Abstract
Introduction
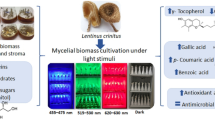
The present work identified and compared intracellular metabolites and metabolic networks in mycelial cultures of Lasiodiplodia theobromae grown under 12 natural light and 24 hours’ dark using a 1 H NMR-based metabolomics approach.
Materials and methods
Fungal cultures were grown in potato dextrose media, and metabolites were extracted by sonication with sodium phosphate-buffered saline (pH = 6.0, 10% D2O, 0.1 mM TSP) from mycelium samples collected every week over four weeks.
Results
Multivariate analyses revealed that the light exposure group showed a positive correlation within beta-hydroxybutyrate, acetoacetate, acetone, betaine, choline, glycerol, and phosphocholine. On the other hand, phenyl acetate, leucine, isoleucine, valine, and tyrosine were positively correlated with dark conditions. Light favored the oxidative degradation of valine, leucine, and isoleucine, leading to the accumulation of choline, phosphocholine, betaine, and ketone bodies (ketogenesis). Ketogenesis, gluconeogenesis, and the biosynthesis of choline, phosphocholine, and betaine, were considered discriminatory routes for light conditions. The light-sensing pathways were interlinked with fungal development, as verified by the increased production of mycelia biomass without fruiting bodies and stress signaling, as demonstrated by the increased production of pigments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Including approximately 3.8 million species, fungi are among the most abundant and diverse microorganisms worldwide. However, only about 120.000 species have been identified (Hawksworth & Lücking, 2017; Góes-Neto et al., 2020). Hence, such biodiversity represents an essential source of primary and secondary metabolites with several biological applications in medical, agricultural, industrial, and environmental areas. As such, it shows the considerable socio-economic value (Lange, 2010; Darwish, 2019). Interestingly, one of the remarkable fungi features is their ability to adapt to different environments by releasing extracellular enzymes that can degrade complex organic compounds into simple molecules that can be readily assimilated during fungal development (Gow & Gadd, 2007; Maggi et al., 2013; Selbmann et al., 2013). Moreover, light quality and quantity are the key factors affecting spore germination, growth, and reproduction in fungi. For example, due to light intensity varying according to season and geographical site, the evaluation and understanding of its effects on fungi development mechanisms is crucial. Despite that, fungi’s biochemical and metabolic responses toward different light-intensity exposure are still not fully understood (Yu & Fischer, 2019).
The Lasiodiplodia genus (Ascomycota, Botryosphaeriaceae) are well-known pathogens of economically important subtropical and tropical crops, known as eliciting agents of black-soot disease. Species of fungus can also endophytically colonize plants and, eventually, produce pycnidia. (Pereira & Ribeiro, 2006; Vijay et al., 2021). Lasiodiplodia theobromae (Pat.) Griffon and Maubl., a common phytopathogen with a broad host range, is an excellent model for studying different growth stages’ underlying biology and biochemistry in response to light. These microorganisms are easy to isolate as they present quick and simple development during in vitro culture (Chen et al., 2013; Zhang et al., 2014; Moubasher et al., 2022). Therefore, the present study applied an NMR-based metabolomics approach to identify the changes in fungal metabolic pathways in response to light exposure and its variations. This technique offers many advantages over other analytical procedures, including quantifying the most abundant compounds in cell extracts with easy sample preparation (Markley et al., 2017; Nagarajan et al., 2022; Porto et al., 2023). This way, this study aimed to compare the intracellular metabolites and metabolic networks in mycelia cultures of L. theobromae incubated under different photoperiod conditions (light and dark). Additionally, the present study is a novel manuscript to focus on L. theobromae metabolome responses to light exposure variations.
2 Materials and methods
2.1 Culture of L. theobromae
Entophytic L. theobromae was isolated from the fruits of Schinus terebinthifolius Raddi, commonly known as the Brazilian pepper tree or aroeira-da-praia (Carlini et al., 2010). The freshly harvested fruits were surface sterilized, cut into small fragments and then, deposited in Petri dishes containing potato-dextrose-agar (PDA). After that, the culture was kept at room temperature for a 12 h photoperiod until fungal outgrowth was observed. Hyphal tips originating from fruit fragments were transferred to a new PDA medium and maintained at room temperature, under the same conditions described above, for 30 days. The fungal species were identified by molecular biology assays, and the absence of fruiting bodies was confirmed by examination of cultures under the light microscope.
The fungal culture was prepared by adding 30 mL of sterilized ultrapure water to a 7-day-old L. theobromae colony and transferring 50 µL aliquots of the hyphal suspension into 64 glass tubes containing 100 mL of potato-dextrose (PD) medium. The culture was incubated at room temperature on an orbital shaker at 160 rpm. The tubes were split into two equal groups (n = 32), with one group kept under dark conditions and the other submitted to a 12 h photoperiod of natural light. Biomasses of eight replicates from each treatment (dark and 12 h photoperiod) were harvested by filtration at the end of the first, second, third, and fourth week of growth, corresponding to different developmental stages of the mycelia. The corresponding mycelial masses were washed with sterilized ultrapure water, frozen in liquid nitrogen, pulverized in a ceramic mortar and pestle, lyophilized, weighed, and stored at 80 °C until required for analysis. The dry mycelium biomass produced each week and under different culture conditions were calculated from the mean masses determined in eight replicate tubes.
2.2 Extraction of metabolites and sample preparation
The intracellular metabolites found in lyophilized mycelia were extracted by adding 800 µL of phosphate-buffered saline (PBS; pH 6.0) containing 10% deuterium dioxide (D2O; 99% purity; Cambridge Isotope Laboratories, Tewksbury, MA, USA) and 0.1 mM sodium 3-(trimethylsilyl) propionate (TSP; Cambridge Isotope Laboratories) to 20 mg of fungal biomass and sonicated for 20 min. Cell debris was removed by centrifugation at 14,000 rpm (Hettich zentrifugen, ROTANTA 460R) for 30 min at 4 °C. The supernatant was transferred to a new tube and submitted to further centrifugation under the abovementioned conditions. An aliquot of the final extract was transferred to a 5 mm NMR tube. To identify the intracellular metabolites produced by mycelia over the four weeks under each culture condition, qualitative control (QC) samples were prepared. The 20 µL aliquots of each of the 32 methanolic extracts obtained from dark-grown cultures were compared with the 20 µL aliquots of each of the 32 extracts obtained from cultures maintained under the lighting photoperiod.
2.3 NMR spectroscopy
Hydrogen NMR analyses were performed in a Bruker UltraShield 400 MHz spectrometer (Billerica, MA, USA) equipped with a multinuclear 5 mm PABBO probe at 300 K. One-dimensional 1 H-NMR spectra were recorded using the noesygppr1d pulse sequence with suppression of the water resonance by pre-saturation. For each sample, 128 transients were collected into 64 K data points with a 90° pulse angle, 19.0 s relaxation delay, 0.05 s mixing time, and 5.11 s acquisition time. Spectra were Fourier transformed, with automatic correction of phase and baseline, using Bruker TopSpin™ NMR data processing software version 3.5. Chemical shifts (δ) were corrected with reference to the TSP signal (0.00 ppm) and expressed in ppm. The identities of metabolite peaks and the assignments of signals were confirmed by comparison of one-dimensional and two-dimensional J-resolved spectral details of QC samples with data available in the Human Metabolome Database (www.hmdb.ca).
2.4 Statistical analysis and chemometrics1q
The dry masses (g) of mycelia harvested at each growth stage were expressed as mean values ± standard deviation (n = 8) and compared using a t-test with the level of statistical significance set at 5%. The 1 H-NMR data were pre-processed using Bruker AMIXTM software version 2018, in which spectra were aligned, normalized, and the region corresponding to D2O was removed. A data matrix comprising 49 observations and 24,971 variables each was generated and exported as a text file to SIMCA™ software version 14 (Umetrics, Umeå, Sweden). Data were submitted to Pareto scaling in order to minimize undesirable noise effects and then submitted to multivariate analysis. The principal component analysis (PCA) and hierarchical cluster analysis (HCA; Ward’s linkage method) were carried out to classify samples according to the developmental stage of the culture. Orthogonal partial least squares - discriminant analysis (OPLS-DA) was performed to examine the differential effects of light and dark on the metabolic profile of L. theobromae. A summary of the main metabolic pathways operative in L. theobromae was constructed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps (https://www.genome.jp/kegg/).
2.5 DNA extraction
Hyphal-tip culture was grown in PD media at 25 ± 2 °C for three days. The mycelial growth was collected using sterile filter paper and washed with autoclaved distilled water. Total DNA was extracted from 20 mg of fresh mycelium, according to Doyle and Doyle (1990), and stored at -20 °C.
2.6 Amplification and sequencing
Total DNA was used as a template for amplification, by Polymerase Chain Reaction (PCR), of the Internal Transcribed Spacer (ITS) and Elongation Factor 1α (EF1α) genomic regions using the primer pairs ITS1, ITS4, 728 F e 986R, respectively. The PCR reactions were performed as follows: 1X MyTaq MasterMix (Bioline®), 0.2 µM of each forward and reverse primers, 1 µL of total DNA (template), and nuclease-free water to a final volume of 25 µL. The cycling conditions were: 2 min at 95 °C as initial denaturation, followed by 35 cycles of denaturation at 95oC for 20 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, with a final extension step at 72 °C for 5 min. The expected size fragments were bidirectionally Sanger sequenced at Macrogen Inc. (South Korea).
2.7 Sequences analysis and phylogeny
The nucleotide sequences were assembled and edited manually using Geneious v.8.1.9 (https://www.geneious.com/). Partial sequences were initially analyzed with the BLASTn algorithm (Altschul et al. 1990) and the NCBI-GenBank non-redundant nucleotide database to determine the species with which they share the most significant identity. A Bayesian phylogenetic tree was reconstructed using concatenated ITS and EF1α partial sequences, downloaded from GenBank, added to the endophytic isolated reported here to support the species identification. The Bayesian inference (BI) was carried out using Mr Bayes v.3.2 (Ronquist et al. 2012) through the CIPRES web portal (Miller et al. 2010), assuming a general time reversible (GTR) nucleotide substitution model with a gamma (G) model of rate heterogeneity and invariable (I) sites. The BI consisted of two replicates with four chains each for 10 million generations and sampling every 1,000 generations. The first 2,500 trees per run were discarded as burn-in. The posterior probabilities (Rannala and Yang 1996) were determined from the majority-rule consensus tree reconstructed with the 15,000 remaining trees.
3 Results and discussion
3.1 Species identification and mycelial growth
No fruiting bodies or spores were detected in the in vitro cultures of the endophytic isolate obtained from S. terebinthifolius. For precise species demarcation based on molecular data, the ITS and EF1α regions were amplified and sequenced. The ITS and EF1α partial nucleotide sequences were 515 and 577 bp in length, respectively, and shred 99.5–100% identity with previously reported L. theobromae isolates in the BLASTn analysis. Additionally, our isolate was grouped with other L. theobromae isolates in the Bayesian phylogenetic tree based on concatenated sequences (ITS and EF1α) (study S26364 deposited in TreeBASE). Therefore, the new isolate was molecularly identified as L. theobromae. The partial sequences reported here were submitted to GenBank under accession nos. MT497468 and MT497469. The quantities of dry mycelia produced each week by dark-grown L. theobromae cultures were significantly lower (p < 0.001) over the four-week period when compared to fungal cultures incubated under the photoperiod (Table 1). However, a growth of 4.7 times between the first and fourth week of cultivation is observed for the 12-hour photoperiod culture, and a growth of 7.9 times for the culture without light exposure. This is related to the fact that the microorganism is using its carbon source for the production of pigments and other secondary metabolites, unlike the light-deprived culture where the carbon source is more available for cellular mass production.
a Mean value ± standard deviation (n = 8).
Light pink mycelia (media) were observed in one-week-old cultures maintained under the lighting photoperiod, whereas those grown in dark conditions were colorless (Fig. S1a). The production of extracellular pigments by four-week-old cultures was confirmed by the grey to black and red coloration exhibited by mycelia and culture media, respectively, regardless of the photoperiod regime applied, but the change in color was more intense in cultures grown under the lighting photoperiod when compared to those maintained in the dark (Fig. S1b).
The influence of light on the growth and sporulation of some fungi species has long been recognized (Yusef & Allam, 1967). Several studies have shown that light of different wavelengths modulates fungal development and can also affect the production of intra- and extra-cellular secondary metabolites, especially pigments (Fanelli et al., 2012; Cheong et al., 2016; Zhang et al., 2021; Dias et al., 2021). Here, the reduced coloration observed in cultures kept under dark conditions can be explained by fungal adaptation to ecological niches considering that the production of pigments is a trait acquired during evolution as a protection against radiation (Schumacher, 2017; Meruvu & Dos Santos, 2021). Moreover, fungi can produce a wide range of pigments that may be classified as carotenoids and polyketides (such as melanin and aromatic phenols) (Lin & Xu, 2020). Carotenoid pigments are synthesized via the mevalonate pathway that is present in all eukaryotes and are generally accumulated in lipid bodies that likely originate from the endoplasmic reticulum (Murphy & Vance, 1999). However, in the absence of lipid bodies, as evidenced by the accumulation of phosphocholine in the cells, these pigments are released into the extracellular environment in the hydrophilic form when they are chemically bounded to carbohydrate groups forming glycosides compounds.
Interestingly, the several types of melanin produced by fungi are generated by the oxidative polymerization of phenolic or indolic intermediates. They may accumulate in cell walls or can also be released into the extracellular environment. In the phylum Ascomycota, melanin is typically synthesized by the pentaketide pathway in which the immediate precursor, 1,8-dihydroxynaphthalene (DHN), is polymerized to DHN-melanin by a laccase. However, some fungal species produce melanin from tyrosine, which is converted into L-dihydroxyphenylalanine (DOPA) by the action of tyrosine 3-mono-oxygenase and subsequently oxidized to dopaquinone. This intermediate undergoes several oxidation, hydroxylation, and decarboxylation reactions to yield dihydroxyindoles, which undergo spontaneous polymerization to form DOPA-melanin (Pombeiro-Sponchiado et al., 2017; Kalra et al., 2020).
3.2 Metabolic fingerprinting of L. theobromae mycelia
Comparing the chemical shifts of the QC sample’s 1 H-NMR analysis with those catalogued in the Human Metabolome Database revealed that the qualitative profiles of the cultures were similar during the entire growth period regardless of the culture conditions (Fig. S2).
Prior to multivariate analysis, variables were scaled according to the Pareto method to reduce the importance of high values and partially preserve the structure of the data (Worley & Powers, 2013). Analysis by PCA increased the interpretability of the large number of variables recorded by reducing the dimensionality of the data set with minimal loss of information. The first six principal components (PC1 to PC6) were calculated based on the culture variation in intracellular fungal metabolites over four weeks. The model presented coefficients of observed variation (R2 = 0.78) and predicted variation (Q2 = 0.66) that were well within limits (R2 ≥ 0.7 and Q2 ≥ 0.4) considered acceptable for the goodness of fit in biological systems (Jiménez-Contreras, 2009). PC1 and PC2 represented 38.7 and 15.1%, respectively, of the variability of the data set and together accounted for 53.8% of the total variability. In addition, the two-dimensional PC1 vs. PC2 score plot (Fig. 1a) revealed that data points relating to mycelia harvested at the same stage of development clustered together in distinct quadrants irrespective of the light/dark regime applied. The clustered points indicated that the metabolomes of the samples were similar, while dispersed points implied distinct differences in metabolomes. In the PC1 vs. PC2 loadings plot (Fig. 1b), data points were separated according to the weightings of the variables (metabolites). Positively correlated variables have approximately the same weighting value and appear near each other, while negatively correlated variables appear diagonally opposite. The HCA dendrogram produced by the Ward method (Fig. 1c) complemented the results obtained by PCA and confirmed the formation of four groups of samples. However, these groups were divided into two subgroups showing that cultures grown under the photoperiod differed in their metabolism from those kept in the dark.
Principal component analysis (PCA) of the of Lasiodiplodia theobromae cultures: (a) PC1 vs. PC2 scores plot showing relationships among the 64 samples obtained from cultures exposed to 12 h photoperiod of natural light (L) and 24 h dark conditions (D); (b) loadings plot showing the correlation between variables (metabolites) in PC1 and PC2; (c) hierarchical cluster analysis (HCA) dendrogram showing the chemical relationships among the 64 samples: the cluster distance between two samples is a measure of similarity
While PCA revealed the variation between groups of samples, the supervised OPLS-DA model captured the variation within groups and separated the variables (metabolites) that were significantly and positively associated with light or dark, as shown in the scores plot (Fig. 2a). According to the S-line plot (Fig. 2c), the intracellular metabolites β-hydroxybutyrate, acetoacetate, acetone, betaine, choline, glycerol, and phosphocholine were positively correlated with light. In contrast, phenylacetate, leucine, isoleucine, valine, and tyrosine were positively correlated with dark conditions. The OPLS-DA model was verified by the leave-one-out cross-validation method, which gave the values R2 = 0.788 and Q2 = 0.734. In the general case, the predictive quality of a model is considered acceptable when R2 > Q2 and the difference between the two coefficients is less than 0.3. More significant differences between R2 and Q2 would indicate that the model suffered from overfitting (Hawkins et al., 2003; Veerasamy et al., 2011), but a difference of < 0.06 was observed in the present study. Moreover, although Q2 was higher than 0.5, permutation tests were obtained using the SIMCA software to ensure that the Q2 value of the model was significant, as shown by the random permutation of the rows in the dataset (Fig. 2b).
Orthogonal partial least squares – discriminant analysis (OPLS-DA) of the 1 H NMR-derived metabolic profiles of Lasiodiplodia theobromae cultures: (a) scores plot showing two separate groups, one containing metabolites produced by cultures exposed to a 12 h photoperiod of natural light (L) and the other containing metabolites produced by cultures exposed to dark (D) conditions; (b) S-line plot showing the metabolites positively correlated with light and dark conditions (R2 = 0.788 and Q2 = 0.734); (c) predictive capacity of the OPLS-DA model as determined by the permutation test
Level variations in the main intracellular metabolites during the growth cycle (Fig. 3) provided many insights into the effects of light on the metabolome of L. theobromae. For example, the branched-chain amino acids (BCAAs) valine, leucine, and isoleucine concentrations were lower in cultures exposed to light compared to their dark-grown counterparts (Fig. 3a-c). Like plants, fungi can synthesize BCAAs, but in this study, the BCAAs were also available in the PDA media (Guedes et al., 2011; Luo et al., 2020). The explanation for the observed difference in levels of BCAAs is that light stimulated the complete oxidation of BCAAs in the mitochondria with the generation of high amounts of energy in the form of adenosine triphosphate (ATP) via oxidative phosphorylation. The first step in BCAA degradation is the formation of branched-chain 2-oxo acids, which suffer irreversible oxidative decarboxylation to acyl-CoA and, ultimately, acetyl-CoA, propionyl-CoA, acetoacetyl-CoA, and acetoacetate (Schertl et al., 2017; Mann et al., 2021). The concentrations of the ketone bodies, β–hydroxybutyrate and acetoacetate (Fig. 3j and k), were positively correlated and increased throughout the growth period regardless of culture conditions, but especially in cultures grown under light conditions. Apparently, both metabolites occurred in similar proportions since β -hydroxybutyrate is formed by the reduction of acetoacetate. Acetone is the breakdown product of acetoacetate, so its concentration also increased during the growth period (Fig. 3l). The decline in the levels of BCAAs, together with those of alanine (Fig. 3n), occurred concomitantly with the augmentation of glucose (Fig. 3m), indicating that depletion of glucose stores during the light-induced accelerated growth triggered endogenous glucose production (gluconeogenesis).
The increased production of acyl-CoA resulting from increased BCAA oxidation under light conditions stimulated the biosynthesis of choline (Fig. 3g), which is a vital metabolite for the synthesis of phosphocholine in eukaryotes (Fig. 3i) to form cellular membrane. The last is a precursor as well as a degradation product of phosphatidylcholine (lecithin), a major component of phospholipids in eukaryotic membranes. In addition, phosphocholine can covalently bind to proteins and glycolipids in a wide spectrum of organisms, and such modified macromolecules play important functions including host defense and virulence (Richter et al., 2016). Hence, the larger mycelial biomass produced by cultures exposed to light rather than dark (Table 1) can be explained by the intensification of phospholipid biosynthesis and the assembly of lipid bilayers in the fungal cells. The choline–betaine pathway is reversible, with one compound being a precursor to the other, depending on the needs of the microorganism. In this studied case, despite betaine (N, N, N-trimethylglycine; Fig. 3h) being a metabolite with robust osmoprotectant properties that confer tolerance to salinity, drought, and other stresses, it served as a precursor in choline production, as can be seen in Fig. 3h and g (Lambou et al., 2013; Ząbek et al., 2017).
The phosphocholine concentration was strongly, but negatively correlated with the levels of its BCAA precursors. The production of phosphocholine peaked during the second week (especially in light-grown cultures) when growth reached its exponential phase. After that, the synthesis of lipid membranes was maximal but decreased from the third week onwards. However, the relative amounts of phosphocholine were higher than those of all other metabolites during the entire growth period, likely due to the degradation of glycerophospholipids as evidenced by the high concentration of glycerol (Fig. 3f) in the mycelia during the third and fourth weeks. Indeed, the concentration of glycerol was strongly and positively correlated with that of phosphocholine. Furthermore, the levels of phenylacetate were much lower in cultures grown under the photoperiod compared with those maintained in the dark (Fig. 3d), mainly because fungi can employ this compound as an alternative carbon source when glucose stores are depleted, a situation that occurs more rapidly under light conditions with the escalation of mycelial growth. Nevertheless, when glucose is abundant, the degradation of phenylacetate is downregulated. The oxidation of phenylacetate in fungi usually proceeds via 2-hydroxyphenylacetate and homogentisate and subsequent conversion to acetate and fumarate. An alternative pathway for phenylacetate oxidation is mediated by phenylacetate-CoA ligase resulting in conversion to benzylpenicillin, an antibiotic used by fungi as protection against bacteria (Middelhoven, 1993; Jami et al., 2018).
Although light stimulates growth via the upregulation of oxidative routes, it threatens fungal survival by inducing the formation and accumulation of highly reactive oxygen species, including H2O2 and O2– cause oxidative damage to biomolecules. To protect against light-induced oxidative stress, some fungi produce pigments such as melanin, which gives the cells a dark brown coloration. Ultra-violet radiation stimulates melanogenesis by triggering oxidative tyrosine degradation and consequent indolic intermediates polymerization (Calvo et al., 2002; Thabet et al., 2014; Zaidi et al., 2014; Pavan et al., 2020). Regardless of their origins, melanin pigments have common structural characteristics that confer the ability to scavenge free radicals and provide natural antioxidant and protective functions under adverse conditions. Evidence suggests that melanin is essential for zoo pathogenic and phytopathogenic microorganisms because of its antioxidant properties (Liu et al., 2022). The statement above explains the lower levels of tyrosine in light-grown mycelia compared with their dark-grown counterparts (Fig. 3e). This amino acid should be a discriminatory metabolite for dark conditions.
It is worth mentioning that the levels of ethanol were high towards the end of the growth cycle regardless of the culture conditions (Fig. 3p). Ethanol is produced from pyruvate (Fig. 3o) in a two-step reaction involving decarboxylation by pyruvate decarboxylase with the formation of acetaldehyde and subsequent reduction to ethanol catalyzed by NADH-dependent alcohol dehydrogenase.
3.3 Discriminatory primary pathways
The identification of the primary intracellular metabolites by 1 H-NMR spectroscopy and subsequent multivariate analysis revealed the pathways that were most affected in L. theobromae cultures by changes in light conditions (Fig. 4). The exposure of fungal cultures to the photoperiod favored primarily oxidative pathways, particularly those involving the degradation of BCAAs, leading to the accumulation of phospholipid membrane components (choline, phosphocholine, and betaine) and ketone bodies. The endogenous synthesis of glucose from non-carbohydrate substrates such as TCA cycle intermediates, alanine, and glycerol was also stimulated by light.
The general scheme of metabolic pathways in Lasiodiplodia theobromae cultures showing the discriminant metabolites produced by oxidative reactions under dark (blue) and 12 h photoperiod of natural light (red) conditions and ethanol produced by reductive reactions (pink). The scheme was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG, 2019) pathway maps
Since ketogenesis and gluconeogenesis were upregulated in cultures grown under light conditions, these pathways are discriminatory. Moreover, the biosynthesis of choline and phosphocholine from BCAAs could also be regarded as discriminatory since the levels of these metabolites increased significantly up to the second week in mycelial cultures exposed to light, following which they declined probably because of the synthesis of phospholipids and the self-assembly of lipid bilayers and vesicles. Interestingly, light did not influence anaerobic glycolysis or fermentation since the end product (ethanol) levels were similar, regardless of the culture conditions.
4 Conclusions
The outcomings presented herein demonstrates the importance of light on the biology of L. theobromae and also provide the groundwork for future studies on this important plant pathogen. 1 H-NMR associated with PCA and OPLS-DA proved to be excellent approach for detecting metabolic changes in L. theobromae cultures grown under light and dark. The PC1 vs. PC2 score plot demonstrated metabolites differences between groups, whereas the loading plot highlighted the metabolites that better expressed such differences. The light-sensing pathways were interlinked not only concerning stress signaling, as demonstrated by the increased production of pigments that serve as protection against oxidative stress, but also with fungal development, as verified by the increased production of mycelia biomass without fruiting bodies. Further, choline, phosphocholine, and betaine emerged as primary discriminatory metabolites. Their accumulation may explain why this isolate is a non-producer of fruiting bodies and why the pigments were released into the media. The light-induced oxidation of tyrosine is an important finding since it suggests that L. theobromae cultures produce DOPA-melanin rather than DHN-melanin. In this context, cultures of L. theobromae could represent a good source of natural pigments for cosmetic, pharmaceutical, and food-processing industries, since microorganisms pigment production is considered an efficient and cost-effective process.
References
Calvo, A. M., Wilson, R. A., Bok, J. W., & Keller, N. P. (2002). Relationship between secondary metabolism and fungal development. Microbiology and molecular biology reviews, 66(3), 447–459. https://doi.org/10.1128/MMBR.66.3.447-459.2002.
Carlini, E. A., Duarte-Almeida, J. M., Rodrigues, E., & Tabach, R. (2010). Antiulcer effect of the pepper trees Schinus terebinthifolius Raddi (aroeira-da-praia) and myracrodruon urundeuva Allemão, Anacardiaceae (aroeira-do-sertão). Revista Brasileira de Farmacognosia, 20, 140–146. https://doi.org/10.1590/S0102-695X2010000200001.
Chen, S. F., Morgan, D., Beede, R. H., & Michailides, T. J. (2013). First report of Lasiodiplodia theobromae associated with stem canker of almond in California. Plant Disease, 97(7), 994–994. https://doi.org/10.1094/PDIS-11-12-1033-PDN.
Cheong, K. K., Strub, C., Montet, D., Durand, N., Alter, P., Meile, J. C., Galindo, S. S., & Fontana, A. (2016). Effect of different light wavelengths on the growth and ochratoxin A production in aspergillus carbonarius and aspergillus westerdijkiae. Fungal Biology, 120(5), 745–751. https://doi.org/10.1016/j.funbio.2016.02.005.
Darwish, A. M. G. (2019). Fungal mycotoxins and natural antioxidants: Two sides of the same coin and significance in food safety. Microbial Biosystems, 4(1), 1–16. https://doi.org/10.21608/mb.2019.37468.
Dias, L. P., Souza, R. K., Pupin, B., & Rangel, D. E. (2021). Conidiation under illumination enhances conidial tolerance of insect-pathogenic fungi to environmental stresses. Fungal Biology, 125(11), 891–904. https://doi.org/10.1016/j.funbio.2021.06.003.
Fanelli, F., Schmidt-Heydt, M., Haidukowski, M., Susca, A., Geisen, R., Logrieco, A., & Mulè, G. (2012). Influence of light on growth, conidiation and fumonisin production by Fusarium verticillioides. Fungal biology, 116(2), 241–248. https://doi.org/10.1016/j.funbio.2011.11.007.
Góes-Neto, A., Marcelino, V. R., Verbruggen, H., da Silva, F. F., & Badotti, F. (2020). Biodiversity of endolithic fungi in coral skeletons and other reef substrates revealed with 18S rDNA metabarcoding. Coral Reefs, 39, 229–238. https://doi.org/10.1007/s00338-019-01880-y.
Gow, N. A., & Gadd, G. M. (Eds.). (2007). Growing fungus. Springer Science & Business Media.
Guedes, R. L. M., Prosdocimi, F., Fernandes, G. R., Moura, L. K., Ribeiro, H. A. L., & Ortega, J. M. (2011, December). Amino acids biosynthesis and nitrogen assimilation pathways: a great genomic deletion during eukaryotes evolution. In BMC genomics (Vol. 12, No. 4, pp. 1–13). BioMed Central. https://doi.org/10.1186/1471-2164-12-S4-S2.
Hawkins, D. M., Basak, S. C., & Mills, D. (2003). Assessing model fit by cross-validation. Journal of chemical information and computer sciences, 43(2), 579–586. https://doi.org/10.1021/ci025626i.
Hawksworth, D. L., & Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiology spectrum, 5(4), 5–4. https://doi.org/10.1128/microbiolspec.FUNK-0052-2016.
Jami, M. S., Martín, J. F., Barreiro, C., Domínguez-Santos, R., Vasco-Cárdenas, M. F., Pascual, M., & García-Estrada, C. (2018). Catabolism of phenylacetic acid in Penicillium rubens. Proteome-wide analysis in response to the benzylpenicillin side chain precursor. Journal of proteomics, 187, 243–259. https://doi.org/10.1016/j.jprot.2018.08.006.
Jiménez-Contreras, E., Torres-Salinas, D., Moreno, R., Baños, R., & López-Cózar, E. (2009). Response surface methodology and its application in evaluating scientific activity. Scientometrics, 79(1), 201–218. https://doi.org/10.1007/s11192-009-0413-3.
Kalra, R., Conlan, X. A., & Goel, M. (2020). Fungi as a potential source of pigments: Harnessing filamentous fungi. Frontiers in Chemistry, 8, 369. https://doi.org/10.3389/fchem.2020.00369.
Lambou, K., Pennati, A., Valsecchi, I., Tada, R., Sherman, S., Sato, H., & Latgé, J. P. (2013). Pathway of glycine betaine biosynthesis in aspergillus fumigatus. Eukaryotic cell, 12(6), 853–863. https://doi.org/10.1128/EC.00348-12.
Lange, L. (2010). The importance of fungi for a more sustainable future on our planet. Fungal Biology Reviews, 24(3–4), 90–92. https://doi.org/10.1016/j.fbr.2010.12.002.
Lin, L., & Xu, J. (2020). Fungal pigments and their roles associated with human health. Journal of Fungi, 6(4), 280. https://doi.org/10.3390/jof6040280.
Liu, R., Meng, X., Mo, C., Wei, X., & Ma, A. (2022). Melanin of fungi: From classification to application. World Journal of Microbiology and Biotechnology, 38(12), 228. https://doi.org/10.1007/s11274-022-03415-0.
Luo, F., Zhou, H., Zhou, X., Xie, X., Li, Y., Hu, F., & Huang, B. (2020). The intermediates in branched-chain amino acid biosynthesis are indispensable for conidial germination of the insect-pathogenic fungus metarhizium robertsii. Applied and environmental microbiology, 86(20), e01682–e01620. https://doi.org/10.1128/AEM.01682-20.
Maggi, O., Tosi, S., Angelova, M., Lagostina, E., Fabbri, A. A., Pecoraro, L., Altobelli, E., Pico, A. A., Savino, E., Branda, E., Turchetti, B., Zotti, M., Vizzini, A., & Buzzini, P. (2013). Adaptation of fungi, including yeasts, to cold environments. Plant Biosystems-An International Journal Dealing with all Aspects of Plant Biology, 147(1), 247–258. https://doi.org/10.1080/11263504.2012.753135.
Mann, G., Mora, S., Madu, G., & Adegoke, O. A. (2021). Branched-chain amino acids: Catabolism in skeletal muscle and implications for muscle and whole-body metabolism. Frontiers in Physiology, 12, 702826. https://doi.org/10.3389/fphys.2021.702826.
Markley, J. L., Brüschweiler, R., Edison, A. S., Eghbalnia, H. R., Powers, R., Raftery, D., & Wishart, D. S. (2017). The future of NMR-based metabolomics. Current opinion in biotechnology, 43, 34–40. https://doi.org/10.1016/j.copbio.2016.08.001.
Meruvu, H., & Dos Santos, J. C. (2021). Colors of life: A review on fungal pigments. Critical reviews in biotechnology, 41(8), 1153–1177. https://doi.org/10.1080/07388551.2021.1901647.
Middelhoven, W. J. (1993). Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi: A literature review and an experimental approach. Antonie van Leeuwenhoek, 63, 125–144. https://doi.org/10.1007/BF00872388.
Moubasher, H. A., Balbool, B. A., Helmy, Y. A., Alsuhaibani, A. M., Atta, A. A., Sheir, D. H., & Abdel-Azeem, A. M. (2022). Insights into asparaginase from endophytic fungus Lasiodiplodia theobromae: Purification, characterization and antileukemic activity. International journal of environmental research and public health, 19(2), 680. https://doi.org/10.3390/ijerph19020680.
Murphy, D. J., & Vance, J. (1999). Mechanisms of lipid-body formation. Trends in biochemical sciences, 24(3), 109–115. https://doi.org/10.1016/S0968-0004(98)01349-8.
Nagarajan, K., Ibrahim, B., Ahmad Bawadikji, A., Lim, J. W., Tong, W. Y., Leong, C. R., Khaw, K. Y., & Tan, W. N. (2022). Recent developments in metabolomics studies of endophytic fungi. Journal of Fungi, 8(1), 28. https://doi.org/10.3390/jof8010028.
Pavan, M. E., López, N. I., & Pettinari, M. J. (2020). Melanin biosynthesis in bacteria, regulation and production perspectives. Applied Microbiology and Biotechnology, 104, 1357–1370. https://doi.org/10.1007/s00253-019-10245-y.
Pereira, A. L., Silva, G. S., & Ribeiro, V. Q. (2006). Caracterização fisiológica, cultural e patogênica de diferentes isolados de lasiodiplodia theobromae. Fitopatologia Brasileira, 31, 572–578. https://doi.org/10.1590/S0100-41582006000600006.
Pombeiro-Sponchiado, S. R., Sousa, G. S., Andrade, J. C., Lisboa, H. F., & Gonçalves, R. C. (2017). Production of melanin pigment by fungi and its biotechnological applications. Melanin, 1(4), 47–75.
Porto, V. A., da Rocha Júnior, E. R., Ursulino, J. S., Porto, R. S., da Silva, M., de Jesus, L. W. O., De Oliveira, J. M., Crispim, A. C., Santos, J. C. C., & de Aquino, T. M. (2023). NMR-based metabolomics applied to ecotoxicology with zebrafish (Danio rerio) as a prominent model for metabolic profiling and biomarker discovery: Overviewing the most recent approaches. Science of The Total Environment, 161737. https://doi.org/10.1016/j.scitotenv.2023.161737.
Richter, K., Mathes, V., Fronius, M., Althaus, M., Hecker, A., Krasteva-Christ, G., & Grau, V. (2016). Phosphocholine–an agonist of metabotropic but not of ionotropic functions of α9-containing nicotinic acetylcholine receptors. Scientific reports, 6(1), 1–13. https://doi.org/10.1038/srep28660.
Schertl, P., Danne, L., & Braun, H. P. (2017). 3-Hydroxyisobutyrate dehydrogenase is involved in both, valine and isoleucine degradation in Arabidopsis thaliana. Plant Physiology, 175(1), 51–61. https://doi.org/10.1104/pp.17.00649.
Schumacher, J. (2017). How light affects the life of Botrytis. Fungal Genetics and Biology, 106, 26–41. https://doi.org/10.1016/j.fgb.2017.06.002.
Selbmann, L., Egidi, E., Isola, D., Onofri, S., Zucconi, L., de Hoog, G. S., Chinaglia, S., Testa, L., Tosi, S., Balestrazzi, A., Lantieri, A., Compagno, R., & Varese, G. C. (2013). Biodiversity, evolution and adaptation of fungi in extreme environments. Plant Biosystems-An International Journal Dealing with all Aspects of Plant Biology, 147(1), 237–246. https://doi.org/10.1080/11263504.2012.753134.
Thabet, S., Simonet, F., Lemaire, M., Guillard, C., & Cotton, P. (2014). Impact of photocatalysis on fungal cells: Depiction of cellular and molecular effects on Saccharomyces cerevisiae. Applied and environmental microbiology, 80(24), 7527–7535. https://doi.org/10.1128/AEM.02416-14.
Veerasamy, R., Rajak, H., Jain, A., Sivadasan, S., Varghese, C. P., & Agrawal, R. K. (2011). Validation of QSAR models-strategies and importance. Int J Drug Des Discov, 3, 511–519.
Vijay, S., Renukadevi, P., Mangammal, P., & Krishnamoorthy, S. V. (2021). Effect of media, temperature and pH on the growth and pycnidial production of Lasiodiplodia theobromae (Pat.), causative of black root rot in mulberry. The Pharma Inn Journal, 10(11), 340–343. https://www.thepharmajournal.com/archives/%3Fyear=2021%26vol=10%26issue=11%26ArticleId=8668.
Worley, B., & Powers, R. (2013). Multivariate analysis in metabolomics. Current metabolomics, 1(1), 92–107.
Yu, Z., & Fischer, R. (2019). Light sensing and responses in fungi. Nature Reviews Microbiology, 17(1), 25–36. https://doi.org/10.1038/s41579-018-0109-x.
Yusef, H. M., & Allam, M. E. (1967). The effect of light on growth and sporulation of certain fungi. Mycopathologia et mycologia applicata, 33(2), 81–89. https://doi.org/10.1007/BF02053439.
Ząbek, A., Klimek-Ochab, M., Jawień, E., & Młynarz, P. (2017). Biodiversity in targeted metabolomics analysis of filamentous fugal pathogens by 1H NMR-based studies. World Journal of Microbiology and Biotechnology, 33, 132. https://doi.org/10.1007/s11274-017-2285-7.
Zaidi, K. U., Ali, A. S., Ali, S. A., & Naaz, I. (2014). Microbial tyrosinases: promising enzymes for pharmaceutical, food bioprocessing, and environmental industry. Biochemistry research international, 2014. https://doi.org/10.1155/2014/854687.
Zhang, Z., Han, X. M., Wei, J. H., Xue, J., Yang, Y., Liang, L., Li, X. J., Guo, Q. M., Xu, Y. H., & Gao, Z. H. (2014). Compositions and antifungal activities of essential oils from agarwood of Aquilaria sinensis (Lour.) Gilg induced by Lasiodiplodia theobromae (Pat.) Griffon. & Maubl. Journal of the Brazilian Chemical Society, 25, 20–26. https://doi.org/10.5935/0103-5053.20130263.
Zhang, H., Wang, G., Yang, Q., Yang, X., Zheng, Y., Liu, Y., & Xing, F. (2021). Effects of Light on the Ochratoxigenic Fungi Aspergillus ochraceus and A. carbonarius. Toxins, 13(4), 251. https://doi.org/10.3390/toxins13040251.
Author information
Authors and Affiliations
Contributions
ACC, SMAC, JRR, RRS constructed experimental design and prepared the manuscript; VAP prepared figures and tables; All authors reviewed and edted the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
This article does not contain any studies involving human or animal models experiment performed by any authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Crispim, A.C., Crispim, S.M.A., Rocha, J.R. et al. Light effects on Lasiodiplodia theobromae metabolome cultured in vitro. Metabolomics 19, 75 (2023). https://doi.org/10.1007/s11306-023-02041-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-023-02041-7