Abstract
Introduction
Climate change is a major concern for the scientific community, demanding novel information about the effects of environmental stressors on living organisms. Metabolic profiling is required for achieving the most extensive possible range of compounds and their concentration changes on stressed conditions.
Objectives
Individuals of the crustacean species Daphnia magna were exposed to three different abiotic factors linked to global climate change: high salinity, high temperature levels and hypoxia. Advanced chemometric tools were used to characterize the metabolites affected by the exposure.
Method
An exploratory analysis of gas chromatography-mass spectrometry (GC–MS) data was performed to discriminate between control and exposed daphnid samples. Due to the complexity of these GC–MS data sets, a comprehensive untargeted analysis of the full scan data was performed using multivariate curve resolution-alternating least squares (MCR-ALS) method. This approach enabled to resolve most of the metabolite signals from interference peaks caused by derivatization reactions. Metabolites with significant changes in their peak areas were tentatively identified and the involved metabolic pathways explored.
Results
D. magna metabolic biomarkers are proposed for the considered physical factors. Metabolites related with energy metabolic pathways including some amino acids, carbohydrates, organic acids and nucleosides were identified as potential biomarkers of the investigated treatments.
Conclusions
The proposed untargeted GC–MS metabolomics strategy and multivariate data analysis tools were useful to investigate D. magna metabolome under environmental stressed conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Metabolomics is concerned with the study of naturally occurring, low molecular weight organic metabolites within a cell or tissue (Griffiths 2008; Harrigan and Goodacre 2003; Lindon et al. 2007). An important application of metabolomics is the study of the interactions of living organisms with their environment, which is defined as environmental metabolomics (Bundy et al. 2009; Miller 2007). A definition proposed for this field is ‘‘the application of metabolomics to the investigation of both free-living organisms obtained directly from the natural environment and of organisms reared under laboratory conditions” (Morrison et al. 2007). Metabolomics presents several advantages for the study of organism–environment interactions and for assessing the health status of organisms in the field (Bundy et al. 2009; Krastanov 2010). Metabolomic measurements report on the actual functional status of biological organism levels, (i.e. cell, tissue) which, in principle, can be mechanistically anchored to effects occurring at lower (i.e. genomic) or higher (phenotypic) levels of biological organization (Fiehn 2001). As such, at present, the number of metabolomic studies applied to environmental problems are growing considerably trying to improve the understanding of organism responses to abiotic stressors (Arbona et al. 2013; Sardans et al. 2011). Metabolomic studies are based on simultaneous measurement of multiple metabolites, using inherently parallel analytical techniques such as nuclear magnetic resonance spectroscopy or mass spectrometry, followed by appropriate statistical analysis (Lindon and Nicholson 2008a, b; Dunn et al. 2011). The application of metabolomic techniques, however, is still limited, due to the inherent problems of separation and identification of unique metabolites and hence metabolic changes (Viant 2008; Bundy et al. 2009; Samuelsson and Larsson 2008; Miller 2007).
To evaluate the effects of stressors on the metabolome, gas chromatography coupled to mass spectrometry (GC–MS) in electron ionization (EI) is a widely used analytical technique as it provides high sensitivity and reproducibility, and permits compound identification using spectral libraries (Steinhauser and Kopka 2007; Vandenbrouck et al. 2010b). The major limitation of GC–MS is the restriction to semi-volatile analytes, and hence the requirement to convert many polar metabolites to volatile derivatives through chemical derivatization processes. Oximation and trimethylsylilation (TMS) reactions of organic compounds are the classical and most widely used derivatization procedures for metabolome analysis by GC–MS (Villas-Boas et al. 2011; Kanani et al. 2008; Yi et al. 2014; Gullberg et al. 2004; De Souza 2013; Little 1999). Derivatization, however, increases the amount of adduct-derivatives and hence the complexity of metabolomics data. Understanding this complex big-data generated from GC–MS is still currently a bottleneck (Vandenbrouck et al. 2010b; Dunn et al. 2013; Wehrens et al. 2014).
In order to maximize the extraction of information, a pipeline for GC–MS metabolomic data analysis was designed and principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA) and multivariate curve resolution-alternating least squares (MCR-ALS) (De Juan et al. 2014; van Stokkum et al. 2009; Tauler 1995) were proposed for the resolution of complex chemical mixtures. This last chemometric tool is a powerful data analysis tool that solves the mixture analysis and interferences co-elution problems in complex natural samples as well as problems derived from GC–MS systems, such as baseline drift, spectral background, noise contributions or low S/N ratio values (Parastar et al. 2012). MCR-ALS has been also used in previous works related with ecological and environmental problems (Navarro-Reig et al. 2015; Malik and Tauler 2015; Alier et al. 2010; Terrado et al. 2009).
Daphnia magna, a freshwater crustacean, is used extensively as an aquatic test species (Ikenaka et al. 2006; Martins et al. 2007; Barata and Baird 2000a) being the object of standardized testing guidelines from the Organization for Economic Co-operation and Development (OECD 2012). Acute and chronic tests of D. magna are among the most frequently performed studies in aquatic toxicology because these animals are relatively easy to culture, have a short lifecycle, and can be maintained at high population densities in relatively small volumes and thus are cost-effective (Barata et al. 2005; De Meester and Vanoverbeke 1999). Daphnia magna is also a keystone species in freshwater food webs, thus it is also an ecological model species to study effects of global climate change. Water temperature, salinity and oxygen levels of freshwater bodies are likely to be affected by climate change (IPCC 2014; Adrian et al. 2009; Coutant 1990).
Here an untargeted GC–MS metabolomics study of D. magna exposed to mild stress provoked by high salinity, increased temperature and low dissolved oxygen is presented.
2 Materials and methods
2.1 Chemicals and standard solutions
Pure metabolites: pimelic acid, malic acid, l-phenylalanine, l-tyrosine, putrescine, dopamine hydrochloride, l-histidine, l-methionine, d-maltose monohydrate, myo-inositol, d-ribose, d-galactose and cytidine used as test standards were purchased from Sigma-Aldrich (St. Louis, MO, USA). d-glucose (U-13C6, 99 %), used as the internal standard (IS), was supplied by Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). Pyridine (anhydrous, 99.8 %), chlorotrimethylsilane (TMCS), methoxyamine hydrochloride (98 %) (MeOX) and N-methyl-N-trimethylsilyl trifluoroacetamide (>98.5 %) (MSTFA), used as derivatizating agents, and the saturated Alkane standard mixture for the performance test of GC systems from C7 to C30 were also obtained from Sigma-Aldrich (St. Louis, MO, USA). Hexane, methanol and chloroform were analytical reagent grade, and sodium chloride (NaCl) salt was supplied by Merck (Darmstadt, Germany).
2.2 Sample preparation
2.2.1 Daphnia magna growth conditions
A single laboratory clone F, which has been the subject of many investigations (Barata and Baird 2000b) was used. Bulk cultures of 10 animals L−1 were maintained in ASTM hard synthetic water (ASTM, 1995) as described by Barata and Baird (2000b). Individual or bulk cultures were fed daily with Chorella vulgaris Beijerinck (5 × 105 cells mL−1, corresponding to 1.8 μg C mL−1) (Barata and Baird 1998). The culture medium was changed every other day, and neonates were removed within 24 h. Photoperiod was set to 14 h light: 10 h dark cycle and temperature at 20 ± 1 °C.
2.2.2 Experimental design
Four days old D. magna juveniles were used for experiment. Experimental animals were obtained from bulk cultures of 100 individuals reared in 10 L of ASTM as described above (“Daphnia magna growth conditions”). Randomly collected animals were exposed to high salinity, elevated temperature and hypoxia treatments in groups of ten individuals in 1.2 L jars filled with 1 L of ASTM hard water. A control treatment of individuals exposed to ASTM hard water maintained at 20 °C under saturating oxygen conditions (≈9 mg L−1) without salt (0 g L−1 NaCl) was also included. Each treatment was replicated five times. Salinity exposed samples were prepared with 5 g L−1 NaCl in ASTM hard water, temperature samples were prepared in a water bath at 25 °C and hypoxia samples at ≤2 mg O2 L−1 were prepared by bubbling liquid nitrogen into the ASTM hard water thus displacing oxygen from it. The oxygen levels were controlled by an oxygen sensor, approximately the levels had to be ≤2 mg L−1 in every jar sample. After 24 h of exposition daphnids were collected from each jar, pooled in an eppendorf, the water was removed and then samples were frozen in liquid N2 and stored at −80°C before metabolite extraction.
2.2.3 Extraction and derivatization of metabolites
In the first part of this work, GC–MS method and derivatization step were optimized to obtain high sensitivity and to maximize the number of detected compounds. Several standard mixtures (i.e. organic acids, sugars, nucleotides, nucleosides and amino acids) were tested to optimize the experimental conditions and the derivatization step in order to ensure a comprehensive and reliable metabolic profiling. An aqueous standard solution (1000 µg mL−1) of each standard was prepared in methanol and stored in the freezer at −20 °C until their use. Working standard solutions of 50 µg mL−1 were obtained by diluting the stock solutions with water. Diluted standard solutions of 5 µg mL−1 were used to optimize experimental conditions and also several derivatization conditions were tested. Metabolite extraction was performed with methanol, water and chloroform and the chemical derivatization was carried out with two agents, methoxyamine (MeOX) in pyridine and N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) with 1 % of trimethylchlorosilane (TMCS) as a catalyst of the reaction, which are very commonly used in metabolomics studies by GC–MS analysis.
Polar metabolites of the whole organism were extracted with 400 µL methanol, sonicated for 15 min, and then 200 µL water and 400 µL chloroform were added before centrifugation at 10,000×g during 15 min, in order to separate the aqueous and the lipid phase. The aqueous phase of every sample was transferred to a new eppendorf and 10 µL of the Internal Standard (IS) [d-glucose (U-13C6, 99 %)] were added at a concentration of 50 µg mL−1. Afterwards, the extract was evaporated to dryness with a Speedvac (Thermo Scientific) at 40 °C during 3 h. When samples were completely dry they were stored at −80 °C until analysis.
Prior the analysis by GC–MS, a derivatization method was performed to create thermally stable derivatives suitable for the analysis (Orata 2012). The initial conditions were adjusted from (Vandenbrouck et al. 2010a) and adapted to the needs of this study, in order to detect the maximum number of metabolites. The first step was methoximation with methoxyamine hydrochloride (MeOX) in pyridine, which was used to stabilize carbonyl radicals of metabolites and to stop charge transfer. Then, trimethylsilylation was carried out with N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) with 1 % of trimethylclorosilane (TMCS) as catalyst. Trimethylsilylation was applied to replace active hydrogens on a wide range of polar compounds with a trimethylsilyl [–Si(CH3)3] group. This derivatization method permits to detect the silylated molecules in the gas chromatography analysis because it increases the volatility of the metabolites. Different MeOX concentrations (20, 30 and 40 mg mL−1), MeOX volumes (30, 65 and 100 µL), MeOX temperatures (20, 30 and 40 °C), MeOX times (90, 525 and 960 min), MSTFA volumes (30, 65 and 100 µL), MSTFA temperatures (20, 30 and 40 °C) and MSTFA times (30, 60 and 90 min) were tested to improve the derivatization reaction and to determine the most suitable conditions for the analysis of D. magna metabolites (Pacchiarotta et al. 2010; Kanani et al. 2008; Azizan et al. 2012; Kanani and Klapa 2007; Strehmel et al. 2008).
After the extraction of polar metabolites, the final protocol of the derivatization conditions was implemented: to the dry residue obtained after extraction from daphnids, 65 µL of 20 µg µL−1 MeOX in pyridine were added. After mixing for 1 min, the mixture was incubated for 90 min at 30 °C. Thereafter, 30 µL of MSTFA (1 % TMCS) were added, vortex mixed for a minute and then incubated for another 30 min at room temperature. Prior to injection, daphnid derivatized extracts were finally filtrated through a 0.22 µm filters (Ultrafree®-MC, Millipore) and, then transferred to a chromatographic vial and 5 µL of hexane were added to obtain a final volume of 100 µL in order to be injected into the GC system.
2.3 GC–MS analysis
A Trace GC–MS Thermo Fisher system operated in EI mode at 70 eV and equipped with a ZB-5MS column (Phenomenex, 30 m × 0.25 mm ID × 0.25 µm) (Vandenbrouck et al. 2010a) was used. The oven temperature program was set at 70 °C and then increased at 10 °C min−1 to 250 °C, and then to 310 °C at 5 °C min−1 and held for 5 min, the delay time was 5.3 min and the time of analysis was 49 min. Other operating conditions were as follows: carrier gas was He, with a flow rate of 0.6 mL min−1; source temperature, 200 °C; interface temperature, 280 °C; split ratio of 1:20; detector voltage, 440 V; 2 µL were injected in the split mode injection. The m/z values were recorded in full scan mode in the range of m/z 60–650 amu. The mass spectrometer was interfaced to a computer workstation running Xcalibur software (version 2.2, Thermo Scientific) for data acquisition and processing. For each detected peak, a linear retention index (RI) (Strehmel et al. 2008) was calculated using GC RI standards (hydrocarbons from C7 to C30 were used as internal standards). Therefore, an n-alkane Standard mixture was prepared dissolved in hexane at a concentration of 10 µg µL−1, and was injected at the same programme conditions as the samples in the GC–MS instrument.
2.4 Data analysis
Original full scan GC–MS data sets “.raw“ from Xcalibur software (version 2.2, Thermo Scientific) were converted to “.cdf“ data files by File Converter tool of Xcalibur Software to be further processed with MATLAB (The Mathworks, Inc., Natick, MA, USA). Every data matrix corresponds to one of the samples analyzed by GC–MS (control and exposed), with 2581 rows (retention times, 49 min run) and the same number of rows equal to 590 columns (m/z values, from 60 to 650 with one unit of mass resolution). Data values in every data matrix were normalized with the peak area value of the Internal Standard (d-glucose U-13C6) to correct for the instrumental intensity drifts among injections, and to scale data internally. A total of 21 individual data matrices [1 blank, 5 controls and 15 exposed (3 treatments × 5 exposed samples)] with dimensions of 2581 × 590 (retention times, m/z values) were obtained and these resulting pre-processed data matrices were analyzed by MCR-ALS.
Additionally, for initial exploration of GC–MS data, a new data set was arranged consisting in a single data matrix where every individual total ion current (TICs) Chromatogram of every control and exposed sample were arranged in the rows of this matrix. Three different single data matrices of TICs were obtained, one for each of the three investigated factors (salinity, temperature and hypoxia), with dimensions of 10 × 2581 [number of samples (5 controls and 5 exposed) × retention times], which were then submitted to PCA and PLS-DA using PLS Toolbox 7.3.1 (Eigenvector Research Inc., Wenatchee, WA, USA).
2.4.1 Initial exploration of GC–MS data
PCA (Wold et al. 1987; Esbensen and Geladi 2010) is an unsupervised technique that assumes no prior knowledge of class information and permits the unbiased comparison of the samples. This allows an initial exploration of the GC–MS TIC data comparing the behavior between control and exposed samples according to salinity, temperature and oxygen levels changes, finding patterns and detecting possible outliers in the samples. Prior to the chemometric analysis, some pretreatment methods were applied using PLS Toolbox 7.3.1 (Eigenvector Research Inc., Wenatchee, WA, USA). The first step was to align the chromatograms to compensate for possible retention time shifts between chromatographic runs using the correlation optimized warping method (Tomasi et al. 2004; Nielsen et al. 1998). After alignment, automatic weighted least squares from PLS Toolbox 7.3.1 (Eigenvector Research Inc., Wenatchee, WA, USA) was applied for automatically removing baseline offsets from TIC data matrices and variables were mean-centered also using PLS Toolbox 7.3.1 (Eigenvector Research Inc., Wenatchee, WA, USA).
Partial least squares discriminant analysis (PLS-DA) (Kalivodová et al. 2015; Wold et al. 2001) is a supervised linear regression technique in which the class for each group of samples is assigned prior to the construction of the sample scores plot, and the maximum separation of the classes (control and exposed samples) is achieved. Variable importance in projection (VIP) scores (Wold et al. 2001) were calculated to recognize the chromatographic regions of interest and to reveal the most influent variables (peak retention times related with metabolite concentration changes) discriminating controls and exposed samples.
2.4.2 MCR-ALS analysis
MCR-ALS method allows for the resolution of the pure elution and mass spectra profiles of all sample constituents in one or multiple samples analyzed by GC–MS, in a similar way to the one previously shown for LC-MS analysis (Farrés et al. 2014) or for other hyphenated chromatographic techniques (Ortiz-Villanueva et al. 2015). From the resolved elution profiles of the constituents, their peak heights or areas can be easily evaluated and compared between runs. Due to the high fragmentation of molecules observed in GC–MS and to the presence of a large number of derivatized subproducts, the proposed MCR-ALS analysis is especially helpful because it resolves and distinguishes the relevant metabolites profiles of sample constituents from the irrelevant contributions of the derivatizing signal, and therefore, it allows for the discrimination of the relevant D. magna elution profiles (metabolites) from the large number of interfering peaks (undesired derivatized compounds).
MCR analysis can be mathematically expressed with the bilinear model shown in the Eq. 1:
where C contains the chromatographic profiles of all the resolved components, ST their resolved (pure) mass spectra, and E is the matrix expressing the error or the variance not explained by the model, related to background and other unknown noise contributions. Every full scan GC–MS gives a data matrix, D, which has the mass spectra at all retention times in its rows, and the chromatograms at all m/z values in its columns.
As shown previously, the solution to Eq. 1 for C and ST is ambiguous if no additional information is available, in other words, this solution contains rotational and scale freedom. This problem is usually referred as the factor analysis ambiguity problem (Tauler 2001). This problem can affect the resolution of the component profiles. Thus, exploratory information should be used as constraints during the resolution process and it can also be used to build good initial estimates of concentration and spectra profiles. Constraints such as non-negativity and others are frequently used in MCR studies (De Juan and Tauler 2007). In the case of GC–MS data, due to the very high spectral selectivity, possible ambiguities in the resolution of elution and spectral profiles are very much diminished or totally eliminated in most of the cases.
Apart from the derivatizing agent blank sample, five control and five exposed D. magna samples were obtained for every investigated treatment: salinity, temperature and hypoxia. This gives a total number of 21 samples analyzed by GC–MS [1 blank, 5 controls and 15 exposed (3 treatments × 5 exposed samples)], giving 21 individual data matrices, each of them of dimensions 2581 retention times (between 5.3 min and 49 min) and 590 m/z values (between 60 and 650 m/z values). These individual GC–MS full scan large sized data matrices were subdivided into eleven different chromatographic time sub-windows to reduce computer storage and execution time requirements. The size limits of these time subwindows were initially obtained considering a sufficient number of retention times (approximately 200 retention times) to cover complete peak clusters and to avoid peak halving. See Table 1 for the details of these eleven time windows and their retention times.
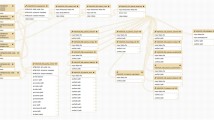
In order to analyze all these data sets, eleven new column-wise augmented data matrices D aug , corresponding to every time window containing the information of one blank derivatizing agent sample, five replicate control samples and five replicate abiotic exposed daphnids samples, were built for every type of treatment (salinity, temperature and hypoxia). A total number of thirty three (3 treatments × 11 time windows) column-wise augmented data matrices were obtained by arranging the corresponding data matrices one on top of each other, containing in all cases the same number of columns (590 m/z values). In this way, all the measured mass spectra were linked across all data (with common m/z values). GC–MS data matrices of a blank sample containing only the derivatizing agent were included in all analyses to determine the influence of derivatization and to describe the components influenced by it. In Fig. 1 these column-wise matrix arrangements are displayed.
GC–MS data sets for blank (1), control (5) and exposed (5) samples. Selection of time and column-wise matrix augmentation for the first time window (w1) Daug to be analyzed by MCR-ALS. Processing steps involve resolution of elution profiles (Caug) of the components present in this time window in the eleven samples [blank (1), control (5) and exposed (5)] and of their pure mass spectra (ST). Statistical evaluation of peak area changes between control and exposed samples via Student’s t test and PLS-DA. Tentative identification of potential biomarkers via NIST mass spectra matching and proposal of possible metabolic pathways involved
Every column-wise augmented data matrix, D aug , corresponding to a time subwindow and containing 11 submatrices (one blank derivatizant, D b ; five control (D 1 …D 5 ) and the five exposed samples (D 6 …D 10 ) according to a specific treatment) can be decomposed according to the bilinear model Eq. 2 (see also this matrix decomposition in Fig. 1):
MCR-ALS applied to this augmented data matrix gave the resolved augmented elution profiles of the resolved components (C aug ) for every sample and a matrix of the pure mass spectra of the resolved components (S T) for all samples.
MCR-ALS analysis requires an initial estimation of the number of components that can be obtained looking at the sizes of the singular values (Golub et al. 2000) of the investigated data matrix. Larger singular values are associated to systematic changes and lower ones to noise and minor contributions. This initial estimation can be afterwards revised according to the explained variances and to the interpretability of the recovered profiles. The number of pre-selected components should describe a sufficient amount of data variance and to include minor contributions such as background and derivatizing agents effects. Only components with reliable chromatographic peak shapes and meaningful metabolite mass spectra are finally considered. MCR-ALS requires also the use of constraints to give meaningful solutions (for instance positive elution and spectra profiles) (Tauler and Barceló 1993; Tauler 1995; De Juan et al. 2010). In this work, constraints of non-negativity to chromatographic and mass spectra profiles and spectra normalization of the pure mass spectra profiles (spectra equal height) were applied. The quality of MCR-ALS models was measured evaluating the lack of fit, which is the difference among the input data D aug and the data reproduced by MCR-ALS (C aug S T), and the percent of explained data variance (R 2).
2.4.3 Biomarkers detection and NIST identification
Resolved elution and spectra profiles can be used then to investigate potential biomarkers of the exposed effects (see Fig. 1). Two different procedures were applied to detect discriminant metabolites. First, a Student’s t test was tested within the data matrix C aug , resolved components showing significant concentration differences between groups (p value lower than 0.05) were finally considered and identified by their corresponding MS spectrum in S T. Secondly, resolved elution profiles were autoscaled to give equal relevance to all metabolites whatever is their total concentration and, focusing more in their relative concentration changes in control and exposed samples. Then, PLS-DA was applied to MCR-ALS autoscaled resolved peak areas for selecting candidate metabolites. Variable importance in projection (VIP) scores (Eriksson et al. 2006) were chosen as selection criteria. Peak areas or elution profiles showing higher VIP scores than 1 were considered discriminant metabolites between control and exposed samples. Metabolites from selected peak areas or elution profiles were identified comparing RIs (from MCR-ALS resolved elution profiles) and mass fragmentation patterns (from MCR-ALS resolved mass spectra profiles) with the standard mass spectral database of the National Institute of Standards and Technology (USA) (NIST 2014) (www.nist.gov/srd/nist1a.htm) and of the Golm metabolome database (GMD) of derivatized compounds (Kopka et al. 2005; Schauer et al. 2005). Two different values for the matching factors of experimental and theoretical mass spectra can be obtained by the NIST Mass Spectral Search 2.2 program distributed with the NIST 2014 library, a match factor for the unknown and the library spectrum (direct match), and a match factor for the unknown and the library spectrum ignoring any peaks in the unknown that are not in the library spectrum, match factor (MF) and reverse match factor (RMF), respectively. For each mass spectrum, 100 hits were retrieved. These matching factors reported are between 0 (no match) and 1000 (perfect match). As a general guide, a value of 900 or greater was considered to be a very good matching; between 800 and 900, a good match; between 700 and 800, a fair match; and less than 600 a poor or very poor match. In the calculation of MF, the experimental spectrum is used as a template, whereas for RMF, the template is the library spectrum. To increase the reliability of the identification, we have included the internal linear RI markers in the evaluation of the library hits, by injecting a saturated Alkane standard mixture from C7 to C30. An advantage of using this MCR-ALS strategy is that it allows for a better comparison of the MS spectra of the resolved peaks with those from the NIST 2014 library, because of the simultaneous use of peak elution retention times and of the presence of the GC–MS fragment ions in the resolved mass spectrum assigned to these peaks.
Finally, as shown in the workflow of Fig. 1, metabolites tentatively identified as potential biomarkers of salinity, temperature and hypoxia treatments were further characterized by Kyoto encyclopaedia of genes and genomes (KEGG) database (www.genome.jp/kegg/ligand.html) (Kanehisa and Goto 2000) to investigate the possible metabolic pathways involved.
2.5 Software
Data analyses were performed using MATLAB 2013a (The Mathworks Inc. Natick, MA, USA) and PLS Toolbox 7.3.1 (Eigenvector Research Inc., Wenatchee, WA, USA) was used for PCA, PLS-DA and variables importance in projection (VIP) scores calculations. MCR-ALS toolbox (Jaumot et al. 2015) were used in MATLAB 2012b (The Mathworks Inc. Natick, MA, USA) for resolution of metabolite profiles from full MS scan augmented data matrices. The tentative identification of the metabolites was carried out by NIST Mass Spectral Search program (version 2.2) (http://chemdata.nist.gov/mass-spc/ms-search/) distributed with NIST 2014 Mass Spectral Library.
3 Results and discussion
3.1 PCA and PLS-DA exploratory and discriminatory analysis of GC–MS TIC of D. magna control and exposed samples
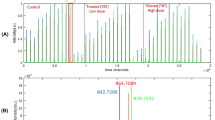
Preliminary analysis of GC–MS TIC data matrices (see Fig. 2) was performed for the three treatments (salinity, Fig. 2a, d, temperature, Fig. 2b, e, and hypoxia, Fig. 2c, f) using PCA and PLS-DA, respectively. In Fig. 2a, b, PC1-PC2 scores plot separates control (red) from exposed (blue) samples. In Fig. 2c (hypoxia treatment) PCA did not distinguish between control and exposed D. magna samples.
PCA scores plots of the GC–MS TICs of D. magna extracts for the salinity (a 5 samples), temperature (b 5 samples) and hypoxia (c 5 samples) treatments. Red are control samples (control conditions) and blue are exposed samples (salinity, temperature or hypoxia). PLS-DA VIP (variables importance in projection) scores plots for GC–MS TIC retention time variables across salinity (d), temperature (e), and hypoxia (f) treatments (Color figure online)
PLS-DA analysis was applied to the same three GC–MS TIC data matrices and VIP scores were calculated for every factor: salinity (Fig. 2d), temperature (Fig. 2e) and hypoxia (Fig. 2f). In the case of salinity a single latent variable was selected explaining 83.5 and 98.31 % of the X- and y-variances respectively. In the case of temperature, two latent variables were selected explaining 56.19 and 97.19 % of the X- and y-variances respectively, and in the case of hypoxia, three latent variables were selected to explain 46.24 and 96.18 % of the X- and y-variances respectively. In all cases, the number of latent variables used for the PLS-DA model were much lower than the number of samples used in the analyses (10 samples). According to these graphs, for salinity and temperature, a small group of retention times were selected to explain most of the variance across control and exposed samples, whereas many retention times were needed to explain the hypoxia treatment.
These two exploratory analysis (PCA and PLS-DA) showed that exposures to salinity and temperature produced larger distinctive effects on the metabolite profile of D. magna samples. However, in the case of low oxygen, effects were not so pronounced and discrimination of samples was difficult. However, although GC–MS retention times with larger VIP scores could already be preliminary investigated, their very large number and the presence of co-eluting chromatographic peak prevented determining precisely metabolite profiles of exposed D. magna samples changed from this preliminary analysis. Further analysis was then attempted using a deeper MCR-ALS analysis of the set of individual full scan GC–MS data matrices for all analyzed samples (control and exposed).
3.2 MCR-ALS analysis of GC–MS full scan data matrices of D. magna control and exposed samples
MCR-ALS investigation of full scan data matrices from GC–MS analysis of control and exposed D. magna samples allowed the simultaneous resolution of the elution and mass spectra profiles of a large number of components. Number of finally resolved peaks, number of resolved peaks in each time window augmented data matrix for each treatment, and explained variances of the MCR-ALS model are summarized in Table 1. Dimensions of the 11 time window augmented data matrices for the three treatments and their dimensions are also given in Table 1.
For all treatments, explained variance (R 2) percentages were higher than 94.8 %, and, in all cases, the number of estimated components was higher than the number of resolved peaks with reliable chromatographic shapes. However, this additional number of estimated components was useful to explain other possible experimental data variance sources such as background and derivatization signals. Examples of results obtained in the MCR-ALS analysis of the GC–MS full scan data are given in Fig. 3 where the representation of resolved concentration and mass spectra profiles for different components are given.
Examples of elution profiles (left) and of corresponding mass spectra profiles (right) resolved by MCR-ALS analysis: a applied to one single control sample in the 5th time window for the salinity treatment, showing in different colors the 8 component resolved elution profiles in this time window (left), and the corresponding mass spectra profiles (right); b applied to the salinity treatment column-wise augmented data matrix showing the resolution of one of the components in the 11 simultaneously analyzed matrices, in the order 1st blank sample, 2nd–6th control samples and 7th–11th exposed samples. And c applied to the same salinity treatment column-wise augmented data matrix than in b but showing the resolution of the derivatizing species too (Color figure online)
In Fig. 3a, results from the application of MCR-ALS analysis on time window 5 for salinity treatment are shown. In this particular case, optimal results were obtained using eight components (in different colors), some of them with highly overlapped (embedded peaks) elution profiles (on the left) and with multiple MS spectra signals (on the right). On the left of Fig. 3b, the 11 elution profiles of one of the resolved components in the different samples (1 blank, 5 control and 5 exposed samples) are given. As it is clearly shown, control samples showed considerably higher peaks than exposed samples. On the right of Fig. 3b the mass spectrum of this resolved component is given. Mass spectrum of this component gives the fragmentation pattern of a metabolite assigned to Arginine [–NH3] (2TMS) derivative according to NIST 2014 MS Spectral Search program (www.nist.gov/srd/nist1a.htm). In Fig. 3c concentration and mass spectra profiles of one component related with the derivatizing agent, MSTFA (m/z = 73) is shown. Additional profiles related to derivatized sub-products are resolved giving peak shapes with an irregular chromatographic shape. All components of this type were finally discarded for their further metabolite identification. Only those MCR-ALS resolved components showing reasonable chromatographic peak shapes (like the one in Fig. 3b) were considered for their identification and linked to D. magna metabolome.
3.3 Tentative metabolite identification
Two strategies were implemented to investigate component profiles changing with the treatments. Prior to the application of these analyses, peak areas of the same MCR-ALS component profiles in every sample and for all components and treatments were arranged in a data table for every treatment. PLS-DA was applied to these three peak area tables of MCR-ALS resolved components in control and exposed samples for each treatment (10 salinity samples × 81 peak areas, 10 temperature samples × 77 peak areas, and 10 hypoxia samples × 70 peak areas). In the case of salinity a single latent variable was selected explaining 67.53 and 96.45 % of the X- and y-variances respectively. In the case of temperature, four latent variables were selected explaining 82.28 and 98.10 % of the X- and y-variances respectively, and in the case of hypoxia, four latent variables were selected to explain 73.07 and 98.33 % of the X- and y-variances respectively. In all cases, the number of latent variables used for the PLS-DA model were much lower than the number of samples used in the analyses.
PLS-DA VIP scores provided the most important variables (resolved peaks) (see Online Resource 1) that help to discriminate between control and exposed samples, with a threshold above 1. For salinity, temperature and hypoxia 50 of the 81 resolved peaks, 27 of the 77 resolved peaks and 24 of the 70 resolved peaks, respectively (Online Resource 2), were determined as potential biomarkers (metabolites with significant concentration changes). Peak areas of MCR-ALS resolved profiles of control and exposed samples were compared using Student’s t test, which confirmed that 66, 26 and 16 peaks were changing significantly (p < 0.05) for salinity, temperature and hypoxia, respectively (Online Resource 2). Finally, only the peaks chosen by both approaches were finally selected to discard false positives. Numbers of selected biomarkers determined for each treatment agreed well with sample discrimination already observed in PCA. As with PCA, there was a good separation of samples of the control vs exposed samples for salinity and temperature treatments and a poor separation for hypoxia treatment.
Tentative identification of metabolites for salinity, temperature and hypoxia treatments using resolved mass spectra and NIST library spectra matching are shown in Online Resource 3, 4 and 5, respectively. Most of the metabolites were identified with high RMF value in their evaluation in NIST library hits. Metabolite identifications for salinity, temperature and hypoxia treatments did not take into account that the derivatization with MSTFA produced different products from the same metabolite. For instance, Lysine (3TMS) and Lysine (4TMS) (Online Resource 3) were found as biomarkers for salinity treatment, both corresponding to the same metabolite (Lysine). Finally, a total number of 74 potential metabolites whose concentrations (peak areas) changed with the treatment were identified, 40, 21 and 13 metabolites for salinity, temperature and hypoxia treatments, respectively.
For the salinity treatment (Online Resource 3) most of the 50 metabolites detected as potential biomarkers were identified with RMF values higher than 715, which can be considered a relatively fair matching factor. For most metabolites the difference between the theoretical and the experimental RI, was lower than 148, except for the resolved peak 52. This peak 52 was identified as Galactose, and it had a difference between experimental and theoretical RI of 205. However, this (Galactose) peak had a very good RMF value (927) showing a very good mass spectral coincidence with the theoretical mass values. For the salinity treatment, most of the identified metabolites were amino acids, carbohydrates, organic acids and nucleosides. Most of these metabolites were down-regulated, except 6 metabolites peak areas, 14, 55, 65, 73, 75 and 76 which were up-regulated.
For the temperature treatment (Online Resource 4), the identification of the 25 metabolites was reached with RMF values higher than 747, considered as fair matches, and with differences between theoretical and experimental RI lower than 52. The theoretical RI value of the resolved peak 76 could not be calculated because of the n-alkane standard only permitted to adjust up to a value of RI of 3000, which corresponds to the alkane C30. Carbohydrates, amino acids and organic acids were the most commonly detected metabolites for changes in the temperature. Only two metabolites were up-regulated: 1,3-diaminopropane and putrescine.
Twelve of the 13 profiles with peak areas changing with the hypoxia treatment were identified (Online Resource 5). This identification of metabolites were reached with a difference of theoretical and experimental RI lower than 34, and with a RMF value higher than 826, considered as good matches. Carbohydrates, amino acids and organic acids were the most commonly detected metabolites. All biomarkers detected were down-regulated, except resolved peak number 17 which was up-regulated.
3.4 Metabolic pathways involved
After identifying potential biomarkers of salinity, temperature and hypoxia treatments, these metabolites were investigated in KEGG database to detect the possible affected metabolic pathways. In Fig. 4, the Venn diagram shows the metabolites identified, commons and unique, in each treatment. Salinity treatment had 24 unique metabolites identified, temperature 5, and hypoxia 6 unique metabolites. By comparing common metabolites for salinity and temperature, 11 metabolites were affected in both treatments. Temperature and hypoxia and, salinity and hypoxia had 2 metabolites in common in each pair of treatments.
Venn diagram of the metabolites detected in every treatment. Salinity factor is represented by 24, temperature by 5 and hypoxia by 6 unique metabolites, respectively. Salinity and temperature have 11 metabolites in common, whereas temperature and hypoxia, and salinity and hypoxia have only 2 metabolites coincident for each
In Table 2 specific enriched KEGG pathways affected by two or more metabolites were depicted. Metabolic pathways were sorted considering the percentage coverage of the pathway (the ratio between the altered metabolites and the total unique metabolites in the KEGG pathway database). The top three KEGG pathways represented in Table 2 coincided with three of the top five pathways described in D. magna exposed to Cd (Poynton et al. 2011). In this previous study, metabolomic changes were determined using different tissues and analytical methods: hemolymph samples and using an FT-ICR mass spectrometer, respectively. This may indicate that many of the obtained altered metabolites were unspecific and probably related with stress. The three tested treatments decreased the relative amount of two to three metabolites from the pyruvate metabolism and Cytrate or Krebs cycle, which are considered critical energy metabolic pathways under hypoxia or normoxia. Indeed individuals of D. magna, exposure to high salinity levels supress energy metabolic rates and oxygen demand (Arnér and Koivisto 1993). Increasing temperature raised up metabolic demands for oxygen in Daphnia, which causes hypoxia (Paul et al. 2004a; Paul et al. 2004b). D. magna is well adapted to hypoxia levels or anoxia, increasing its hemoglobin content and under severe conditions switching to anaerobic metabolism (Paul et al. 1998). This means that the three studied environmental stressors had in common several features related to hypoxia and as a result shared several metabolites and metabolomic pathways. Specific metabolomic effects of salinity included increased levels of glycerol and trehalose, which are related to cell osmoregulation (Diamant et al. 2001).
4 Concluding remarks
Detection and identification of Daphnia magna metabolites whose concentrations suffered changes during the exposition to the three abiotic factors studied (salinity, temperature and hypoxia) was achieved. Changes on metabolite GC–MS peak areas of controls and exposed samples were statistically assessed for their discrimination. 74 metabolites were identified as possible biomarkers of the studied effects. For salinity treatment 40 metabolites were identified, for temperature and hypoxia treaments 21 and 13 metabolites were identified, respectively. The three treatments shared effects in several metabolites related with pyruvate metabolism and Cytrate or Krebs cycle, which are linked to energy metabolic pathways under hypoxia. Apart from common metabolites altered, salinity treatment had impact in glycerol and trehalose, which are related to cell osmoregulation.
The methodology proposed in this work confirmed the usefulness of the combination of GC–MS and multivariate data analysis tools for metabolomics studies of D. magna, exposed to mild stress of salinity, temperature and hypoxia. MCR-ALS analysis is a powerful tool to resolve directly the chromatographic signals (elution and spectra profiles) of most of the constituents (metabolites) present in complex biological samples, and differentiate them from the large number of GC–MS interfering derivatizing signals.
References
Adrian, R., O’Reilly, C. M., Zagarese, H., Baines, S. B., Hessen, D. O., Keller, W., et al. (2009). Lakes as sentinels of climate change. Limnology and Oceanography, 54(6 part 2), 2283–2297.
Alier, M., Felipe, M., Hernández, I., & Tauler, R. (2010). Trilinearity and component interaction constraints in the multivariate curve resolution investigation of NO and O3 pollution in Barcelona. Analytical and Bioanalytical Chemistry, 399(6), 2015–2029. doi:10.1007/s00216-010-4458-1.
Arbona, V., Manzi, M., Ollas, C., & Gómez-Cadenas, A. (2013). Metabolomics as a Tool to Investigate Abiotic Stress Tolerance in Plants. International Journal of Molecular Sciences, 14(3), 4885.
Arnér, M., & Koivisto, S. (1993). Effects of salinity on metabolism and life history characteristics of Daphnia magna. Hydrobiologia, 259(2), 69–77. doi:10.1007/BF00008373.
Azizan, K. A., Baharum, S. N., Ressom, H. W., & Noor, N. M. (2012). GC–MS analysis and PLS-DA validation of the trimethyl silyl-derivatization techniques. American Journal of Applied Sciences, 9(7), 1124–1136.
Barata, C., & Baird, D. J. (1998). Phenotypic plasticity and constancy of life-history traits in laboratory clones of Daphnia magna straus: Effects of neonatal length. Functional Ecology, 12(3), 442–452. doi:10.1046/j.1365-2435.1998.00201.x.
Barata, C., & Baird, D. J. (2000a). Determining the ecotoxicological mode of action of chemicals from measurements made on individuals: Results from instar-based tests with Daphnia magna Straus. Aquatic Toxicology, 48(2–3), 195–209. doi:10.1016/S0166-445X(99)00038-7.
Barata, C., & Baird, D. J. (2000b). Determining the ecotoxicological mode of action of toxicants from measurements on individuals: results from short duration chronic tests with Daphnia magna Straus. Aquatic Toxicology, 48, 195–209.
Barata, C., Carlos Navarro, J., Varo, I., Carmen Riva, M., Arun, S., & Porte, C. (2005). Changes in antioxidant enzyme activities, fatty acid composition and lipid peroxidation in Daphnia magna during the aging process. Comparative Biochemistry and Physiology B: Biochemistry and Molecular Biology, 140(1), 81–90. doi:10.1016/j.cbpc.2004.09.025.
Bundy, J. G., Davey, M. P., & Viant, M. R. (2009). Environmental metabolomics: A critical review and future perspectives. Metabolomics, 5(1), 3–21. doi:10.1007/s11306-008-0152-0.
Coutant, C. C. (1990). Temperature-oxygen habitat for freshwater and coastal striped bass in a changing climate. Transactions of the American Fisheries Society, 119(2), 240–253. doi:10.1577/1548-8659.
De Juan, A., Jaumot, J., & Tauler, R. (2014). Multivariate curve resolution (MCR). Solving the mixture analysis problem. Analytical Methods, 6(14), 4964. doi:10.1039/c4ay00571f.
De Juan, A., Rutan, S. C., & Tauler, R. (2010). Two-way data analysis: multivariate curve resolution—iterative resolution methods. Comprehensive Chemometrics, 2, 325–344.
De Juan, A., & Tauler, R. (2007). Factor analysis of hyphenated chromatographic data: Exploration, resolution and quantification of multicomponent systems. Journal of Chromatography A, 1158(1–2), 184–195. doi:10.1016/j.chroma.2007.05.045.
De Meester, L., & Vanoverbeke, J. (1999). An uncoupling of male and sexual egg production leads to reduced inbreeding in the cyclical parthenogen Daphnia. Proceedings of the Royal Society B: Biological Sciences, 266(1437), 2471–2477.
De Souza, D. (2013). Detection of polar metabolites through the use of gas chromatography–mass spectrometry. In U. Roessner & D. A. Dias (Eds.), Metabolomics tools for natural product discovery: Methods in molecular biology (Vol. 1055, pp. 29–37). Totowa: Humana Press.
Diamant, S., Eliahu, N., Rosenthal, D., & Goloubinoff, P. (2001). Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. Journal of Biological Chemistry, 276(43), 39586–39591. doi:10.1074/jbc.M103081200.
Dunn, W. B., Broadhurst, D. I., Atherton, H. J., Goodacre, R., & Griffin, J. L. (2011). Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chemical Society Reviews, 40(1), 387–426.
Dunn, W., Erban, A., Weber, R. J. M., Creek, D. J., Brown, M., Breitling, R., et al. (2013). Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics, 9(Suppl.1), 44–66. doi:10.1007/s11306-012-0434-4.
Eriksson, L., Johansson, E., Kettaneh-Wold, N., Trygg, J., Wikström, C., & Wold, S. (2006). Multi- and megavariate data analysis: Part I: Basic principles and applications. Umeå: Umetrics.
Esbensen, K. H., & Geladi, P. (2010). Principal component analysis: concept, geometrical interpretation, mathematical background, algorithms, history, practice. Comprehensive Chemometrics, 2, 211–226.
Farrés, M., Piña, B., & Tauler, R. (2014). Chemometric evaluation of Saccharomyces cerevisiae metabolic profiles using LC–MS. Metabolomics, 11(1), 210–224. doi:10.1007/s11306-014-0689-z.
Fiehn, O. (2001). Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative and Functional Genomics, 2(3), 155–168. doi:10.1002/cfg.82.
Golub, G., Sølna, K., & Van Dooren, P. (2000). Computing the SVD of a general matrix product/quotient. SIAM Journal on Matrix Analysis and Applications, 22(1), 1–19. doi:10.1137/S0895479897325578.
Griffiths, W. J. (2008). Metabolomics, metabonomics, and metabolite profiling. Cambridge: Royal Society of Chemistry.
Gullberg, J., Jonsson, P., Nordstrom, A., Sjostrom, M., & Moritz, T. (2004). Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Analytical Biochemistry, 331(2), 283–295. doi:10.1016/j.ab.2004.04.037.
Harrigan, G. G., & Goodacre, R. (2003). Metabolic profiling: its role in biomarker discovery and gene function analysis. Boston: Kluwer Academic.
Ikenaka, Y., Eun, H., Ishizaka, M., & Miyabara, Y. (2006). Metabolism of pyrene by aquatic crustacean Daphnia magna. Aquatic Toxicology, 80(2), 158–165. doi:10.1016/j.aquatox.2006.08.005.
IPCC. (2014). Climate change 2013: The physical science basis: Working group I contribution to the fifth assessment report of the Intergovernmental Panel on Climate Change. New York: Cambridge University Press.
Jaumot, J., de Juan, A., & Tauler, R. (2015). MCR-ALS GUI 2.0: New features and applications. Chemometrics and Intelligent Laboratory Systems, 140, 1–12. doi:10.1016/j.chemolab.2014.10.003.
Kalivodová, A., Hron, K., Filzmoser, P., Najdekr, L., Janečková, H., & Adam, T. (2015). PLS-DA for compositional data with application to metabolomics. Journal of Chemometrics, 29(1), 21–28. doi:10.1002/cem.2657.
Kanani, H., Chrysanthopoulos, P. K., & Klapa, M. I. (2008). Standardizing GC–MS metabolomics. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 871(2), 191–201. doi:10.1016/j.jchromb.2008.04.049.
Kanani, H., & Klapa, M. I. (2007). Data correction strategy for metabolomics analysis using gas chromatography-mass spectrometry. Metabolic Engineering, 9(1), 39–51. doi:10.1016/j.ymben.2006.08.001.
Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research, 28(1), 27–30. doi:10.1093/nar/28.1.27.
Kopka, J., Schauer, N., Krueger, S., Birkemeyer, C., Usadel, B., Bergmuller, E., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics, 21(8), 1635–1638. doi:10.1093/bioinformatics/bti236.
Krastanov, A. (2010). Metabolomics—the state of art. Biotechnology and Biotechnological Equipment, 24(1), 1537–1543. doi:10.2478/V10133-010-0001-y.
Lindon, J. C., & Nicholson, J. K. (2008a). Analytical technologies for metabonomics and metabolomics, and multi-omic information recovery. TrAC Trends in Analytical Chemistry, 27(3), 194–204. doi:10.1016/j.trac.2007.08.009.
Lindon, J. C., & Nicholson, J. K. (2008b). Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Annual Review of Analytical Chemistry, 1(1), 45–69. doi:10.1146/annurev.anchem.1.031207.113026.
Lindon, J. C., Nicholson, J. K., & Holmes, E. (2007). The handbook of metabonomics and metabolomics. Oxford: Elsevier.
Little, J. L. (1999). Artifacts in trimethylsilyl derivatization reactions and ways to avoid them. Journal of Chromatography A, 844(1–2), 1–22. doi:10.1016/S0021-9673(99)00267-8.
Malik, A., & Tauler, R. (2015). Exploring the interaction between O3 and NOx pollution patterns in the atmosphere of Barcelona, Spain using the MCR-ALS method. Science of the Total Environment, 517, 151–161. doi:10.1016/j.scitotenv.2015.01.105.
Martins, J., Oliva Teles, L., & Vasconcelos, V. (2007). Assays with Daphnia magna and Danio rerio as alert systems in aquatic toxicology. Environment International, 33(3), 414–425. doi:10.1016/j.envint.2006.12.006.
Miller, M. G. (2007). Environmental metabolomics: A SWOT analysis (strengths, weaknesses, opportunities, and threats). Journal of Proteome Research, 6(2), 540–545. doi:10.1021/pr060623x.
Morrison, N., Bearden, D., Bundy, J. G., Collette, T., Currie, F., Davey, M. P., et al. (2007). Standard reporting requirements for biological samples in metabolomics experiments: Environmental context. Metabolomics, 3(3), 203–210. doi:10.1007/s11306-007-0067-1.
Navarro-Reig, M., Jaumot, J., García-Reiriz, A., & Tauler, R. (2015). Evaluation of changes induced in rice metabolome by Cd and Cu exposure using LC-MS with XCMS and MCR-ALS data analysis strategies. Analytical and Bioanalytical Chemistry, 407(29), 8835–8847. doi:10.1007/s00216-015-9042-2.
Nielsen, N.-P. V., Carstensen, J. M., & Smedsgaard, J. (1998). Aligning of single and multiple wavelength chromatographic profiles for chemometric data analysis using correlation optimised warping. Journal of Chromatography A, 805(1–2), 17–35. doi:10.1016/S0021-9673(98)00021-1.
OECD. (2012). OECD Guidelines for the testing of chemicals. In OECD (Ed.), Daphnia magna Reproduction Test (211). Paris: OECD.
Orata, F. (2012). Derivatization reactions and reagents for gas chromatography analysis. Rijeka: INTECH Open Access Publisher.
Ortiz-Villanueva, E., Jaumot, J., Benavente, F., Piña, B., Sanz-Nebot, V., & Tauler, R. (2015). Combination of CE-MS and advanced chemometric methods for high-throughput metabolic profiling. Electrophoresis, 36(18), 2324–2335. doi:10.1002/elps.201500027.
Pacchiarotta, T., Nevedomskaya, E., Carrasco-Pancorbo, A., Deelder, A. M., & Mayboroda, O. A. (2010). Evaluation of GC-APCI/MS and GC-FID as a complementary platform. Journal of Biomolecular Techniques, 21(4), 205–213.
Parastar, H., Jalali-Heravi, M., Sereshti, H., & Mani-Varnosfaderani, A. (2012). Chromatographic fingerprint analysis of secondary metabolites in citrus fruits peels using gas chromatography-mass spectrometry combined with advanced chemometric methods. Journal of Chromatography A, 1251, 176–187. doi:10.1016/j.chroma.2012.06.011.
Paul, R. J., Colmorgen, M., Pirow, R., Chen, Y. H., & Tsai, M. C. (1998). Systemic and metabolic responses in Daphnia magna to anoxia. Comparative Biochemistry and Physiology A: Molecular and Integrative Physiology, 120(3), 519–530. doi:10.1016/S1095-6433(98)10062-4.
Paul, R. J., Lamkemeyer, T., Maurer, J., Pinkhaus, O., Pirow, R., Seidl, M., et al. (2004a). Thermal acclimation in the microcrustacean Daphnia: A survey of behavioural, physiological and biochemical mechanisms. Journal of Thermal Biology, 29(7–8 SPEC. ISS), 655–662. doi:10.1016/j.jtherbio.2004.08.035.
Paul, R. J., Zeis, B., Lamkemeyer, T., Seidl, M., & Pirow, R. (2004b). Control of oxygen transport in the microcrustacean Daphnia: Regulation of haemoglobin expression as central mechanism of adaptation to different oxygen and temperature conditions. Acta Physiologica Scandinavica, 182(3), 259–275. doi:10.1111/j.1365-201X.2004.01362.x.
Poynton, H. C., Taylor, N. S., Hicks, J., Colson, K., Chan, S., Clark, C., et al. (2011). Metabolomics of microliter hemolymph samples enables an improved understanding of the combined metabolic and transcriptional responses of Daphnia magna to cadmium. Environmental Science and Technology, 45(8), 3710–3717.
Samuelsson, L. M., & Larsson, D. G. J. (2008). Contributions from metabolomics to fish research. Molecular BioSystems, 4(10), 974–979. doi:10.1039/b804196b.
Sardans, J., Peñuelas, J., & Rivas-Ubach, A. (2011). Ecological metabolomics: overview of current developments and future challenges. Chemoecology, 21(4), 191–225. doi:10.1007/s00049-011-0083-5.
Schauer, N., Steinhauser, D., Strelkov, S., Schomburg, D., Allison, G., Moritz, T., et al. (2005). GC–MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters, 579(6), 1332–1337. doi:10.1016/j.febslet.2005.01.029.
Steinhauser, D., & Kopka, J. (2007). Methods, applications and concepts of metabolite profiling: primary metabolism. EXS, 97, 171–194.
Strehmel, N., Hummel, J., Erban, A., Strassburg, K., & Kopka, J. (2008). Retention index thresholds for compound matching in GC–MS metabolite profiling. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 871(2), 182–190. doi:10.1016/j.jchromb.2008.04.042.
Tauler, R. (1995). Multivariate curve resolution applied to second order data. Chemometrics and Intelligent Laboratory Systems, 30(1), 133–146. doi:10.1016/0169-7439(95)00047-X.
Tauler, R. (2001). Calculation of maximum and minimum band boundaries of feasible solutions for species profiles obtained by multivariate curve resolution. Journal of Chemometrics, 15(8), 627–646. doi:10.1002/cem.654.
Tauler, R., & Barceló, D. (1993). Multivariate curve resolution applied to liquid chromatography-diode array detection. Trends in Analytical Chemistry, 12(8), 319–327. doi:10.1016/0165-9936(93)88015-W.
Terrado, M., Barceló, D., & Tauler, R. (2009). Quality assessment of the multivariate curve resolution alternating least squares method for the investigation of environmental pollution patterns in surface water. Environmental Science and Technology, 43(14), 5321–5326. doi:10.1021/es803333s.
Tomasi, G., Van Den Berg, F., & Andersson, C. (2004). Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. Journal of Chemometrics, 18(5), 231–241. doi:10.1002/cem.859.
van Stokkum, I. H. M., Mullen, K. M., & Mihaleva, V. V. (2009). Global analysis of multiple gas chromatography–mass spectrometry (GC/MS) data sets: A method for resolution of co-eluting components with comparison to MCR-ALS. Chemometrics and Intelligent Laboratory Systems, 95(2), 150–163. doi:10.1016/j.chemolab.2008.10.004.
Vandenbrouck, T., Jones, O. A., Dom, N., Griffin, J. L., & De Coen, W. (2010). Mixtures of similarly acting compounds in Daphnia magna: From gene to metabolite and beyond. Environment International, 36(3), 254–268. doi:10.1016/j.envint.2009.12.006.
Viant, M. R. (2008). Recent developments in environmental metabolomics. Molecular BioSystems, 4(10), 980–986. doi:10.1039/b805354e.
Villas-Boas, S. G., Smart, K. F., Sivakumaran, S., & Lane, G. A. (2011). Alkylation or Silylation for Analysis of Amino and Non-Amino Organic Acids by GC–MS? Metabolites, 1(1), 3–20. doi:10.3390/metabo1010003.
Wehrens, R., Weingart, G., & Mattivi, F. (2014). MetaMS: An open-source pipeline for GC–MS-based untargeted metabolomics. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 966, 109–116. doi:10.1016/j.jchromb.2014.02.051.
Wold, S., Esbensen, K., & Geladi, P. (1987). Principal component analysis. Chemometrics and Intelligent Laboratory Systems, 2(1–3), 37–52. doi:10.1016/0169-7439(87)80084-9.
Wold, S., Sjöström, M., & Eriksson, L. (2001). PLS-regression: A basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems, 58(2), 109–130. doi:10.1016/S0169-7439(01)00155-1.
Yi, L., Shi, S., Yi, Z., He, R., Lu, H., & Liang, Y. (2014). MeOx-TMS derivatization for GC–MS metabolic profiling of urine and application in the discrimination between normal C57BL/6 J and type 2 diabetic KK-Ay mice. Analytical Methods, 6(12), 4380. doi:10.1039/c3ay41522h.
Acknowledgments
The research leading to these results received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013) / ERC Grant Agreement No. 320737.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Elba Garreta-Lara, Bruno Campos, Carlos Barata, Silvia Lacorte and Romà Tauler declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Garreta-Lara, E., Campos, B., Barata, C. et al. Metabolic profiling of Daphnia magna exposed to environmental stressors by GC–MS and chemometric tools. Metabolomics 12, 86 (2016). https://doi.org/10.1007/s11306-016-1021-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-016-1021-x