Abstract
The practice of exercise can change biological markers. Thus, the use of saliva to define possible biomarkers has increased with the advances in “omics” techniques. Therefore, this study analyzed the salivary proteomic profile in young adults before and after interval exercise. Five adults, without comorbidities and with normal salivary flow, aged between 20 and 35 years (3 men and 2 women) were selected. The collection of stimulated (SS) and non-stimulated (NSS) saliva was done before (BE) and after (AE) exercise, which consisted of interval exercise with 5 min walking on a treadmill and 5 min at rest, during a total of 40 min. The proteins were extracted individually and processed by label-free proteomics. There was no significant difference between BE and AE salivary flows. For SS, 69 and 60 proteins were found BE (12 exclusives) and AE (3 exclusives), respectively. Ten subunits of immunoglobulins (8 more than twofold), as well as Lysozyme C and Cystatin-S, were increased AE compared to BE. Six hemoglobin’s isoforms were decreased AE compared to BE. For NSS, 76 and 65 proteins were found BE (18 exclusives) and AE (7 exclusives), respectively. Albumin was increased AE, while Basic salivary proline-rich protein 1 and 2 decreased. Apolipoprotein A-I was identified exclusively AE compared to BE. The salivary proteome after interval exercise suggests increase of immunity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regular physical activity is associated with several general health benefits, including a protective action against metabolic diseases, such as obesity, diabetes mellitus and hypertension [1, 2]. The practice of exercise produces reactive oxygen species (ROS) in cells and body fluids. To neutralize these molecules, the body is stimulated to respond with the antioxidant system, which includes some enzymatic and non-enzymatic mechanisms, uric acid, vitamins and minerals [3]. Some studies also suggest that the variation in the duration, type and intensity of the exercise can affect, for example, oxidative stress markers and the levels of pro-inflammatory biomarkers in urine, blood or/and saliva [4, 5, 6].
In the study of Hajizadeh Maleki et al. [4], participants walked on a treadmill for 25–30 min, 3 days a week for 12 weeks. In the next 12 weeks, the time was increased for 40–45 min, during 3 days per week as well. Blood samples were collected before the beginning of the research and at different periods of follow-up. The authors observed that low and moderate-intensity training decreased the levels of IL-1β, IL-6, IL-8 and TNF-α in the blood for a period of 12–24 weeks; however, their concentrations return to the baseline values after 30 days without exercise [4].
Body fluids play a fundamental role as a tool for the analysis of the biomarkers. One of the main fluids applied in sports medicine is saliva, since the collection is non-invasive, without pain or discomfort, differently from what occurs in blood analysis [7]. Salivary secretion is controlled by the autonomic nervous system; so sympathetic activation results in the secretion of saliva rich in proteins, while parasympathetic activation is responsible for water and ionic content [8]. Therefore, the secretion and composition of saliva may vary throughout the day and by different stimuli [9, 10].
Studies have shown that the practice of exercise alters the expression of several salivary biomarkers, due to the activation of the sympathetic nervous system [6, 8, 11]. The concentration of salivary protein may increase with the sympathetic stimulation of the salivary glands by catecholamines [12], which are more expressed above the anaerobic threshold [11, 13]. Cycling for 15 min increases the saliva viscosity immediately after exercise [13], as a result of increase in the concentration of proteins, such as MUC5B [13, 14]. Other proteins—amylase, lactoferrin, lysozyme—also increase due to exercise, with a reduction of the level 30 min after the end of the activity [10, 15]. These proteins are known for their antimicrobial effect and role in the formation of the acquired enamel pellicle [16]. In addition, studies have shown that exercise may alter the immune system, depending on its volume, intensity, and nature of the activity [17, 18]. Exercise at moderate intensity increases IgA secretion after the immediate practice; however, these values are reduced to baseline levels after 2–24 h [19].
Considering the aforementioned parameters, it is expected that exercise may modulate the innate and acquired immune system and the response to neuroendocrine stress, which is represented by the hypothalamic–pituitary–adrenal axis (HHA) and the adreno-medullary sympathetic axis (SAM) [20, 21]. One of the main markers for their activation is cortisol [21, 22]. Cortisol can directly impact immune cells, since it induces lymphopenia under high-intensity physical exercises. In contrast, at low intensity, there is no change or, in some cases, a reduction in the concentration of cortisol [23]. Therefore, cortisol tends to combat the stress of exercise, from the axes HHA, modulating the immune system [24]. Instead of analyzing salivary cortisol, the proteomic determination of saliva may show proteins involved in the immune system after exercise, which will help to disseminate the importance of the exercise and to standardize biomarkers able to confirm the benefits of exercise on the oral and general health.
Accordingly, there are no data on quantitative and comparative protein changes in stimulated (SS) and unstimulated (NSS) saliva from the collection before and after exercise under controlled conditions. In view of this, the present study compared the main proteins present in stimulated and unstimulated saliva of young adults, before and after 40 min of interval exercise (low to moderate intensity), using a proteomic approach, to find possible quantitative and qualitative changes in biomarkers expressed in the saliva that reflect the effects of exercise on oral and general health.
Materials and methods
Ethical aspects
This study was submitted to the local Institutional Ethics Committee (No. 29182720.3.0000.5417). SS and NSS, before (BE) and after (AE) interval exercise, were collected after approval and the signature of the informed consent form. The study was carried out in accordance with the Declaration of Helsinki.
Volunteers’ information
Five young adults, aged between 20 and 35 years, from both genders (3 men and 2 women), with good oral and general health, non-smokers, non-pregnant and without any systemic condition, were included. Volunteers presenting gingivitis, active caries or erosive lesions were excluded [25]. SS and NSS were collected BE and AE. In addition, all volunteers practiced aerobic exercise twice a week.
Saliva collection and exercise
The volunteers were previously instructed to have the last meal and brush their teeth at least 30 min (the mean was 40–60 min) before saliva collection. The collection of SS and NSS saliva was performed strictly as previously described [26, 27]. Saliva collection was done between 4:00 pm and 6:00 pm The volunteers rinsed their mouths with 5 mL of deionized water before collection. For the collection of NSS, the volunteers spat out all the saliva in tubes immersed in ice for 10 min. For the collection of SS, the volunteers chewed a Parafilm® (5 cm) and all saliva produced by the stimulation was spat out in tubes immersed in ice for 5 min.
The volunteers were submitted to exercise protocol, under interval exercise method, on a treadmill (Inbramed ATL, Inbrasport, Porto Alegre, Brazil) with a preferred speed for 5 min walking (medium speed of 4.6 ± 0.4 km/h) and 5 min at rest, during a total period of 40 min. To determine the preferred speed, the volunteers performed three efforts on a linear 10-m circuit 5 min before the first saliva collection. The mean speed of these efforts was assumed as the exercise intensity (4.6 ± 0.4 km h−1). The time of each effort was assessed through a stopwatch (Timex, model 85103). After the exercise, the volunteers remained seated for 5 min and the saliva collection was performed by repeating the same steps described above for SS and NSS. In total, saliva was collected twice (before the first cycle of exercise and after the last cycle of exercise).
After collection, the salivary flow was measured and the saliva samples were centrifuged at 4500×g for 15 min at 4 °C to remove possible debris such as insoluble material, cells and food debris. The supernatant from each sample was frozen at − 80 °C until proteomics analysis. These procedures were based also on previous studies [26, 27, 28].
Shotgun label-free quantitative proteomic analysis
SS and NSS, BE and AE, were extracted and prepared individually. For this, 1 mL of saliva was transferred to new tubes and an extraction solution containing 6 M urea, 2 M thiourea in 50 mM Ammonium Bicarbonate, pH 7.8, in a volume similar to the samples (1:1) was used. Samples were vortexed for 10 min at 4 °C, sonicated for 5 min and centrifuged at 20.817×g for 10 min at 4 °C for twice. The procedures for the preparation of saliva samples and shotgun proteomic analysis were performed exactly as described [27].
Briefly, after extraction, the samples were concentrated with Amicon tubes (Amicon Ultra-15 Centrifugal Filter Units—Merck Millipore®, Tullagreen, County Cork, Ireland) and centrifuged at 4.500×g at 4 °C to a volume of approximately 150 μL. The total protein quantification was performed by Bradford method (Bio-Rad Bradford Assays, Hercules, California, USA) and the samples were then reduced (5 mM dithiothreitol, Bio Rad Laboratories, Canada) and alkylated (10 mM iodoacetamide, GE Healthcare, Little Chalfont, Buckinghamshire UK). In sequence, the samples were digested for 14 h at 37 °C by the addition of 2% (w/w) trypsin (Thermo Scientific Pierce Trypsin Protease, Rockford, lL, USA). Digestion was stopped by the addition of 5% formic acid solution. Samples were desalted and purified using C18 spin columns (Thermo Scientific, Rockford, Illinois, USA). A 1-μL aliquot of each sample was removed for protein quantification by the Bradford method (Bio-Rad Bradford Assays, Hercules, California, USA). Samples were then resuspended in 3% acetonitrile and 0.1% formic acid for nano LC–ESI–MS/MS.
Data acquisition was performed by Xevo G2 QTof mass spectrometer coupled to the nanoACQUITY Ultra Performance Liquid Chromatography (both from Waters, Manchester, UK) controlled by MassLynx v.4.1 (Waters, Manchester, UK). Data collection was in data-independent acquisition mode (LC–MSE), the mass ranges from 50 to 2000 m/z, and assays were performed exactly as previously described [25, 27]. All samples were analyzed individually, thus totalizing 5 analyses for each condition. The proteins were identified using the software's ion counting algorithm, and a search was performed on the Homo sapiens database (revised only, UniProtKB/Swiss-Prot) downloaded in December 2020 from UniProtKB (http://www.uniprot.org). Maximum missed cleavages by trypsin allowed up to 1, variable modifications by oxidation and a false discovery rate value at maximum 4%.
For the label-free quantitative analysis, PLGS was used for analyzing nine raw MS files from each group. Proteins identified with a confidence score higher than 95% were included. The identical peptides from each duplicate by sample were pooled according to mass accuracy (< 10 ppm) and the retention time tolerance < 0.25 min, using the clustering software included in the PLGS. Normalization was automatically implemented by the software (default parameters). The difference in expression between the groups was analyzed by dependent t test (p < 0.05). The following relevant comparisons were performed: SS AE vs BE; NSS AE vs BE; BE SS vs NSS; AE SS vs NSS.
Bioinformatics analysis
The proteins were analyzed by their access number by UNIPROT, and repeated proteins, reverse proteins and fragments were excluded. Gene ontology was evaluated according to the ClueGo® plugins of the Cytoscape® 3.8.2 Software. The functional distribution of proteins identified with differential expression in the comparison SS, AE vs BE, was done. Protein categories was based on gene ontology (GO) annotation of the broad biological process, molecular function, immune system process and cell component. Terms of significance (κ = 0.04) and distribution were according to the percentage of the number of associated genes. The number of access of the proteins was provided by UNIPROT. The mass spectrometric proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier (PXD028085).
Results
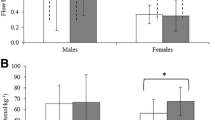
All participants (ages between 20 and 35 years), 3 male and 2 females, with a mean body mass of 74.6 ± 12.4 kg and mean height of 1.68 ± 0.08 m, completed the study protocol. The mean of the individual body mass index was 26.60 ± 5.15 kg/ cm2 (2 normal, 2 overweight and 1 obese). Table 1 shows the mean (± SD) of NSS and SS flows, BE and AE.
The total mean amounts of protein recovered from NSS for BE and AE were 31.9 and 28.6 μg, respectively, while from SS for BE and AE were 28.7 and 28.0 μg, respectively. For the quantitative proteomics analysis, all samples were standardized to 28.0 µg of proteins.
For the comparison of SS, AE vs BE, the total number of proteins identified were 60 and 69 proteins, respectively, among which 57 proteins were common to both conditions, such as Statherin and Histatin-1 and -3 (Fig. 1A). Twelve proteins were identified exclusively BE, such as Neutrophil defensin 1 and -3. Three proteins were identified exclusively AE such as Haptoglobin and Lactotransferrin. In the differently expressed proteins, 32 and 13 proteins were increased and decreased, respectively, in AE compared to BE. Among the down-regulated proteins AE were: 4 isoforms of amylase, Salivary acidic proline-rich phosphoprotein ½, Immunoglobulin kappa constant and 6 isoforms of hemoglobin. On the other hand, 10 isoforms of immunoglobulins, Lysozyme C, Albumin, Mucin-7, Cystatin-S, Submaxillary gland androgen-regulated protein 3B, Carbonic anhydrase 6, 5 isoforms of cystatins and 2 isoforms of proline-rich protein were up-regulated AE compared to BE (Table 2).
Venn diagram with the total number of proteins identified in stimulated (SS) and non-stimulated (NSS) saliva before interval exercise (BE) and after interval exercise (AE). A comparison between AE vs BE in the stimulated salivary flow (SS). B comparison between AE vs BE in non-stimulated salivary flow (NSS). C comparison between SS vs NSS, BE. D comparison between SS vs NSS, AE
Figure 2 shows the functional analysis according to the biological process by Gene Ontologies (GO) with the most significant term, for the comparison of SS, AE vs BE. The categories with the high percentages of genes were retina homeostasis (31.9%) and phagocytosis, engulfment (21.3%) (Fig. 2). In the functional analysis according to the molecular function process by Gene Ontologies (GO) with the most significant term, for the comparison of SS, AE vs BE, it was shown the highest percentage of genes was hemoglobin alpha binding (55.6%) (Fig. 3). The functional analysis according to the cell component process by Gene Ontologies (GO) with the most significant term was also done in this comparison. The category with the highest percentage of genes was immunoglobulin complex, circulating secretory (66.7%) (Fig. 4). In the functional analysis according to the immune system process by Gene Ontology (GO) with the most significant term, for the comparison of SS, AE vs BE, it was shown that the category with the highest percentage of genes was immunoglobulin complex, circulating (46.2%) (Fig. 5).
Graphic of the functional distribution of proteins identified with differential expression in the SS after interval exercise (AE) vs. before interval exercise (BE). Protein categories based on the Gene Ontology (GO) annotation of the broad Biological Process. Terms of significance (κ = 0.04) and distribution according to the percentage of the number of associated genes. The access number for the proteins was provided by UNIPROT. Genetic ontology was evaluated according to the ClueGo® pluggins of the Cytoscape® 3.8.2 software
Graphic of the functional distribution of proteins identified with differential expression in the SS after interval exercise (AE) vs. before interval exercise (BE). Protein categories based on the Gene Ontology (GO) annotation of the broad Molecular Function. Terms of significance (κ = 0.04) and distribution according to the percentage of the number of associated genes. The access number for the proteins was provided by UNIPROT. Genetic ontology was evaluated according to the ClueGo® pluggins of the Cytoscape® 3.8.2 software
Graphic of the functional distribution of proteins identified with differential expression in the SS after interval exercise (AE) vs. before interval exercise (BE). Protein categories based on the Gene Ontology (GO) annotation of the broad Cellular Component. Terms of significance (κ = 0.04) and distribution according to the percentage of the number of associated genes. The access number for the proteins was provided by UNIPROT. Genetic ontology was evaluated according to the ClueGo® pluggins of the Cytoscape® 3.8.2 software
Graphic of the functional distribution of proteins identified with differential expression in the SS after interval exercise (AE) vs. before interval exercise (BE). Protein categories based on the Gene Ontology (GO) annotation of the broad Immune System Process. Terms of significance (κ = 0.04) and distribution according to the percentage of the number of associated genes. The access number for the proteins was provided by UNIPROT. Genetic ontology was evaluated according to the ClueGo® pluggins of the Cytoscape® 3.8.2 software
When NSS was analyzed, AE vs BE, the total number of proteins identified were 65 and 76, respectively, with 58 proteins common to both conditions such as Salivary acidic proline-rich phosphoprotein ½, Carbonic anhydrase 6, 15 isoforms of immunoglobulins, 5 isoforms of cystatins, Histatin-1 and -3, Lactotransferrin, Neutrophil defensin 1 and -3, 4 isoforms of amylase, Mucin-7, Lysozyme C, Statherin and Submaxillary gland androgen-regulated protein 3B (Fig. 1B). Eighteen proteins were identified exclusively BE, such as BPI proteins fold-containing family B member 1 and Torsin-2A, while 7 proteins were identified exclusively AE, such as Apolipoprotein AI, Proline-rich protein 4 and Serotransferrin. In the differentially expressed proteins, only Albumin was increased AE, while 3 proteins were decreased, such as Basic salivary proline-rich protein 1 and -2 (Table 3).
For the comparison BE, SS vs NSS, the total number of proteins identified were 70 and 76, respectively, with 54 proteins common to both conditions such as Albumin, Carbonic anhydrase 6, Histatin-1 and -3, Cystatin-D and 5 isoforms of immunoglobulins. Sixteen proteins were identified exclusively SS, such as Apolipoprotein A-I and 6 isoforms of hemoglobin. Twenty-two proteins were identified exclusively NSS, such as BPI fold-containing family B member 1, 3 isoforms of immunoglobulins and Lactotransferrin (Fig. 1C). In the differentially expressed proteins, 10 proteins were increased in the SS, such as Salivary acidic proline-rich phosphoprotein ½, Neutrophil defensin 1 and -3, Basic salivary proline-rich protein 2 and 4 isoforms of amylases, while 27 proteins were decreased, such as 4 isoforms of cystatins, 7 isoforms of immunoglobulins, Submaxillary gland androgen-regulated protein 3B, Statherin, BPI fold-containing family A member 2, Prolactin-inducible protein, Basic salivary proline-rich protein 1, Mucin-7 (decreased nearly fivefold) and Lysozyme C (decreased more than fivefold) (Table 4).
As for the comparison AE, SS vs NSS, the total number of proteins identified were 63 and 65, respectively, with 52 proteins being identified in common to both conditions, such as Prolactin-inducible protein, Lactotransferrin, Basic salivary proline-rich protein 2, Histatin-1, Alpha-amylase 1C and Cystatin-D. Eleven proteins were identified exclusively in the SS, such as 7 isoforms of hemoglobin. On the other hand, 13 proteins were identified exclusively in the NSS, such as Apolipoprotein A-I, 4 isoforms of immunoglobulins, Proline-rich protein 4, Serotransferrin and Neutrophil defensin 1 and -3 (Fig. 1D). In the quantitative analysis, 10 proteins were increased in the SS, such as Salivary acidic proline-rich phosphoprotein ½, Basic salivary proline-rich protein 1, 2 isoforms of immunoglobulins, Statherin, Carbonic anhydrase 6 and Alpha-amylase 2B, while 27 proteins were decreased, such as 6 isoforms of immunoglobulins, Alpha-amylase 1A and -1B, Mucin-7, Albumin, Lysozyme C, Submaxillary gland androgen-regulated protein 3B, Histatin-3 and 4 isoforms of cystatins (Table 5).
Discussion
The search for markers through saliva has numerous advantages, mainly because its collection is not invasive, as in blood analysis, for example. In addition, saliva provides the identification of an arsenal of proteins and consequently biomarkers typical for the oral cavity and related to systemic health or disease either [29, 30]. Little is known about the difference in protein profile between the different salivary flows (stimulated and unstimulated) before and after exercise. Our study highlights important information about changes in salivary proteomic profile after interval exercise (low to moderate intensity), which may have an impact on oral and general health.
SS brings more information than NSS with this respect. When BE and AE were compared in the SS, it was observed an increase in many proteins involved with responses to the immune system. Among these findings, various immunoglobulins were increased AE compared to BE, among which 8 were increased more than twofold (Table 2). It is known that the practice of exercise, in general, provides great health benefits [31]. Although our results were not from a longer follow-up after exercise, our data suggest that the practice of exercise, under interval method, even if it is just a walk (as done in the present study), may provide important changes in immunity. This finding was supported by the results of the most affected genes. In the biological process, 15% of these proteins were involved with humoral antimicrobial response and 21.3% were related to phagocytosis activity. Checking the great interaction of these proteins involved with immune responses, almost 50% were involved in immunoglobulin complex circulating (Fig. 5). The immunoglobulin complex has the function of binding to the antigen if a suitable antigen is available. In addition, this complex is usually found in areas of mucosa, blood or lymph [32]. It is important to bear in mind that the increase of immunity was seen under the interval exercise tested in the present study, since it has been shown that the practice of exercise under high intensity for prolonged periods can decrease immunity [18], while the regular exercise of moderate intensity is beneficial, improving immunity throughout life [33] and stimulating immune functions [34]. On the other hand, longer follow-up after physical exercise shall be evaluated in future studies.
The practice of exercise also has a beneficial influence on oral health. Mucin-7, Lysozyme C, Albumin, cystatins and proline-rich proteins (PRPs) were increased in stimulated saliva AE in comparison to BE (Table 2). Mucin-7 is an acid-resistant protein, which was identified in the acquired pellicle after episodes of erosive challenges [35]. In addition, it participates in important processes related to immune response, such as antimicrobial activity, and is involved with essential processes for mucosal protection and wound healing [36]. This protein has a protective capacity, promoting the elimination of bacteria in the oral cavity, assisting chewing, speech and swallowing [37]. In this perspective, Lysozyme C, also increased AE, has a bacteriological function, mainly in tissues and body fluids associated with the monocyte–macrophage system, which increases the activity of immune agents; in addition, it is related to antimicrobial humoral response (defense responses to gram-positive and gram-negative bacteria) and participates in inflammatory responses [37]. In addition, in a previous study, Lysozyme was increased in saliva after 30 min of steady-state cycling exercise [38].
On the other hand, six isoforms of the hemoglobin (Hemoglobin subunit alpha, subunit delta, subunit epsilon, subunit gamma-1 and gamma-2) were found reduced AE compared to BE (Table 2). The decrease in these proteins may be due to the vasoconstriction in response to the sympathetic stimulation caused by the practice of exercise [39]. Exercise increases sympathetic vasoconstrictive responsiveness through a nitric oxide-dependent mechanism. When the exercise is prolonged, there are additional constricting adjustments in the less active tissues [40]. This constricting action allows the correct redistribution of blood to satisfy the needs of active muscles [41]. Among the processes of the most affected genes evaluated from the bioinformatics analysis, we verified that in the biological processes 12.8% of these proteins were involved with oxygen-carrying activity (Fig. 2). We believe that this condition is directly related to the identified hemoglobin, since the most affected processes of molecular function with the highest percentage (55.6%) were related to hemoglobin bonds, mainly of the alpha subunit (Fig. 3).
These six isoforms of hemoglobin have already been identified in saliva in previous studies, where the salivary proteome was evaluated [27]. It is important to consider that these proteins may also have come from the crevicular fluid, where the hemoglobin alpha, beta and delta subunits have also been previously identified [42]. Another important point is that hemoglobin was reduced in SS compared to NSS, which is expected, since during stimulation, the parasympathetic action is predominant [8, 16, 43].
Regarding NSS, it was observed a decrease in basic proline-rich proteins and an increase in albumin in the results. In addition, albumin has been increased in saliva after moderate exercise and high-intensity sports [44]. However, no data have been identified in the literature on the increase of albumin in saliva under low-intensity exercises. Thus, to the best of our knowledge, this is the first study to report this finding. The increase in albumin in saliva seems to be related to physical activity regardless of the intensity. Interesting, lipids used as energy source during exercise are included in blood circulation bounded to albumin [45], which should be considered in future studies.
Based on our results, it is clear that the interval exercise did not have the same clear impact on NSS as it did for SS. Although NSS is considered the most important flow for the formation of the acquired pellicle and the protection of dental surface and mucosa [16, 43], during exercise the stimulated saliva seems to provide better data for assessing the protein profile of saliva and to correlate the findings with general health. One of the reasons is related to the time of collection, SS was collected 15 min after the end of the exercise, allowing those proteins from blood be found in saliva. Other important point to take into consideration is the role of glands in the production of SS and NSS, while parotid is very important under mechanical stimulation, submandibular has a great contribution for NSS (Proctor 2016), which may impact in its proteomic profile. These findings should be confirmed in future studies.
On the other hand, Apolipoprotein A-I was identified exclusively AE when compared to BE in NSS (Table 3) and when compared SS vs NSS AE (Table 5). This protein participates in the reverse transport of cholesterol from tissues to the liver for excretion, promoting the efflux of cholesterol from tissues and acting as a cofactor for lecithin cholesterol acyltransferase (information from UNIPROT). In addition, Apolipoprotein A-I binds to HDL (High-Density Lipoproteins), which removes excess cholesterol and takes it back to the liver, to be eliminated from the body [46]. Thus, even with low to moderate exercise, it is possible to observe that the lipid metabolism is activated, with possible increase in HDL level. Apolipoprotein A-I has also been identified in resistance training [47].
When we compared SS and NSS, BE, we observed that PRPs, amylases, Neutrophil defensin 1 and 3 were increased in SS when compared to NSS. Statherin, cystatins isoforms, immunoglobulins isoforms, Prolactin-inducible protein, Mucin-7, Lysozyme C and Basic salivary proline-rich protein 1 were reduced in SS when compared to NSS. Statherin is a salivary protein that stabilizes the calcium saturation by inhibiting the precipitation of calcium phosphate salts under neutral pH. It also modulates the formation of hydroxyapatite crystals on the tooth surface [37]. Regarding proline-rich proteins (PRPs), these are a heterogeneous group of proteins that are exclusively secreted by the parotid gland. The acidic PRPs inhibit the formation of hydroxyapatite and the basic PRPs strongly bind to the diet's tannins, avoiding their toxic effects by blocking their intestinal absorption [48]. In addition, PRPs act as highly potent inhibitors of the growth of calcium phosphate crystals under neutral pH, providing a protective and repairing environment for tooth enamel [37].
In general, proteins involved with antimicrobial and acid-resistant protection were reduced in SS when compared to NSS before exercise, which is expected, since in SS, the parasympathetic stimulation is predominant, with consequent high release of water and ions [8, 49] (Table 4).
The same findings were shown, when we compared the different salivary flows AE. We observed that isoforms of immunoglobulins, PRPs and Statherin were increased in the SS compared to NSS. On the other hand, amylases, Albumin, Mucin-7, Lysozyme C, cystatins, Histatin-3 and isoforms of immunoglobulins were decreased in SS compared to NSS. In addition, Apolipoprotein A-I, isoforms of immunoglobulins, Neutrophil defensin 1 and 3, PRPs and Serotransferrin were identified exclusively in the NSS. Therefore, regardless of performing exercise (BE or AE), it is expected to find a low number of proteins in SS compared to NSS. Finally, the identification of amylases is very important, because these proteins are known as biomarkers of stress, anxiety and depression [50]. In our study, these proteins were reduced in saliva under stimulated flow after physical exercise, which demonstrates the benefits of physical activity. Furthermore, variations of amylase levels in saliva may be a response, especially in cases of intense training, to the need of individual ingests food energy after exercise [51, 52].
In our study, SS seems to be better at differentiating the effect of exercise on the salivary proteomic profile compared to NSS. On the other hand, in real life NSS is predominant over SS, which only happens during stimulation induced by meals (less than 1–2 h day−1) [49, 53, 54]. This should be considered in the interpretation of the findings and in planning new studies. The stimulation of salivary flow after exercise should be considered in future studies. Moreover, it is interesting to further evaluate the effect of interval exercise for long-term periods, to verify how long the increased immunity-related proteins remain in saliva and their impact on oral and systemic conditions. In addition, these findings can still be evaluated in future studies with a larger number of participants.
From the results of this preliminary study, we can conclude that proteomic changes are more evident for stimulated than non-stimulated saliva and that the tested exercise protocol induced an increase of proteins related to immunity, which shall be confirmed in future studies with higher number of participants.
Availability of data and materials
The mass spectrometric proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier (PXD028085).
Code availability
Data set identifier PXD028085 in the PRIDE.
References
Nuzum H, Stickel A, Corona M, Zeller M, Melrose RJ, Wilkins SS (2020) Potential benefits of physical activity in MCI and dementia. Behav Neurol. https://doi.org/10.1155/2020/7807856
Warburton DER, Bredin SSD (2017) Health benefits of physical activity: a systematic review of current systematic reviews. Curr Opin Cardiol 32(5):541–556. https://doi.org/10.1097/HCO.0000000000000437
Finkler M, Lichtenberg D, Pinchuk I (2014) The relationship between oxidative stress and exercise. J Basic Clin Physiol Pharmacol 25(1):1–11. https://doi.org/10.1515/jbcpp-2013-0082
Hajizadeh Maleki B, Tartibian B, Mooren FC, FitzGerald LZ, Kruger K, Chehrazi M, Malandish A (2018) Low-to-moderate intensity aerobic exercise training modulates irritable bowel syndrome through antioxidative and inflammatory mechanisms in women: results of a randomized controlled trial. Cytokine 102:18–25. https://doi.org/10.1016/j.cyto.2017.12.016
Shi M, Wang X, Yamanaka T, Ogita F, Nakatani K, Takeuchi T (2007) Effects of anaerobic exercise and aerobic exercise on biomarkers of oxidative stress. Environ Health Prev Med 12(5):202–208. https://doi.org/10.1265/ehpm.12.202
Souza AV, Giolo JS, Teixeira RR, Vilela DD, Peixoto LG, Justino AB, Caixeta DC, Puga GM, Espindola FS (2019) Salivary and plasmatic antioxidant profile following continuous, resistance, and high-intensity interval exercise: preliminary study. Oxid Med Cell Longev 2019:5425021. https://doi.org/10.1155/2019/5425021
Malamud D (2011) Saliva as a diagnostic fluid. Dent Clin N Am 55(1):159–178. https://doi.org/10.1016/j.cden.2010.08.004
Proctor GB, Carpenter GH (2007) Regulation of salivary gland function by autonomic nerves. Auton Neurosci 133(1):3–18. https://doi.org/10.1016/j.autneu.2006.10.006
Bosch JA, Veerman EC, de Geus EJ, Proctor GB (2011) Alpha-amylase as a reliable and convenient measure of sympathetic activity: don’t start salivating just yet! Psychoneuroendocrinology 36(4):449–453. https://doi.org/10.1016/j.psyneuen.2010.12.019
Ligtenberg AJ, Brand HS, van den Keijbus PA, Veerman EC (2015) The effect of physical exercise on salivary secretion of MUC5B, amylase and lysozyme. Arch Oral Biol 60(11):1639–1644. https://doi.org/10.1016/j.archoralbio.2015.07.012
de Oliveira VN, Bessa A, Lamounier RP, de Santana MG, de Mello MT, Espindola FS (2010) Changes in the salivary biomarkers induced by an effort test. Int J Sports Med 31(6):377–381. https://doi.org/10.1055/s-0030-1248332
Bortolini MJS, De Agostini GG, Reis IT, Lamounier RPMS, Blumberg JB, Espindola FS (2009) Total protein of whole saliva as a biomarker of anaerobic threshold. Res Q Exerc Sport 80(3):604–610
Ligtenberg AJ, Liem EH, Brand HS, Veerman EC (2016) The effect of exercise on salivary viscosity. Diagnostics (Basel). https://doi.org/10.3390/diagnostics6040040
Mulic A, Tveit AB, Songe D, Sivertsen H, Skaare AB (2012) Dental erosive wear and salivary flow rate in physically active young adults. BMC Oral Health 12:8. https://doi.org/10.1186/1472-6831-12-8
Gillum T, Kuennen M, McKenna Z, Castillo M, Jordan-Patterson A, Bohnert C (2017) Exercise increases lactoferrin, but decreases lysozyme in salivary granulocytes. Eur J Appl Physiol 117(5):1047–1051. https://doi.org/10.1007/s00421-017-3594-0
Buzalaf MA, Hannas AR, Kato MT (2012) Saliva and dental erosion. J Appl Oral Sci 20(5):493–502. https://doi.org/10.1590/s1678-77572012000500001
Kruger K, Mooren FC, Pilat C (2016) The immunomodulatory effects of physical activity. Curr Pharm Des 22(24):3730–3748. https://doi.org/10.2174/1381612822666160322145107
Simpson RJ, Kunz H, Agha N, Graff R (2015) Exercise and the regulation of immune functions. Prog Mol Biol Transl Sci 135:355–380. https://doi.org/10.1016/bs.pmbts.2015.08.001
Sari-Sarraf V, Reilly T, Doran DA, Atkinson G (2007) The effects of single and repeated bouts of soccer-specific exercise on salivary IgA. Arch Oral Biol 52(6):526–532. https://doi.org/10.1016/j.archoralbio.2006.11.016
Herman JP, Cullinan WE (1997) Neurocircuitry of stress: central control of the hypothalamo-pituitary–adrenocortical axis. Trends Neurosci 20(2):78–84. https://doi.org/10.1016/s0166-2236(96)10069-2
Maruyama Y, Kawano A, Okamoto S, Ando T, Ishitobi Y, Tanaka Y, Inoue A, Imanaga J, Kanehisa M, Higuma H, Ninomiya T, Tsuru J, Hanada H, Akiyoshi J (2012) Differences in salivary alpha-amylase and cortisol responsiveness following exposure to electrical stimulation versus the Trier Social Stress Tests. PLoS ONE 7(7):e39375. https://doi.org/10.1371/journal.pone.0039375
de Vries WR, Bernards NT, de Rooij MH, Koppeschaar HP (2000) Dynamic exercise discloses different time-related responses in stress hormones. Psychosom Med 62(6):866–872. https://doi.org/10.1097/00006842-200011000-00017
Leicht CA, Goosey-Tolfrey VL, Bishop NC (2018) Exercise intensity and its impact on relationships between salivary immunoglobulin A, saliva flow rate and plasma cortisol concentration. Eur J Appl Physiol 118(6):1179–1187. https://doi.org/10.1007/s00421-018-3847-6
Rutherfurd-Markwick K, Starck C, Dulson DK, Ali A (2017) Salivary diagnostic markers in males and females during rest and exercise. J Int Soc Sports Nutr 14:27. https://doi.org/10.1186/s12970-017-0185-8
Ventura T, Cassiano LPS, Souza ESCM, Taira EA, Leite AL, Rios D, Buzalaf MAR (2017) The proteomic profile of the acquired enamel pellicle according to its location in the dental arches. Arch Oral Biol 79:20–29. https://doi.org/10.1016/j.archoralbio.2017.03.001
Jasim H, Olausson P, Hedenberg-Magnusson B, Ernberg M, Ghafouri B (2016) The proteomic profile of whole and glandular saliva in healthy pain-free subjects. Sci Rep 6:39073. https://doi.org/10.1038/srep39073
Ventura T, Ribeiro NR, Dionizio AS, Sabino IT, Buzalaf MAR (2018) Standardization of a protocol for shotgun proteomic analysis of saliva. J Appl Oral Sci 26:e20170561. https://doi.org/10.1590/1678-7757-2017-0561
Winck FV, Prado Ribeiro AC, Ramos Domingues R, Ling LY, Riano-Pachon DM, Rivera C, Brandao TB, Gouvea AF, Santos-Silva AR, Coletta RD, Paes Leme AF (2015) Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci Rep 5:16305. https://doi.org/10.1038/srep16305
Buzalaf MAR, Ortiz AC, Carvalho TS, Fideles SOM, Araujo TT, Moraes SM, Buzalaf NR, Reis FN (2020) Saliva as a diagnostic tool for dental caries, periodontal disease and cancer: is there a need for more biomarkers? Expert Rev Mol Diagn 20(5):543–555. https://doi.org/10.1080/14737159.2020.1743686
Wang X, Kaczor-Urbanowicz KE, Wong DT (2017) Salivary biomarkers in cancer detection. Med Oncol 34(1):7. https://doi.org/10.1007/s12032-016-0863-4
Ruegsegger GN, Booth FW (2018) Health benefits of exercise. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a029694
Gaudet P, Livstone MS, Lewis SE, Thomas PD (2011) Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform 12(5):449–462. https://doi.org/10.1093/bib/bbr042
Simpson RJ, Lowder TW, Spielmann G, Bigley AB, LaVoy EC, Kunz H (2012) Exercise and the aging immune system. Ageing Res Rev 11(3):404–420. https://doi.org/10.1016/j.arr.2012.03.003
Romeo J, Warnberg J, Pozo T, Marcos A (2010) Physical activity, immunity and infection. Proc Nutr Soc 69(3):390–399. https://doi.org/10.1017/S0029665110001795
Delecrode TR, Siqueira WL, Zaidan FC, Bellini MR, Moffa EB, Mussi MC, Xiao Y, Buzalaf MA (2015) Identification of acid-resistant proteins in acquired enamel pellicle. J Dent 43(12):1470–1475. https://doi.org/10.1016/j.jdent.2015.10.009
Crosara KTB, Zuanazzi D, Moffa EB, Xiao Y, Machado M, Siqueira WL (2018) Revealing the amylase interactome in whole saliva using proteomic approaches. Biomed Res Int 2018:6346954. https://doi.org/10.1155/2018/6346954
UniProt C (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49(D1):D480–D489. https://doi.org/10.1093/nar/gkaa1100
Kunz H, Bishop NC, Spielmann G, Pistillo M, Reed J, Ograjsek T, Park Y, Mehta SK, Pierson DL, Simpson RJ (2015) Fitness level impacts salivary antimicrobial protein responses to a single bout of cycling exercise. Eur J Appl Physiol 115(5):1015–1027. https://doi.org/10.1007/s00421-014-3082-8
Jendzjowsky NG, DeLorey DS (2012) Short-term exercise training augments sympathetic vasoconstrictor responsiveness and endothelium-dependent vasodilation in resting skeletal muscle. Am J Physiol Regul Integr Comp Physiol 303(3):R332-339. https://doi.org/10.1152/ajpregu.00053.2012
Just TP, DeLorey DS (2016) Exercise training and alpha1-adrenoreceptor-mediated sympathetic vasoconstriction in resting and contracting skeletal muscle. Physiol Rep 4(3):1. https://doi.org/10.14814/phy2.12707
Pagan LU, Gomes MJ, Okoshi MP (2018) Endothelial function and physical exercise. Arq Bras Cardiol 111(4):540–541. https://doi.org/10.5935/abc.20180211
Silva-Boghossian CM, Colombo AP, Tanaka M, Rayo C, Xiao Y, Siqueira WL (2013) Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS ONE 8(10):e75898. https://doi.org/10.1371/journal.pone.0075898
Vukosavljevic D, Custodio W, Buzalaf MA, Hara AT, Siqueira WL (2014) Acquired pellicle as a modulator for dental erosion. Arch Oral Biol 59(6):631–638. https://doi.org/10.1016/j.archoralbio.2014.02.002
Crewther BT, Lowe TE, Ingram J, Weatherby RP (2010) Validating the salivary testosterone and cortisol concentration measures in response to short high-intensity exercise. J Sports Med Phys Fit 50(1):85–92
Noland RC (2015) Exercise and regulation of lipid metabolism. Prog Mol Biol Transl Sci 135:39–74. https://doi.org/10.1016/bs.pmbts.2015.06.017
Tosheska Trajkovska K, Topuzovska S (2017) High-density lipoprotein metabolism and reverse cholesterol transport: strategies for raising HDL cholesterol. Anatol J Cardiol 18(2):149–154. https://doi.org/10.14744/AnatolJCardiol.2017.7608
Wilund KR, Colvin PL, Phares D, Goldberg AP, Hagberg JM (2002) The effect of endurance exercise training on plasma lipoprotein AI and lipoprotein AI:AII concentrations in sedentary adults. Metabolism 51(8):1053–1060. https://doi.org/10.1053/meta.2002.33356
Padiglia A, Orru R, Boroumand M, Olianas A, Manconi B, Sanna MT, Desiderio C, Iavarone F, Liori B, Messana I, Castagnola M, Cabras T (2018) Extensive characterization of the human salivary basic proline-rich protein family by top-down mass spectrometry. J Proteome Res 17(9):3292–3307. https://doi.org/10.1021/acs.jproteome.8b00444
Proctor GB (2016) The physiology of salivary secretion. Periodontol 2000 70(1):11–25. https://doi.org/10.1111/prd.12116
Chojnowska S, Ptaszynska-Sarosiek I, Kepka A, Knas M, Waszkiewicz N (2021) Salivary biomarkers of stress, anxiety and depression. J Clin Med. https://doi.org/10.3390/jcm10030517
Feeney EL, Leacy L, O’Kelly M, Leacy N, Phelan A, Crowley L, Stynes E, de Casanove A, Horner K (2019) Sweet and umami taste perception differs with habitual exercise in males. Nutrients. https://doi.org/10.3390/nu11010155
Gauthier AC, Guimaraes RF, Namiranian K, Drapeau V, Mathieu ME (2020) Effect of physical exercise on taste perceptions: a systematic review. Nutrients. https://doi.org/10.3390/nu12092741
Humphrey SP, Williamson RT (2001) A review of saliva: normal composition, flow, and function. J Prosthet Dent 85(2):162–169. https://doi.org/10.1067/mpr.2001.113778
Matsuo R, Garrett JR, Proctor GB, Carpenter GH (2000) Reflex secretion of proteins into submandibular saliva in conscious rats, before and after preganglionic sympathectomy. J Physiol 527(Pt 1):175–184. https://doi.org/10.1111/j.1469-7793.2000.00175.x
Acknowledgements
The authors thank FAPESP for financial support Proc. FAPESP 2020/02175-6 and Proc. FAPESP 2019/21797-0. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. All authors gave their final approval and agreed to be accountable for all aspects of the work.
Funding
The authors thank FAPESP for financial support Proc. FAPESP 2020/02175-6 and Proc. FAPESP 2019/21797-0.
Author information
Authors and Affiliations
Contributions
TMOV and KOS: participated in the data acquisition, methodology, formal analysis, interpretation of the data and wrote the original draft. ASB and LTGT: participated in the data acquisition and methodology. MARB, FAB, CAK-F and MHF: participated in the methodology and collaboration with the interpretation of the data and draft revision of the manuscript. ACM: designed the study, participated in the data acquisition, methodology, formal analysis, interpretation of the data, wrote the original draft and supervision. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
This study was approved to the local Institutional Ethics Committee (No. 29182720.3.0000.5417). The study was carried out in accordance with the Declaration of Helsinki.
Consent to participate
The collection was done after approval and the signature of the informed consent form.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ventura, T.M.O., Santos, K.O., Braga, A.S. et al. Salivary proteomic profile of young adults before and after the practice of interval exercise: preliminary results. Sport Sci Health 18, 983–997 (2022). https://doi.org/10.1007/s11332-021-00883-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11332-021-00883-z