantings within seven years. Biogeochemistry 88:89–101
Squartini A (2003) Functional ecology of the rhizobium-legume symbiosis. In: Pinton R, Varanini Z, Nannipieri P (eds) The rhizosphere: biochemistry and organic substances at the soil plant interface. Marcel Dekker, New York, pp. 297–326
Sun F, Zhang X, Zhang Q, Liu F, Zhang J, Gong J (2015) Seagrass (Zostera marina) colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Applied & Environmental Microbiology 81:6901–6914
Wang FM, Li ZA, Xia HP, Zou B, Li NY, Liu J, Zhu WX (2010) Effects of nitrogen-fixing and non-nitrogen-fixing tree species on soil properties and nitrogen transformation during forest restoration in southern China. Soil Sci Plant Nutri 56:297–306
Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JE (2012) Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1664
Yu Y, Wang H, Liu J, Wang Q, Shen T, Guo W, Wang R (2012) Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River estuary. Eur J Soil Biol 49:12–21
Zedler JB (2000) Progress in wetland restoration ecology. Trends Ecol Evol 15:402–407
Zhang JY, Ma KM, Fu BJ (2010) Wetland loss under the impact of agricultural development in the Sanjiang Plain, NE China. Environ Monit Assess 166:139–148
Acknowledgments
This research was supported by the National Natural Science Foundation of China (41101230, 40771203 & 40871243), Shanghai Science and Technology Committee (10231201600) and Shanghai Key Laboratory of Bio-Energy Crops (10DZ2271800).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor:Yi-ping Chen
Electronic supplementary material
ESM 1
(DOC 1021 kb)
Rights and permissions
About this article
Cite this article
Chen, X., Yang, J., Zhu, X. et al. N-fixing trees in wetland restoration plantings: effects on nitrogensupply and soil microbial communities. Environ Sci Pollut Res 23, 24749–24757 (2016). https://doi.org/10.1007/s11356-016-7454-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-7454-y