Abstract
Genomic selection methods are particularly useful for traits that are difficult or expensive to measure. We investigated the impact of using predictor growth traits and/or genomic information to increase the breeding value (BV) predictive accuracies for target scarcely recorded wood quality traits in an open-pollinated Eucalyptus grandis population. The performance of single- and multiple-trait single-step genomic best linear unbiased prediction and conventional pedigree-based models were compared in terms of the predictive accuracies (PA) of estimated BV for the target traits. We also derived the contributions of the BV for candidate trees to better understand our results. The inclusion of predictor traits in both, the training and the validation sets, together with genomic information, improved the PA (up to 17.7%) for pulp yield and cellulose. However, significant improvements in PA were not observed when predictor traits were recorded only in the training set or when the impact of genomic information alone was assessed. Changes in the PA were explained by the variations in the maternal contributions, contribution/s from all the predictor/s trait/s, and from genotyped trees. We conclude that there is not a “universal” rule regarding the use of genomic information and records on predictor traits. However, assessing the contributions to the BV of validation trees may help to better design how to benefit from predictor traits in forest tree breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The availability of high throughput genotyping of single nucleotide polymorphism (SNP) made it possible to generate genomic-enable predictions of breeding values (BVs) by means of the genomic selection (GS) approach (Meuwissen et al. 2001). The GS employs genotyped and phenotyped individuals in a training (or reference) population to predict BVs of selection candidates in a test population that are also genotyped but not necessary phenotyped. Therefore, the GS methodology is particularly relevant for traits that are difficult or expensive to measure (e.g., Calus and Veerkamp 2011).
The efficiency of GS depends on the accuracy of predicted genomic BVs (Pszczola et al. 2013). Among other factors, such as the number of markers employed, the relatedness between the training and validation populations, and the genetic architecture of the target trait, the size of the training population significantly impacts on the accuracy of genomic predictions (Hayes et al. 2009). For instance, in a Picea abies population, Chen et al. (2018) observed that the predictive accuracy rapidly increased with increasing sizes of the training set for growth and wood quality traits. However, obtaining large training sets is unfeasible for many important traits in forest tree breeding, where cost considerations or operational problems result in a limited amount of records available. This is particularly true for wood quality traits (Apiolaza et al. 1999; Schimleck et al. 2019), branch architecture traits (Shepherd et al. 2002), novel traits related to climate-change, such as drought resistance indices (Zas et al. 2020) and water use efficiency (Baltunis et al. 2008), and chemical traits related to pest and pathogens resistance (Erbilgin 2019). For instance, in their review of non-destructive evaluation technologies for wood properties, Schimleck et al. (2019) emphasize the difficulty and time-consuming nature of measuring various wood quality traits, particularly those relevant to the economics of the pulp and paper industry, such as pulp yield.
One strategy to improve the prediction accuracies of BVs for these scarcely recorded traits has been the use of one or more predictor traits in the context of so-called multiple-trait (MT) models (Henderson and Quaas 1976; Schaeffer 1984; Mrode 2005). To be useful, predictor traits need to be easily recordable, inexpensive to measure, heritable, and most importantly, genetically correlated with the target scarcely recorded trait/s (Pszczola et al. 2013). The use of predictor trait/s in MT models opens up the possibility to assemble larger training sets (Arojju et al. 2020). Simulation studies have shown that MT genomic best linear unbiased prediction (GBLUP) models, a GS method that combines a MT model with data from marker panels (VanRaden 2008), can produce more accurate breeding value estimates for scarcely recorded traits when a predictor trait is included (Calus and Veerkamp 2011; Guo et al. 2014). These benefits have also been empirically assessed in farm animals (Pszczola et al. 2013) and several crop species (Schulthess et al. 2016; Fernandes et al. 2017; Sun et al. 2017), and trees (Lenz et al. 2020). Lenz et al. (2020) showed in Norway spruce (Picea abies (L.) Karst.) that incorporating the height-to-diameter ratio as predictor trait in the MT-GBLUP model resulted in increased accuracy for the prediction of weevil attacks, a trait that is both difficult and costly to measure.
Despite these promising results, the large size of forest tree breeding populations, which typically have thousands of progenies from multiple tested parents, hinders the implementation of the GBLUP approach on a large scale due to logistical issues and high genotyping costs (Isik 2014). Therefore, the main limitation of the MT-GBLUP method is that only records from genotyped trees can be included in the evaluation, resulting in a waste of phenotypic records of trees that has not been genotyped. In these situations, the single-step GBLUP method (ssGBLUP) has been proposed as an alternative and more practical approach (Legarra et al. 2009; Misztal et al. 2009; Aguilar et al. 2010; Christensen and Lund 2010). This approach fits data from both non-genotyped and genotyped individuals in a single round of the genetic evaluation and thus, has the benefit of including historical phenotypic data without the concerns of the availability of DNA samples. The ssGBLUP method has been employed in forest tree breeding in both single-trait (ST) (Cappa et al. 2017; Klápště et al. 2018; Thavamanikumar et al. 2020; Ukrainetz and Mansfield 2020; Quezada et al. 2022; Thumma et al. 2022) and MT (Ratcliffe et al. 2017; Cappa et al. 2018; Jurcic et al. 2021; Mphahlele et al. 2021; Callister et al. 2021) genetic evaluations.
In this study, we investigated the impact of using easily-recorded predictor traits as an inexpensive way to improve the accuracy of BVs of scarcely recorded traits in a forest tree breeding context. In particular, we assayed improvements in the predictive accuracy of BVs for scarcely recorded wood quality traits when adding phenotypic records on predictor growth traits. To investigate the impact of the addition of genomic information on the accuracy of BVs, we fitted both ssGBLUP models and the conventional pedigree-based ABLUP model. Data comes from a first-generation open-pollinated progeny trial of Eucalyptus grandis (Hill ex Maiden). Timely, we also developed and present theoretical results regarding the contributions to the BVs of candidate trees that allowed a better understanding of our results.
Materials and methods
Plant materials and phenotypic measurements
Data for this study was obtained from a progeny trial of Eucalyptus grandis (Hill ex Maiden) as part of the breeding program of the company Forestal Oriental S.A. This trial was planted following a randomized incomplete block design with eight replications and 10 incomplete blocks within replications and four trees per plot in November of 2012 in Sánchez Grande, Río Negro Department, Uruguay (32° 48' 49" S 57° 39' 40" W). This population consisted of 125 open-pollinated (OP) families, where 85 families were from a third generation of breeding cycle, ten from a second generation of breeding cycle and the remaining 30 families were introductions from the E. grandis breeding program of the Instituto Nacional de Investigación Agropecuaria (INIA, Uruguay). All living trees (3159) were measured at four years old for the following growth traits: diameter at breast height (1.3 m, DBH, [cm]) and total tree height (HT, [m]). In order to measure wood chemical and physical traits (i.e., wood quality traits), near-infrared (NIR) spectroscopy was employed on 1214 trees from 92 of the 125 OP families at age five. This represents the range of variation in growth traits (DBH and HT), with an average of 13.2 trees per family (ranging from 7 to 28). The wood quality traits were pulp yield (PY, [%]), cellulose (CEL, [%]), extractive (EXT, [%]), and wood density (WD, [kg.m-3]). A calibration model that included samples of E. grandis and E. dunnii between 7 and 11 years old for wood chemical traits (PY, CEL and EXT), and only E. grandis between 3 and 6 years old for the wood physical WD trait, was developed using a partial least-squares algorithm in the R-package “pls” (Mevik and Wehrens 2007). This method has been used as a fast method for the estimation of many wood properties using NIR spectra collected from milled wood chips (e.g., Raymond and Schimleck 2011). Prior to the analyses, all trait values were standardized (mean = zero and variance = 1). The number of phenotyped trees and summary statistics in their original scale for each studied trait are summarized in the Online Resource 1.
Molecular markers
Genomic DNA was extracted from lyophilized young leaves using the standard CTAB method (Doyle and Doyle, 1987), and quality and quantity were confirmed by agarose (1%) gel electrophoresis and spectrometry using Nanodrop equipment (Thermo Fisher Scientific, Waltham, MA, USA). The DNA samples were genotyped at Genexa - ADN Evolutivo, Inc. (Montevideo, Uruguay), with a recommended concentration of 15 ng, using the AxiomTM Eucalyptus Genotyping Array (Axiom Euc72K, https://www.thermofisher.com/order/catalog/product/551134; accessed on 28 April 2023). Notably, this array has an average reproducibility rate of 99.8%. A total sample of 548 trees originated from 40 families, with a range of 7 to 27 trees per family, were genotyped. The final marker data included 37,229 (37K) SNP markers filtered by minor allele frequency (MAF) ≥ 0.05 and Call Rate (CR) ≥ 90%. All the 548 genotyped trees were assessed for wood quality traits (i.e., PY, CEL, EXT, and WD). The total number of phenotyped trees with at least one genotyped half-sib was 1,048 (out of 3,159) (i.e., 33.2%) (see Online Resource 2 for a summary). The distribution of DBH, HT, PY, CEL, EXT, and WD traits for genotyped and non-genotyped trees is presented in Online Resource 3. Genotyped trees showed the same trait distribution as non-genotyped trees for all traits.
Pedigree validation
Pedigree errors were corrected using the available 37K SNP markers for the 548 sampled trees from the 40 OP families, as described by Muñoz et al. (2014). Specifically, pedigree validation involved comparing the expected (pedigree) and realized (molecular) genetic relationship coefficients. The G-matrix calculated using the first method proposed by VanRaden (2008) was used to obtain the realized relationships. Trees that showed relationships close to 0 with their half-siblings were identified using a customized R-script. These trees were reassigned to the maternal family in which they showed relationships close to 0.25 with all the members of the family, resulting in the reassignment of ten trees to the appropriate maternal family. In one case, the correct maternal family could not be assigned, so the mother was assigned a value of 0. In addition, eight duplicate trees were identified and removed. The Online Resource 4 shows the distribution of the realized pairwise relationship coefficients using the corrected pedigrees.
Statistical analysis
The following individual-tree mixed models were fitted:
-
(1)
Single-trait mixed model (ST) (only for target wood quality trait):
where y is the vector of original individual-tree records, β is the vector of fixed effects for genetic groups formed according to the degree of improvement (levels: third and second generations of breeding cycle, or introductions); r is the vector of random replicate effects distributed as \(\boldsymbol{r}\sim \boldsymbol{N}\left(\textbf{0},\boldsymbol{I}{\sigma}_r^2\right)\), where I is the identity matrix and \({\sigma}_r^2\) is the replicate variance; b is the vector of random incomplete block effects distributed as \(\boldsymbol{b}\sim \boldsymbol{N}\left(\textbf{0},\boldsymbol{I}{\sigma}_b^2\right)\), where \({\sigma}_b^2\) is the incomplete block within replication variance; u is a vector of random effects that represents the additive genetic effects (or breeding values) distributed as \(\boldsymbol{u}\sim \boldsymbol{N}\left(\textbf{0},\boldsymbol{A}{\sigma}_u^2\right)\) where A is the average numerator relationship matrix derived from the pedigree (Henderson 1984) and containing the additive genetic relationships among all trees: 125 mothers without records plus 3159 offspring, and \({\sigma}_a^2\) is the additive genetic variance. Finally, e is the vector of random errors distributed as \(\boldsymbol{e}\sim \boldsymbol{N}\left(\textbf{0},\boldsymbol{I}{\sigma}_e^2\right)\) where \({\sigma}_e^2\) is the error variance. The X, Zr, Zb and Zu are all incidence matrices for their respective effects.
-
(B)
Two-trait mixed model (BT) (one predictor growth trait and one target wood quality trait):
where yi and yj are the vectors of individual tree phenotypes for trait i (i = DBH or TH) and trait j (j = PY, CEL, EXT, or WD); \({\boldsymbol{\upbeta}}^{\prime }=\left[{\boldsymbol{\upbeta}}_i^{\prime },{\boldsymbol{\upbeta}}_j^{\prime}\right]\) is the vector of fixed effect of genetic groups for traits i and j; the random vector of replicate effects in \({\boldsymbol{r}}^{\prime }=\left[{\boldsymbol{r}}_i^{\prime },{\boldsymbol{r}}_j^{\prime}\right]\) is distributed as r~N(0, Σr ⨂ I), where Σr is the (co) variance matrix of replicate effects for the two traits with dimension 2 × 2; the random vector of incomplete block effects in \({\boldsymbol{b}}^{\prime }=\left[{\boldsymbol{b}}_i^{\prime },{\boldsymbol{b}}_j^{\prime}\right]\) is distributed as b~N(0, Σb ⨂ I), where Σb is the (co) variance matrix of incomplete block effects with dimension 2 × 2; the random vector of additive genetic effects in \({\boldsymbol{u}}^{\prime }=\left[{\boldsymbol{u}}_i^{\prime },{\boldsymbol{u}}_j^{\prime}\right]\) is distributed as u~N(0, Σu ⨂ A), where Σu is the (co) variance matrix of genetic effects of order 2 × 2 and A as defined above. Finally, \({\boldsymbol{e}}^{\prime }=\left[{\boldsymbol{e}}_i^{\prime },{\boldsymbol{e}}_{j}^{\prime}\right]\) is the vector of random errors distributed as e~N(0, R0 ⨂ I) where R0 is the error (co) variance matrix for the two traits with dimension 2 × 2. We assumed an unstructured (co) variance matrix for the replicate (Σr), incomplete block (Σb), genetic (Σu) and error (R0) effects. The matrices Xi and Xj, \({\boldsymbol{Z}}_{r_i}\), and \({\boldsymbol{Z}}_{r_j}\), \({\boldsymbol{Z}}_{b_i}\) and \({\boldsymbol{Z}}_{r_j}\), and \({\boldsymbol{Z}}_{u_i}\), and \({\boldsymbol{Z}}_{u_j}\), relate the phenotype to the means of the genetic group effects in β, replicate effects in r, incomplete block effects in b, and the additive genetic effects in u. The apostrophe indicates the transpose operation.
-
(C)
Three-trait mixed model (TT) (both predictor growth traits and one target wood quality trait):
where yDBH, yHT and yj are the vectors of individual tree phenotypes for the growth traits DBH and HT and the jth target wood quality trait (j = PY, CEL, EXT, or WD); \({\boldsymbol{\upbeta}}^{\prime }=\left[{\boldsymbol{\upbeta}}_{DBH}^{\prime },{\boldsymbol{\upbeta}}_{HT}^{\prime },{\boldsymbol{\upbeta}}_j^{\prime}\right]\) is the vector of fixed effects genetic groups for the growth trait DBH and HT, and the wood quality trait j; the random vector of replicate effects in \({\boldsymbol{r}}^{\prime }=\left[{\boldsymbol{r}}_{DBH}^{\prime },{\boldsymbol{r}}_{HT}^{\prime },{\boldsymbol{r}}_j^{\prime}\right]\) is distributed as r~N(0, Σr ⨂ I), where Σr is the (co) variance matrix of replicate effects for the three traits with dimension 3 × 3; the random vector of incomplete block effects in \({\boldsymbol{b}}^{\prime }=\left[{\boldsymbol{b}}_{DBH}^{\prime },{\boldsymbol{b}}_{HT}^{\prime },{\boldsymbol{b}}_j^{\prime}\right]\) is distributed as b~N(0, Σb ⨂ I), where Σb is the (co) variance matrix of incomplete block effects with dimension 3 × 3; the random vector of additive genetic effects in \({\boldsymbol{u}}^{\prime }=\left[{\boldsymbol{u}}_{DBH}^{\prime },{\boldsymbol{u}}_{HT}^{\prime },{\boldsymbol{u}}_j^{\prime}\right]\) is distributed as u~N(0, Σu ⨂ A), where Σu is the (co) variance matrix of genetic effects of order 3 × 3 and A as defined above. Finally, \({\boldsymbol{e}}^{\prime }=\left[{\boldsymbol{e}}_{DBH}^{\prime },{\boldsymbol{e}}_{HT}^{\prime },{\boldsymbol{e}}_j^{\prime}\right]\) is the vector of random errors distributed as e~N(0, R0 ⨂ I) where R0 is the error (co) variance matrix for the three traits with dimension 3 × 3. We assumed an unstructured (co) variance matrix for the replicate (Σr), incomplete block (Σb), genetic (Σu) and error (R0) effects. The matrices XDBH, XHT and Xj, \({\boldsymbol{Z}}_{r_{DBH}}\), \({\boldsymbol{Z}}_{r_{HT}}\) and \({\boldsymbol{Z}}_{r_j}\), \({\boldsymbol{Z}}_{b_{DBH}}\), \({\boldsymbol{Z}}_{b_{HT}}\) and \({\boldsymbol{Z}}_{r_j}\), and \({\boldsymbol{Z}}_{u_{DBH}}\), \({\boldsymbol{Z}}_{u_{HT}}\) and \({\boldsymbol{Z}}_{u_j}\), relate the observation to the means of the genetic group effects in β, replicate effects in r, incomplete block effects in b, and the additive genetic effects in u.
In order to fit the ssGBLUP models, the pedigree-based relationship A-matrix of ST models [1], and MT models [2] and [3] were replaced by the combined pedigree- and marker-based relationship H-matrix, of the same dimension as the A-matrix. Actually, only the inverse of H is needed to fit the ssGBLUP models. Therefore, the inverse of the H-matrix (H−1) was obtained as follows (Legarra et al. 2009; Misztal et al. 2009; Aguilar et al. 2010; Christensen and Lund 2010):
where λ scales the differences between genomic and pedigree-based information, G−1 is the inverse of the genomic relationship matrix, and \({\boldsymbol{A}}_{22}^{-1}\) is the inverse of the pedigree-based relationship matrix for the genotyped individuals. In all our analyses, the scale parameter was set to λ = 0.95. Based on the recent work of Jurcic et al. (2021), an identity by descent (IBD) measure was used to calculate the G-matrix (Han and Abney 2013) implemented in the IBDLD v3.14 software (Han and Abney 2011), using the “GIBDLD” option (see details in Jurcic et al. (2021)). G-matrix was scaled to have the same diagonal and off-diagonal averages as the corresponding A-matrix, as previously described Christensen et al. (2012) (eq. (4)).
The narrow-sense heritability \({\hat{h}}^2\) for each trait was estimated as:
where \({\hat{\sigma}}_{u_i}^2\) is the estimated additive genetic variance for the trait i, and \({\hat{\sigma}}_{e_i}^2\) the estimated error variance for the trait i from the three-trait ssGBLUP models (eq. (3)). The genetic \(\left({\hat{r}}_{A_{ij}}\right)\) and residual \(\left({\hat{r}}_{R_{ij}}\right)\) correlations between traits i and j were estimated as:
\({\hat{r}}_{A_{ij}}=\frac{{\hat{\sigma}}_{u_{ij}}}{\sqrt{{\hat{\sigma}}_{u_{ii}}\times {\hat{\sigma}}_{u_{jj}}}}\) ; \({\hat{r}}_{R_{ij}}=\frac{{\hat{\sigma}}_{e_{ij}}}{\sqrt{{\hat{\sigma}}_{e_{ii}}\times {\hat{\sigma}}_{e_{jj}}}}\)
where \({\hat{\sigma}}_{a_{ij}}\) and \({\hat{\sigma}}_{e_{ij}}\) are the estimated additive genetic and residual covariance between trait i and j respectively.
Variance components and their functions (heritabilities and correlations) for the ABLUP and the ssGBLUP three-trait models (eq. (3)) were estimated in R (www.r-project.org) with the function remlf90 from the package ‘breedR’ (Muñoz and Sanchez 2020), that uses the Expectation-Maximization (EM) algorithm followed by one round of an Average Information (AI) algorithm to compute the standard errors (Chateigner et al. 2020). The remlf90 function in the R-package ‘breedR’ is based on the REMLF90 (for the EM algorithm) and AIREMLF90 (for the AI algorithm) of the BLUPF90 family (Misztal et al. 2018). The program preGSf90, also from the BLUPF90 family (Misztal et al. 2018), was used to build-up the inverse of the H-matrix.
Predictive accuracy and bias of estimated breeding values
The performance of the different statistical models was evaluated by means of a 10-fold cross-validation study. This analysis involved the partition of the dataset in ten folds, where one randomly chosen subsample at a time was used as the validation set and the remaining nine as the training set (Hastie et al. 2009). In each round, the validation set comprised approximately 120 trees selected at random from the 1240 phenotyped for the wood quality traits studied. These selected trees came from almost the same number of mothers. In addition, each fold had similar number of genotyped individuals. The wood quality phenotypes of these selected trees of the validation set were hidden in each round to fit the models, and the prediction target was the estimated BVs of these trees for the wood quality traits under different scenarios regarding the availability of records (see next section). The measure of performance was the predictive accuracy (PA), calculated in each round as the Pearson correlation between the prediction target and the corresponding (and hidden) adjusted phenotypes, scaled by the square root of the narrow-sense heritability. Adjusted phenotypes were obtained for each tree and trait by subtracting the estimated replicate and incomplete block effects from the original phenotype. For the cross-validation analysis, the variance components were fixed across all the folds to the variance components obtained with all the available trees with phenotypic data from the respective three-trait ssGBLUP model.
In addition to the PA, the prediction bias (PB) was also calculated in each fold as the slope of the regression of the estimated BVs of trees in the validation set on their respective adjusted phenotype. A slope equal to one is consistent with no bias, whereas a value greater or smaller than one indicates inflated or deflated predictions, respectively.
For each trait and model, PA and PB measures were averaged over all the folds. However, as the means of each fold are highly correlated (> 0.75 for PA) across the models assayed, paired t-tests (p-value < 0.05) were used to statistically assess differences in their PA and PB (Schrauf et al. 2021). BLUPF90 family software (Misztal et al. 2018) were used for fitting the models. A customized R-script was written to automate the cross-validation analyses, and is available from the corresponding author on reasonable request.
Scenarios analyzed
Several scenarios were assayed in this study. These scenarios were generally defined in terms of phenotypic data available on predictor and target traits in the training and validation sets of the cross-validation analyses, following the methodology outlined by Pszczola et al. (2013) closely.
Training sets were defined based on the availability of phenotypic records on the wood quality traits and predictor traits. Specifically, we defined four different training sets: (1) records on only one wood quality trait (WQi, i = PY, CEL, EXT, WD) were available; (2) records on one wood quality trait and DBH as a predictor trait were available; (3) records on one wood quality trait and HT as a predictor trait were available; and (4) records on one wood quality trait and both DBH and HT as predictor traits (i.e., DBH and HT) were available.
Combined with these training sets, four validation sets were defined based on the availability of phenotypic records on predictor traits from the trees to be validated. These validation sets included: (1) records on predictor traits were not available (NA); (2) only records on DBH were available; (3) only records on HT were available; and (4) records on both DBH and HT predictor traits were available.
The combination of these four training and four validation sets gives a total of 16 scenarios. However, as in Pszczola et al. (2013), the nine most practical were analyzed in this study (Table 1 and Online Resource 5). First, we selected all those scenarios where all the phenotypic records were available in the training set irrespective of whether or not records on predictor traits were available for the validation sets. Second, we analyzed all the scenarios where the predictor traits were not available for the trees in the validation set. Finally, we excluded those scenarios where records on the predictor growth traits were available only for the trees in the validation set but not in the training set (refer to Table 1 and Online Resource 5 for details). For each of these nine scenarios, either an ABLUP or ssGBLUP model was fitted and used to estimate the breeding values of the trees in the validation set.
These nine scenarios allowed us to assess changes in predictive accuracies and bias of the estimated breeding values for the different target traits resulting from the data addition of (1) genomic information; (2) records on predictor traits from the trees in the training set; (3) records on predictor traits from trees in both, the training and the validation sets; (4) records on predictor traits from the trees in the validation set, and (5) genomic information and records on the two predictor traits in both, the training and the validation sets. Please note that in cases where additional records for the predictor growth trait/s (DBH and/or HT) were included (items 2, 3, and 5), the training sets had a larger numbers of trees.
Contributions to the breeding values of validation trees
To better understand the changes in the predictive accuracy across the different scenarios, we derived the formulas regarding the contributions to estimated breeding values of validation trees without records on the target wood quality traits. This approach has been used in animal breeding to decompose the breeding value of individuals in single-trait ABLUP (VanRaden and Wiggans 1991) and ssGBLUP (Aguilar et al. 2010; Lourenco et al. 2015; Abdollahi-Arpanahi et al. 2021) models, as well as in two-trait ABLUP models (Schaeffer 1984).
Working out the mixed model equations, VanRaden and Wiggans (1991) showed that the estimated breeding value of individual i (\({\hat{u}}_i\)) under a ST-ABLUP model can be written as:
\({\hat{u}}_i=\) w1 PAv + w2 YD + w3 PC
where PAv is the parent average, YD is the yield deviation (phenotypes adjusted for all the effects other than additive genetic effects), and PC is the progeny contribution. In turn, w1, w2 and w3 are coefficients that sum to one and weight the amount of information available. For example, the estimated breeding value of an individual that has neither been recorded for the trait nor has been progeny tested relies entirely on its parent average. Conversely, the progeny contribution is the determinant source of information for progeny tested individuals. Extensions of the formulae for MT models and ssGBLUP models can be consulted in Mrode (2005) and Lourenco et al. (2015), respectively.
Following the same approach, in the Appendix, we derived the contributions to the breeding value of a tree with only one known parent (open-pollinated tree), that has not been recorded for the target trait and that has not been progeny tested under single- and multiple-trait ABLUP and ssGBLUP models. According to the specific model, the estimated breeding value comprised the maternal contribution, the contributions of predictor traits (Schaeffer 1984), and the contributions from other genotyped trees. Explicitly, the equations for the breeding value of tree i without a record on the target wood quality trait s (s = PY, CEL, EXT, WD), \({\hat{u}}_{i_s}\), under either ST- or MT- (i.e., including the predictor growth trait/s p = DBH and/or HT) ABLUP or ssGBLUP models, are as follows:
where ST, BT, and TT stand for single-, two-, and three-trait models, respectively, \({\hat{u}}_{m_p}\) is the breeding value of the mother of individual i for the predictor trait p, and \({\beta}_{{\textrm{BT}}_{p_s}}\)and \({\beta}_{{\textrm{TT}}_{p_s}}\) are the partial regression coefficients of the target trait s on the predictor growth traits p for the BT- and TT- trait ABLUP and ssGBLUP models (Eq. (9) in the Appendix).
The partial regression coefficients were further derived following Wermuth (1992, Chapter 2) as:
where \({\sigma}_u^{p-s}\) indicate the off-diagonal entries between the predictor trait p and wood quality trait s in the inverse of the additive genetic covariance matrix \(\left({\boldsymbol{\varSigma}}_u^{-1}\right)\) and \({\sigma}_u^{ss}\) are the diagonal entries corresponding to the wood quality trait s. In turn, \({\sigma}_{u_{p-s\ or\ p}}\) indicate the covariance between the breeding values of predictor trait p and the wood quality trait s, and \({\sigma}_{u_p}^2\) is the variance of the predictor trait p from the covariance matrix for additive genetic random effects (Σu). Notice that while the partial regression coefficients of the BT models are directly proportional to the additive genetic correlations between the predictor and the target trait, this may no longer be the case for the TT models. Here, partial regression coefficients depend on all the genetic (co) variance components of the predictor and target traits.
For the ssGBLUP models, the contribution to the breeding value of the tree i in the validation set from other genotyped trees (j) under either ST (Eq. (6) in the Appendix) or MT (Eq. (12) in the Appendix) models are the following:
where hij are the off-diagonal entries of the H−1 matrix between the tree i and the remaining genotyped individuals j, and hii are the diagonal elements of the H−1 matrix corresponding to the tree i.
Results
Functions of the variance component estimates (heritability, genetic correlations, and partial regression coefficients)
Table 2 displays the estimates of narrow-sense heritability for each studied trait, the genetic and residual correlations between growth traits (DBH and HT) and wood quality traits (PY, CEL, EXT, and WD), and the partial regression coefficients obtained from the MT (three-trait) ssGBLUP model. Overall, there were clear differences in the heritability estimates between the growth traits (average 0.084) and wood quality traits (average 0.318). Predictor traits were sensibly less heritable.
Genetic correlation estimates between growth traits (DBH and HT) and wood quality traits varied from low to moderate (absolute values < 0.426) and were generally negative (average across traits −0.116; range −0.426 to 0.282), except for PY and CEL (0.038 and 0.282, respectively). PY and WD showed higher correlation with DBH than with HT (PY −0.344 vs. 0.038, respectively for DBH and HT; WD −0.426 vs. −0.073, respectively for DBH and HT), whereas CEL and EXT showed higher correlation with HT than with DBH (CEL 0.282 vs. −0.034, respectively for HT and DBH; EXT −0.273 vs. −0.100 for HT and DBH, respectively).
On average, partial regression coefficients increased (in absolute value) when switching from a two- to a three-trait model for PY and CEL traits, but these coefficients showed smaller changes for EXT and WD. This indicates that there is generally a greater increase in the magnitude of contributions from the predictor growth traits for PY and CEL traits compared to EXT and WD traits. Moreover, we observed changes in the sign of some of these partial regression coefficients when switching from a two- to a three-trait model for the EXT and WD traits, suggesting that these contributions of the predictor traits may also change the direction.
Predictive accuracy and bias of estimated breeding values
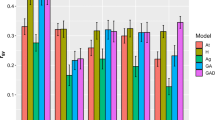
Predictive accuracies (PA) of estimated breeding values under the different scenarios studied from the pedigree-based (ABLUP) and combined pedigree-genomic (ssGBLUP) evaluation models are shown in Online Resource 6. Overall, averaging across traits and single- (ST) and multiple-trait (MT) models, ssGBLUP models showed slightly higher PA than ABLUP (0.455 vs. 0.447, respectively). These differences between ABLUP and ssGBLUP models in PA were different across all the nine scenarios and traits studied, with variations between −1.36 and 7.66% (Fig. 1). Dissecting the results across the different wood quality traits, PY showed a slightly higher relative increment in PA (average across scenarios 4.86%) when fitting the ssGBLUP model over the estimates obtained under the classical ABLUP model than the other three traits (average across scenarios: CEL 0.12%, EXT 1.58%, and WD 1.54%).
Relative performance (in percent) in predictive accuracy from the combined pedigree-genomic ssGBLUP model over the classical pedigree-based ABLUP model of trees in the validation sets under the nine different scenarios studied. Abbreviations used for the scenarios and traits are described, respectively, in the Table 1 and text. All these differences were not statistically significant (p-value > 0.05)
When records on one or two predictor traits were added only to the training set, the PA generally decreased with respect to the scenario that did not include those predictor trait/s, although in most cases non-significantly (p-values > 0.05), either under the ABLUP or the ssGBLUP model (Fig. 2 and Online Resource 6). These results varied across the wood quality traits: while EXT and WD showed the largest significant decrease (from −0.40 to −11.34%, Fig. 2), PY and CEL showed no significant change in PA (from 0.24 to 4.74%, Fig. 2). These decreases in PA varied also depending on whether DBH, HT, or both predictor traits were added to the training sets. For instance, for CEL and EXT, the highest decrease in PA was observed when the two predictor growth traits available were included jointly in the training sets. In contrast, for WD and PY, decreases in PA were largest when either DBH or HT, respectively, was included in the training sets (Fig. 2).
Relative performance (in percent) in predictive accuracy over the single-trait model of trees in the validation sets from the two- and three-trait ABLUP and ssGBLUP models when predictor traits were added only to the training sets. Abbreviations used for the scenarios and traits are described, respectively, in the Table 1 and text. Significance are noted as not statistically significant (ns; p-value > 0.05), *statistically significant 0.01 < p-value < 0.05, **statistically highly significant (p-value <0.01)
When records from predictor traits from the trees in the validation sets were also included, statistically significant increments were observed (Figs. 3 and 4 and Online Resource 6) under both ABLUP and ssGBLUP models for PY (up to 16.71%) and CEL (up to 19.23%) traits. However, non-significant differences were observed for EXT (up to 5.82%), and a significant decrease in PA was observed for WD. These results can be revealed by comparing the performance of MT against ST models, on the one hand. On the other hand, the results can be revealed by comparing the PA when predictor traits records are added on the validation sets over datasets that included the same predictor traits only in the training population, i.e., within MT models. When ST and MT models were compared to assess the impact of adding records on predictor traits in both training and validation sets, the latter showed greater PA for PY and CEL but not for EXT and WD (Fig. 3, and Online Resource 6). For instance, the maximum increment were for the CEL trait, being from 0.409 to 0.465 (13.69%, Fig. 3). When the comparison was made to assess the addition of predictor traits records on the validation sets, PA was higher for PY and CEL, showed no difference for EXT, and again decreased for WD (Fig. 4 and Online Resource 6). The maximum increment was also for the CEL trait, being for PA from 0.390 to 0.465 (19.23%, Fig. 4).
Relative performance (in percent) in predictive accuracy over the single-trait model of trees in the validation sets from the two- and three-trait ABLUP and ssGBLUP models when predictor traits were added in trees from both the training and the validation sets. Abbreviations used for the scenarios and traits are described, respectively, in the Table 1 and text. Significance are noted as not statistically significant (ns; p-value > 0.05), *statistically significant 0.01 < p-value < 0.05, **statistically highly significant (p-value <0.01)
Relative performance (in percent) in predictive accuracy over the respective (two-trait or three-trait) model with recorded growth predictor traits only in the training set of trees in the validation sets from the two- and three-trait ABLUP and ssGBLUP models when predictor traits were added in trees from the training and the validation sets. Abbreviations used for the scenarios and traits are described, respectively, in the Table 1 and text. Significance are noted as not statistically significant (ns; p-value > 0.05), *statistically significant 0.01 < p-value < 0.05, **statistically highly significant (p-value <0.01)
These increments or reductions on PA also varied across wood quality traits depending on whether one or both predictor traits were added to the training and validation sets (Fig. 3 and Fig. 4). Highest predictive accuracies for PY and CEL traits were observed when both predictor traits were added to the training and validation sets, and this was also the case for EXT when we added predictor traits on the validation sets (Fig. 4). For instance, for the trait PY under the ssGBLUP model, adding records of DBH increased the PA from 0.464 to 0.474 (2.16%, Online Resource 6 and Fig. 3), while adding records of both DBH and HT increased the PA from 0.464 to 0.521 (12.28%, Fig. 3).
Finally, we compared the three-trait ssGBLUP models to the single-trait ABLUP, i.e., when records on the predictor growth traits were added in both the training and validation sets along with genomic information. The results showed that the predictive accuracies significantly increased for PY (17.77%) and CEL (12.49%) traits. In contrast, no significant differences were found for EXT, and a strong and significant decrease (−42.56%) was observed for WD (Fig. 5 and Online Resource 6).
Relative performance (in percent) in predictive accuracy over the classical pedigree-based ABLUP single-trait model of trees in the validation sets when both recorded predictor growth traits are added in both training and validation sets and genomic information are used together. Significance are noted as not statistically significant (ns; p-value > 0.05), *statistically significant 0.01 < p-value < 0.05, **statistically highly significant (p-value <0.01)
Regarding prediction bias (PB), the regression coefficients across the nine scenarios and four target traits studied ranged from 0.744 to 1.445 (average absolute deviation from 1 was equal to 0.135) for the traditional ABLUP model, and from 0.751 to 1.419 (average absolute deviation from 1 was equal to 0.107) for the ssGBLUP model (Online Resource 7), showing slightly lower bias in the ssGBLUP models as compared to ABLUP. However, these differences between both models in the PB values were not statistically significant. When one and/or two predictor traits were added to the training set but not to the validation set, both ABLUP and ssGBLUP generally showed significantly smaller biases for all traits, with a more stringent reduction when the genomic information was available (ssGBLUP). However, when both predictor traits were added to the training and validation sets, smaller bias were obtained for PY (no significant, p-values > 0.05), EXT, and WD but not for CEL, where bias generally increased (only significant, p-values < 0.05, when we compared the ST model with the TT model) in both ABLUP and ssGBLUP models.
Discussion
The increment of the productivity and adaptability of future forest breeding populations will require recurrent selection on a great number of traits that are expensive, difficult, and/or time-demanding to record. These traits will be certainly scarcely recorded. In this context, genomic selection and multiple-trait models are becoming important tools in forest tree breeding. In this study, we investigated how the addition of predictor traits data impacts the predictive accuracy of breeding values for scarcely recorded traits. Data from an E. grandis breeding population with 548 genotyped trees and 3159 trees phenotyped for two growth traits was used to test improvements on the predictive accuracy of the estimated breeding values of four scarcely recorded wood quality traits under different scenarios. Our main findings indicated that improvements in accuracy and a reduction in bias can be generally obtained by adding records on predictor traits from candidate trees, but not always. However, the addition of genomic information did not improve the predictive accuracy significantly compared to the standard ABLUP models. To the best of our knowledge, the impact of utilizing predictor growth traits along with genomic information to enhance the accuracy of estimated breeding values for scarcely recorded wood quality traits have not been previously studied in forest tree species
Heritability and correlations
Heritability and correlation estimates for the growth and wood quality traits studied in our E. grandis population (Table 2) are in the line with those reported previously for E. grandis (Mphahlele et al. 2020) and other Eucalyptus species (Hamilton et al. 2009; Stackpole et al. 2011; Makouanzi et al. 2018). However, Harrand et al. (2009) previously reported higher heritability estimates in an E. grandis population at age 4.5, varying from 0.08 to 0.34 for diameter and from 0.09 to 0.37 for height. Additive genetic correlations for the wood quality traits studied had a relatively high standard error, likely associated with the low number of assessed trees for these wood quality traits (Table 2).
The literature suggests that beneficial scenarios for including predictor traits under multiple-trait models involve a relatively low heritable target trait that is complemented with records on an intensively recorded and genetically correlated predictor trait, which exhibits a large heritability (Schulthess et al. 2016; Sun et al. 2017). This ideal scenario was not generally met in the data used in this study, where the two predictor growth traits showed lower heritability than the target wood quality traits, and where genetic correlations ranged from low to moderate (Table 2). However, the addition of more than one predictor trait may have helped to counteract this setting in some traits. In fact, we observed an improvement in the predictive accuracy of the estimated breeding values of the target traits PY and CEL traits when we added records on both predictor traits (DBH and HT) in the datasets.
ssGBLUP vs. ABLUP
Both ABLUP and ssGBLUP models were fitted in the nine scenarios studied (Table 1 and Online Resource 5). In theory, a better predictive performance would be expected when adding genomic information (ssGBLUP models). This improved performance would be attributed to the ability of genomic models to account for, and predict, differences in the Mendelian sampling deviation among full- or half-sib family members (Ashraf et al. 2016), and depends on the connectedness between the training and validation sets (Pszczola et al. 2012). However, in our study, the ssGBLUP models did not outperformed (at least with statistical significance) the ABLUP models in terms of the predictive accuracy (PA) of the estimated breeding values (Online Resource 6 and Fig. 1). This was likely due to the relative simple half-sib family structure of the genotyped trees. Indeed, the studied population includes a large proportion of unrelated trees among the genotyped ones, resulting in small coefficients of genomic relationship between individuals in the training and validation sets.
The genotyping effort seems to be a critical factor in determining the superior performance of ssGBLUP over ABLUP, as shown in Eucalyptus (Cappa et al. 2014) and conifers (Ratcliffe et al. 2017; Ukrainetz and Mansfield 2020). In our study, even though 45.14% of the individuals analyzed for the target wood quality traits were genotyped (Online Resource 2), a relatively low proportion of trees with genomic information over the total number of trees (17.30%) was analyzed. Ukrainetz and Mansfield (2020) did not observe differences in predictive accuracy between ABLUP and ssGBLUP when the genotyping effort was 20% in growth and wood quality traits of a half-sib and full-sib Pinus contorta Dougl. ex. Loud. population. To overcome this problem, Bartholomé et al. (2016) in Pinus pinaster, suggested that the inclusion of genotypes and phenotypes of the parental population in the training set of prediction models is an important factor for improving the accuracy of the progenies and outperform the ABLUP results.
Scarcely recorded traits, such as wood quality traits and disease-related traits, have been focus of research studies in Eucalyptus species and conifers populations. However, there is considerable disagreement on whether the accuracy of breeding values from genomic models (i.e., ssGBLUP and GBLUP) increases (Tan et al. 2017; Mphahlele et al. 2020; Paludeto et al. 2021; Quezada et al. 2022), decreases (Resende et al. 2017; Cappa et al. 2019), or does no change at all (Ratcliffe et al. 2017; Lenz et al. 2020; Klápště et al. 2020; Thumma et al. 2022), compared to the accuracy values obtained from the pedigree-based ABLUP model. In a nutshell, the increase, decrease, or no change in the accuracy of breeding values from ssGBLUP and ABLUP models reported in these previous works could be due to several factors. These include (1) different forest species (Eucalypts vs. conifers) with different genome size (larger in conifers) and genetic structures (half-sib, full-sib), genotyped with different effort (Ratcliffe et al. 2017; Ukrainetz and Mansfield 2020) including or not genotyped parents (Bartholomé et al. 2016), and different effective population size (Resende et al. 2012); and (2) different analyzed traits (wood quality- and disease-related traits) with different genetic architectures (Grattapaglia and Resende 2011) and accuracy of phenotypic measurements. Moreover, the use of different validation methods used to compare the accuracy from genomic approaches to classical pedigree-based could also be a factor (Putz et al. 2018).
As discussed by Putz et al. (2018), the accuracy validation methods play a critical role in comparing alternative genetic model evaluations. In this study, the accuracies were calculated from the correlation between the estimated breeding values for target traits in the validated trees and their respective adjusted phenotypes divided by the square root of heritability. In any case, as stated by Misztal (2016), the properties of each particular validation method can be clarified analyzing the contributions to the estimated breeding values for validation trees as we did in this study.
Inclusion of predictor trait/s in the training set
In this study, we observed equal or lower performance in the predictive accuracy (PA) of breeding values of the target scarcely recorded wood quality traits when fitting the multiple-trait (MT) models (two- or three-trait) compared to the single-trait (ST) models, in the scenarios where records on predictor traits were only available in the training sets (Fig. 2). When connecting these results to the contributions to the breeding value of a target validation tree, it is important to note that when the phenotypic records of the predictor growth trait/s in the MT (two- and three-trait) models are not available for a validation tree, the factor/s that multiplies the partial regression coefficient becomes zero. For instance, in the ABLUP models, this means that the equation of the estimated breeding value of a given tree for a target wood quality trait (\({\hat{u}}_{i_s}\)) is equal, in both the ST- and MT-ABLUP models, to the contribution of its mother (\(\frac{1}{2}{\hat{u}}_{m_s}\)) (maternal contribution). As a consequence, differences observed in PA between the ST and MT models are only due to the different maternal contributions that arise from the MT models. For the ssGBLUP models, in addition to the maternal contribution, we have the contribution of the genomic information from the predictor growth traits of genotyped trees. However, our empirical dataset showed that these contributions from the genotyped trees were of low magnitude, as evidenced by the lack of differences in PA between the ABLUP and ssGBLUP models (see discussion above).
Inclusion of predictor trait/s in both, the training and validation sets
When predictor growth traits were recorded on trees of both, the training and validation sets, the performance of the MT-ABLUP and -ssGBLUP models over the ST models, in terms of PA, varied depending on the inclusion of either one or two predictor traits (Fig. 3). The inclusion of a single predictor trait in the models did not increase PA for the target wood quality traits studied, except for the target trait CEL and the growth predictor trait HT (significant increments 5.87% and 4.38% for ABLUP and ssGBLUP, respectively). Furthermore, significant decreases in the PA for the two-trait models were observed for the EXT trait with DBH and WD with DBH and with HT. These results can be attributed to the low genetic correlations between these predictor and target traits (e.g., Jia and Jannink 2012) (absolute values < 0.426, Table 2), which are proportional to the partial regression coefficients in the two-trait models (\({\beta}_{BT_{p_s}}\)) (see equations above). In other words, in the two-trait models, the covariance (i.e., the off-diagonal elements of the (co) variance matrix of genetic effects, Σu) between the predictor and target traits is the same in the numerator of the estimated genetic correlations (\({\hat{r}}_{A_{ij}}\)) and in the partial regression coefficients (\({\beta}_{BT_{p_s}}\)), and thus, both are proportional. Rambolarimanana et al. (2018) also reported a similar predictive ability between the ST approach and the two-trait approach in Eucalyptus robusta. Specifically, these authors showed that the cross-validation predictive ability of wood chemical traits (total lignin and holo-cellulose) from a two-trait models with one growth trait as predictor (volume), did not differ significantly from that achieved by the ST model. The authors explained these results by the weak genetic correlation between the growth and wood quality traits studied.
When the two predictor growth traits were jointly included in the model, significant gains in PA were observed for the target traits PY and CEL compared to the ST models, while there were no changes at all for EXT and there was a significant loss for WD (Fig. 3). The different performance of these three-trait models can be related to the change in the value of the partial regression coefficients when switching from a two-trait to a three-trait model. The magnitude of \({\beta}_{TT_{p_s}}\), which is not proportional to the genetic correlation in the case of the three-trait model, increased considerably for the target traits PY and CEL (Table 2). However, for the EXT trait, the \({\beta}_{TT_{p_s}}\) decreased for DBH (from \({\beta}_{{\textrm{BT}}_{DBH}}\) = 0.159, to \({\beta}_{{\textrm{TT}}_{DBH}}\) = -0.008, Table 2), while remained unchanged for HT (from \({\beta}_{{\textrm{BT}}_{HT}}\) = 0.504, to \({\beta}_{{\textrm{TT}}_{HT}}\) = 0.508, Table 2). For the WD trait, there were small changes in the magnitude of \({\beta}_{TT_{p_s}}\), but this partial regression coefficient changed sign for HT (from \({\beta}_{{\textrm{BT}}_{HT}}\) = 0.149 to \({\beta}_{{\textrm{TT}}_{HT}}\) = -0.188, as shown in Table 2). This change in sign could have contributed to the observed decrease in PA compared to the other scenarios analyzed (Fig. 3). These results highlight that even if the genetic covariance (or correlation) between a predictor trait and a target trait is low the inclusion of another predictor trait may change the PA.
Previous studies have already reported a lack of improvement in the predictive accuracy of the target trait breeding values when information from the predictor traits was added only to the training set. On the contrary, they reported an increased predictive accuracy when predictor growth traits were recorded on both, training and validation sets. This has been reported in animal breeding, specifically in beef (Pszczola et al. 2013) and dairy cattle (Manzanilla-Pech et al. 2020), as well as in plant breeding for crops such as sorghum (Fernandes et al. 2017; Velazco et al. 2019), barley (Bhatta et al. 2020), perennial ryegrass (Arojju et al. 2020), and wheat (Rutkoski et al. 2016; Sun et al. 2017; Lado et al. 2018; Gaire et al. 2022).
In the scenarios where predictor growth traits were included in trees from both the training and validation sets, different results were obtained from those previous works regarding the superiority of two-trait (BT) models or three-trait (TT) models over single-trait models. The TT models have been found superior to the BT models in comparison to the ST, as reported in previous studies on animal breeding (Manzanilla-Pech et al. 2020) and plant breeding (e.g., Gaire et al. 2022). However, other works concluded that if the aim is to improve the predictive accuracy of one target scarcely recorded trait, a model with just one predictor trait will be more accurate than those that incorporate more than one predictor trait (e.g., Lenz et al. 2020). Our results suggest a gain in accuracy of a particular target trait due to the inclusion of a second predictor trait in training and validation sets.
The patterns observed when comparing performance of the MT models with records on predictor traits in the training and validation sets to that of the MT models with records on predictor traits only in the training set (Fig. 4) were similar to those observed when comparing the performance of the MT models over the ST models (Fig. 3). These results are expected because when the validation trees do not have records for predictor trait/s, the contributions to the breeding values of the target traits is the same as in the ST models. However, generally the PA gains from the MT models with records of growth traits in the training and validation sets were higher when those values were compared with the MT models (Fig. 4) compared to those from the ST model (Fig. 3). This could be due to the fact that in the comparisons depicted in Fig. 4, the maternal contribution is the same for both, the trees with records on predictor traits in the training and validation sets and for the trees with records on predictor traits only in the training set, whereas in the comparisons depicted in Fig. 3, the maternal contributions are different for the ST and MT models. In addition, our results showed that these additional PA gains varied between traits (Fig. 4). We observed higher PA gains for the PY and CEL traits and no differences for EXT. Meanwhile for WD, the negative effect on PA due to the inclusion of growth traits, especially DBH, was confirmed.
Inclusion of both genomic and predictor traits in training and validation sets
To study the impact on the predictive accuracies and bias of the estimated breeding values of the different target traits when both genomic information and recorded predictor traits were added on both, the training and validation sets, results of the MT-ssGBLUP model were compared to those of the ST-ABLUP model (Fig. 5 and Online Resource 6, and Online Resource 7). Our results showed a large and significant increment in PA for PY (17.77%) and CEL (12.49%). As expected, these increments on the PA of these target wood quality traits were slightly higher than when considering separately the inclusion of genomic information through the ssGBLUP models (Fig. 1) or the addition of predictor growth traits in the training and validation sets (Fig. 3). In this study, the addition of predictor growth traits in the training and validation sets had a greater impact on the PA of the estimated breeding values of the target wood quality traits than the inclusion of genomic information through the ssGBLUP models. As we discussed before, to improve the contribution of genomic information, we think it is necessary to increase the number of genotyped trees (i.e., greater genotyping effort) and/or to include genotyped and phenotyped parents in the training set.
While our study provides promising results for the use of genomic information and predictor traits on both the training and validation sets for the PY and CEL wood quality traits in this particular population of E. grandis, no gains in PA were observed for EXT, and there were even losses for WD. Therefore, there are some limitations to consider when applying these findings to other tree species and traits. For example, as previously discussed, the effectiveness of the gain in PA in this context may depend on the relatedness between the training and validation populations, the amount of genotypes available, and the genetic (co) variance components of the predictor and target traits. Moreover, difficulties may arise in applying our approach in real-world breeding programs, including the instability of genetic correlations over time due to varying levels of emphasis placed on different traits in the selection index. However, we believe that there are opportunities to build on our findings in future research. For instance, as high-throughput field phenotyping (HTFP) platforms become widely adopted in forest genetic trials (Ludovisi et al. 2017; Solvin et al. 2020), traits collected from these HTFP platforms and genetically correlated with the scarcely recorded target trait could be considered as predictor traits to improve rates of genetic gain for the target traits in GS (Sun et al. 2017). Future research could also explore the use of other modeling approaches, such as structural equation models, to enhance the accuracy of predicted breeding values for target scarcely traits in forest tree species.
Conclusion
Our study using the E. grandis population showed that adding records on predictor growth traits in trees of both, training and validation sets, together with genomic information, improves the predictive accuracy of estimated breeding values of scarcely recorded PY and CEL traits. However, when records on predictor traits were recorded only in the training set, the accuracy showed no increase for the target wood quality traits. For this dataset, the inclusion of genomic information did not significantly increase the predictive accuracy of the studied wood quality traits. These changes in the predictive accuracies were explained by the variations in three components: maternal contributions, contribution/s from all the predictors traits, and contributions from genotyped trees. The relative weights of these contributions depend on the estimated (co) variance components. We conclude then that there is not a “universal” rule regarding the use of genomic information and records on predictor traits. Success depends on the relatedness between the training and validation populations, the amount of genotypes available, and all the genetic (co) variance components of the predictor and target traits.
References
Abdollahi-Arpanahi R, Lourenco D, Misztal I (2021) Detecting effective starting point of genomic selection by divergent trends from best linear unbiased prediction and single-step genomic best linear unbiased prediction in pigs, beef cattle, and broilers. J Anim Sci 99:skab243. https://doi.org/10.1093/jas/skab243
Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ (2010) Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci 93:743–752. https://doi.org/10.3168/jds.2009-2730
Apiolaza LA, Burdon RD, Garrick DJ (1999) Effect of univariate subsampling on the efficiency of bivariate parameter estimation and selection using half-sib progeny tests. For Genet 6:79–87
Arojju SK, Cao M, Trolove M, Barrett BA, Inch C, Eady C, Stewart A, Faville MJ (2020) Multi-trait genomic prediction improves predictive ability for dry matter yield and water-soluble carbohydrates in perennial ryegrass. Front. Plant Sci 11:1197. https://doi.org/10.3389/fpls.2020.01197
Ashraf B, Edriss V, Akdemir D, Autrique E, Bonnett D, Crossa J, Janss L, Singh R, Jannink JL (2016) Genomic prediction using phenotypes from pedigreed lines with no marker data. Crop Sci 56:957–964. https://doi.org/10.2135/cropsci2015.02.0111
Baltunis BS, Martin TA, Huber DA, Davis JM (2008) Inheritance of foliar stable carbon isotope discrimination and third-year height in Pinus taeda clones on contrasting sites in Florida and Georgia. Tree Genet Genomes 4:797–807. https://doi.org/10.1007/s11295-008-0152-2
Bartholomé J, Van Heerwaarden J, Isik F, Boury C, Vidal M, Plomion C, Bouffier L (2016) Performance of genomic prediction within and across generations in maritime pine. BMC Genom 17:604. https://doi.org/10.1186/s12864-016-2879-8
Bhatta M, Gutierrez L, Cammarota L, Cardozo F, Germán S, Gómez-Guerrero B, Pardo MF, Lanaro V, Sayas M, Castro AJ (2020) Multi-trait genomic prediction model increased the predictive ability for agronomic and malting quality traits in barley (Hordeum vulgare L.). G3 Genes Genom Genet 10:1113–1124. https://doi.org/10.1534/g3.119.400968
Callister AN, Bradshaw BP, Elms S, Gillies RA, Sasse JM, Brawner JT (2021) Single-step genomic BLUP enables joint analysis of disconnected breeding programs: an example with Eucalyptus globulus Labill. G3 Genes Genom Genet 11(10). https://doi.org/10.1093/g3journal/jkab253
Calus MPL, Veerkamp RF (2011) Accuracy of multi-trait genomic selection using different methods. Genet Sel Evol 43:26. https://doi.org/10.1186/1297-9686-43-26
Cappa EP, de Lima BM, da Silva-Junior OB, Garcia CC, Mansfield SD, Grattapaglia D (2019) Improving genomic prediction of growth and wood traits in Eucalyptus using phenotypes from non-genotyped trees by single-step GBLUP. Plant Sci 284:9–15. https://doi.org/10.1016/J.PLANTSCI.2019.03.017
Cappa EP, El-Kassaby YA, Garcia MN, Villalba PV, Klápště J, Marcucci Poltri SN (2014) Joint use of phenotypic, pedigree and genomic information in genetic evaluation: an example in Eucalyptus grandis. In: 2014 IUFRO Forest Tree Breeding Conference. Prague, Czech Republic, p 92
Cappa EP, El-Kassaby YA, Muñoz F, Garcia MN, Villalba PV, Klápště J, Marcucci Poltri SN (2017) Improving accuracy of breeding values by incorporating genomic information in spatial-competition mixed models. Mol Breed 37:125. https://doi.org/10.1007/s11032-017-0725-6
Cappa EP, El-Kassaby YA, Muñoz F, Garcia MN, Villalba PV, Klápště J, Marcucci Poltri SN (2018) Genomic-based multiple-trait evaluation in Eucalyptus grandis using dominant DArT markers. Plant Sci 271:27–33. https://doi.org/10.1016/J.PLANTSCI.2018.03.014
Chateigner A, Lesage-Descauses MC, Rogier O et al (2020) Gene expression predictions and networks in natural populations supports the omnigenic theory. BMC Genom 21:416. https://doi.org/10.1186/s12864-020-06809-2
Chen ZQ, Baison J, Pan J, Karlsson B, Andersson B, Westin J, García-Gil MR, Wu HX (2018) Accuracy of genomic selection for growth and wood quality traits in two control - pollinated progeny trials using exome capture as genotyping platform in Norway spruce. BMC Genom 19:946. https://doi.org/10.1186/s12864-018-5256-y
Christensen OF, Lund MS (2010) Genomic prediction when some animals are not genotyped. Genet Sel Evol 42:1–8. https://doi.org/10.1186/1297-9686-42-2
Christensen OF, Madsen P, Nielsen B, Ostersen T, Su G (2012) Single-step methods for genomic evaluation in pigs. Animal 6:1565–1571. https://doi.org/10.1017/S1751731112000742
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Erbilgin N (2019) Phytochemicals as mediators for host range expansion of a native invasive forest insect herbivore. New Phytol 221:1268–1278. https://doi.org/10.1111/nph.15467
Fernandes SB, Dias KOG, Ferreira DF, Brown PJ (2017) Efficiency of multi-trait, indirect, and trait-assisted genomic selection for improvement of biomass sorghum. Theor Appl Genet 131:747–755. https://doi.org/10.1007/s00122-017-3033-y
Gaire R, de Arruda MP, Mohammadi M, Brown-Guedira G, Kolb FL, Rutkoski J (2022) Multi-trait genomic selection can increase selection accuracy for deoxynivalenol accumulation resulting from fusarium head blight in wheat. Plant Genome 15:e20188. https://doi.org/10.1002/tpg2.20188
Grattapaglia D, Resende MDV (2011) Genomic selection in forest tree breeding. Tree Genet Genomes 7:241–255. https://doi.org/10.1007/s11295-010-0328-4
Guo G, Zhao F, Wang Y, Zhang Y, Du L, Su G (2014) Comparison of single-trait and multiple-trait genomic prediction models. BMC Genet 15:30. https://doi.org/10.1186/1471-2156-15-30
Hamilton MG, Raymond CA, Harwood CE, Potts BM (2009) Genetic variation in Eucalyptus nitens pulpwood and wood shrinkage traits. Tree Genet Genomes 5:307–316. https://doi.org/10.1007/s11295-008-0179-4
Han L, Abney M (2011) Identity by descent estimation with dense genome-wide genotype data. Genet Epidemiol 35:557–567. https://doi.org/10.1002/gepi.20606
Han L, Abney M (2013) Using identity by descent estimation with dense genotype data to detect positive selection. Eur J Hum Genet 21:205–211. https://doi.org/10.1038/ejhg.2012.148
Harrand L, Hernández JJV, Upton JL, Valverde GR (2009) Genetic parameters of growth traits and wood density in Eucalyptus grandis progenies planted in argentina. Silvae Genet 58:11–19
Hastie T, Tibshirani R, Friedman JH (2009) The elements of statistical learning: data mining, inference, and prediction. Springer, New York
Hayes BJ, Visscher PM, Goddard ME (2009) Increased accuracy of artificial selection by using the realized relationship matrix. Genet Res (Camb) 91:47–60. https://doi.org/10.1017/S0016672308009981
Henderson CR (1976) A simple method for computing the inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 32:69–83
Henderson CR (1984) Applications of linear models in animal breeding. University of Guelph, Guelph
Henderson CR, Quaas RL (1976) Multiple trait evaluation using relatives’ records. J Anim Sci 43:1188–1197. https://doi.org/10.2527/jas1976.4361188x
Isik F (2014) Genomic selection in forest tree breeding: the concept and an outlook to the future. New for 45:379–401. https://doi.org/10.1007/s11056-014-9422-z
Jia Y, Jannink JL (2012) Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics 192:1513–1522. https://doi.org/10.1534/genetics.112.144246
Jurcic EJ, Villalba PV, Pathauer PS et al (2021) Single-step genomic prediction of Eucalyptus dunnii using different identity-by-descent and identity-by-state relationship matrices. Heredity (Edinb) 127:176–189. https://doi.org/10.1038/s41437-021-00450-9
Klápště J, Dungey HS, Graham NJ, Telfer EJ (2020) Effect of trait’s expression level on single-step genomic evaluation of resistance to Dothistroma needle blight. BMC Plant Biol 20:205. https://doi.org/10.1186/s12870-020-02403-6
Klápště J, Suontama M, Dungey HS et al (2018) Effect of hidden relatedness on single-step genetic evaluation in an advanced open-pollinated breeding program. J Hered 109:802–810. https://doi.org/10.1093/jhered/esy051
Lado B, Vázquez D, Quincke M, Silva P, Aguilar I, Gutiérrez L (2018) Resource allocation optimization with multi-trait genomic prediction for bread wheat (Triticum aestivum L.) baking quality. Theor Appl Genet 131:2719–2731. https://doi.org/10.1007/s00122-018-3186-3
Legarra A, Aguilar I, Misztal I (2009) A relationship matrix including full pedigree and genomic information. J Dairy Sci 92:4656–4663. https://doi.org/10.3168/jds.2009-2061
Lenz PRN, Nadeau S, Mottet MJ et al (2020) Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway spruce. Evol Appl 13:76–94. https://doi.org/10.1111/eva.12823
Lourenco DAL, Fragomeni BO, Tsuruta S, Aguilar I, Zumbach B, Hawken RJ, Legarra A, Misztal I (2015) Accuracy of estimated breeding values with genomic information on males, females, or both: an example on broiler chicken. Genet Sel Evol 47:56. https://doi.org/10.1186/s12711-015-0137-1
Ludovisi R, Tauro F, Salvati R, Khoury S, Mugnozza Scarascia G, Harfouche A (2017) UAV-based thermal imaging for high-throughput field phenotyping of black poplar response to drought. Front Plant Sci 8:1681. https://doi.org/10.3389/fpls.2017.01681
Makouanzi G, Chaix G, Nourissier S, Vigneron P (2018) Genetic variability of growth and wood chemical properties in a clonal population of Eucalyptus urophylla × Eucalyptus grandis in the Congo. South For a J For Sci 80:151–158. https://doi.org/10.2989/20702620.2017.1298015
Manzanilla-Pech CIV, Gordo D, Difford GF et al (2020) Multitrait genomic prediction of methane emissions in Danish Holstein cattle. J Dairy Sci 103:9195–9206. https://doi.org/10.3168/jds.2019-17857
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mevik B-H, Wehrens R (2007) The pls Package: principal component and partial least squares regression in R. J Stat Softw 18:1–23
Misztal I (2016) Is genomic selection now a mature technology? J Anim Breed Genet 133:81–82. https://doi.org/10.1111/jbg.12209
Misztal I, Legarra A, Aguilar I (2009) Computing procedures for genetic evaluation including phenotypic, full pedigree, and genomic information. J Dairy Sci 92:4648–4655. https://doi.org/10.3168/jds.2009-2064
Misztal I, Tsuruta S, Lourenco D et al (2018) Manual for BLUPF90 family of programs. University of Georgia, Athens, USA
Mphahlele MM, Isik F, Hodge GR, Myburg AA (2021) Genomic breeding for diameter growth and tolerance to Leptocybe Gall wasp and Botryosphaeria/Teratosphaeria fungal disease complex in Eucalyptus grandis. Front Plant Sci 12:638969. https://doi.org/10.3389/fpls.2021.638969
Mphahlele MM, Isik F, Mostert-O’Neill MM, Reynolds SM, Hodge GR, Myburg AA (2020) Expected benefits of genomic selection for growth and wood quality traits in Eucalyptus grandis. Tree Genet Genomes 16:1–12. https://doi.org/10.1007/s11295-020-01443-1
Mrode R (2005) Linear models for the prediction of animal breeding values, 2nd edn. CABI Publishing, Wallingford, Oxfordshire, UK
Muñoz F, Sanchez L (2020) breedR: Statistical methods for forest genetic resources analysts. In: R package version, pp 12–14 https://github.com/famuvie/breedR
Muñoz PR, Resende MFR, Huber DA, Quesada T, Resende MD, Neale DB, Wegrzyn JL, Kirst M, Peter GF (2014) Genomic relationship matrix for correcting pedigree errors in breeding populations: Impact on genetic parameters and genomic selection accuracy. Crop Sci 54:1115–1123. https://doi.org/10.2135/cropsci2012.12.0673
Paludeto JGZ, Grattapaglia D, Estopa RA, Tambarussi EV (2021) Genomic relationship–based genetic parameters and prospects of genomic selection for growth and wood quality traits in Eucalyptus benthamii. Tree Genet Genomes 17:38. https://doi.org/10.1007/s11295-021-01516-9
Pszczola M, Strabel T, Mulder HA, Calus MPL (2012) Reliability of direct genomic values for animals with different relationships within and to the reference population. J Dairy Sci 95:389–400. https://doi.org/10.3168/jds.2011-4338
Pszczola M, Veerkamp RF, de Haas Y, Wall E, Strabel T, Calus MPL (2013) Effect of predictor traits on accuracy of genomic breeding values for feed intake based on a limited cow reference population. Animal 7:1759–1768. https://doi.org/10.1017/S175173111300150X
Putz AM, Tiezzi F, Maltecca C, Gray KA, Knauer MT (2018) A comparison of accuracy validation methods for genomic and pedigree-based predictions of swine litter size traits using Large White and simulated data. J Anim Breed Genet 135:513. https://doi.org/10.1111/jbg.12302
Quezada M, Aguilar I, Balmelli G (2022) Genomic breeding values’ prediction including populational selfing rate in an open-pollinated Eucalyptus globulus breeding population. Tree Genet Genomes 18:10. https://doi.org/10.1007/s11295-021-01534-7
Rambolarimanana T, Ramamonjisoa L, Verhaegen D, Leong Pock Tsy JM, Jacquin L, Cao-Hamadou TV, Makouanzi G, Bouvet JM (2018) Performance of multi-trait genomic selection for Eucalyptus robusta breeding program. Tree Genet Genomes 14:71. https://doi.org/10.1007/s11295-018-1286-5
Ratcliffe B, Gamal El-Dien O, Cappa EP, Porth I, Klápště J, Chen C, El-Kassaby YA (2017) Single-step BLUP with varying genotyping effort in open-pollinated Picea glauca. G3 Genes Genom Genet 7:935–942. https://doi.org/10.1534/g3.116.037895
Raymond CA, Schimleck LR (2011) Development of near infrared reflectance analysis calibrations for estimating genetic parameters for cellulose content in Eucalyptus globulus. Can J For Res 32:170–176. https://doi.org/10.1139/x01-174
Resende MDV, Resende MFR Jr, Sansaloni CP et al (2012) Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol 194:116–128. https://doi.org/10.1111/j.1469-8137.2011.04038.x
Resende RT, Resende MDV, Silva FF, Azevedo CF, Takahashi EK, Silva-Junior OB, Grattapaglia D (2017) Assessing the expected response to genomic selection of individuals and families in Eucalyptus breeding with an additive-dominant model. Heredity (Edinb) 119:245–255. https://doi.org/10.1038/hdy.2017.37
Rutkoski J, Poland J, Mondal S, Autrique E, Pérez LG, Crossa J, Reynolds M, Singh R (2016) Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3 Genes Genom Genet 6:2799–2808. https://doi.org/10.1534/g3.116.032888
Schaeffer LR (1984) Sire and cow evaluation under multiple trait models. J Dairy Sci 67:1567–1580. https://doi.org/10.3168/jds.S0022-0302(84)81479-4
Schimleck L, Dahlen J, Apiolaza LA, Downes G, Emms G, Evans R, Moore J, Pâques L, Van den Bulcke J, Wang X (2019) Non-destructive evaluation techniques and what they tell us about wood property variation. Forests 10:728. https://doi.org/10.3390/f10090728
Schrauf MF, de los Campos G, Munilla S (2021) Comparing genomic prediction models by means of cross validation. Front Plant Sci 12:2648. https://doi.org/10.3389/fpls.2021.734512
Schulthess AW, Wang Y, Miedaner T, Wilde P, Reif JC, Zhao Y (2016) Multiple-trait- and selection indices-genomic predictions for grain yield and protein content in rye for feeding purposes. Theor Appl Genet 129:273–287. https://doi.org/10.1007/s00122-015-2626-6
Shepherd M, Cross M, Dieters MJ, Henry R (2002) Branch architecture QTL for Pinus elliottii var. elliottii × Pinus caribaea var. hondurensis hybrids. Ann For Sci 59:617–625. https://doi.org/10.1051/forest:2002047
Solvin TM, Puliti S, Steffenrem A (2020) Use of UAV photogrammetric data in forest genetic trials: measuring tree height, growth, and phenology in Norway spruce (Picea abies L. Karst.). Scand J For Res 35:322–333. https://doi.org/10.1080/02827581.2020.1806350
Stackpole DJ, Vaillancourt RE, Alves A et al (2011) Genetic variation in the chemical components of Eucalyptus globulus. Wood G3(1):151–159. https://doi.org/10.1534/g3.111.000372
Sun J, Rutkoski J, Poland JA, Crossa J, Jannink JL, Sorrells ME (2017) Multitrait, random regression, or simple repeatability model in high-throughput phenotyping data improve genomic prediction for wheat grain yield. Plant Genome 10. https://doi.org/10.3835/plantgenome2016.11.0111
Tan B, Grattapaglia D, Martins GS et al (2017) Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol 17:110. https://doi.org/10.1186/s12870-017-1059-6
Thavamanikumar S, Arnold RJ, Luo J, Thumma BR (2020) Genomic studies reveal substantial dominant effects and improved genomic predictions in an open-pollinated breeding population of Eucalyptus pellita. G3 Genes GenomGenet 10:3751–3763. https://doi.org/10.1534/g3.120.401601
Thumma BR, Joyce KR, Jacobs A (2022) Genomic studies with preselected markers reveal dominance effects influencing growth traits in Eucalyptus nitens. G3 Genes Genom Genet 12:jkab363. https://doi.org/10.1093/g3journal/jkab363
Ukrainetz NK, Mansfield SD (2020) Prediction accuracy of single-step BLUP for growth and wood quality traits in the lodgepole pine breeding program in British Columbia. Tree Genet Genomes 16:1–13. https://doi.org/10.1007/s11295-020-01456-w
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423. https://doi.org/10.3168/jds.2007-0980
VanRaden PM, Wiggans GR (1991) Derivation, calculation, and use of national animal model information. J Dairy Sci 74:2737–2746. https://doi.org/10.3168/jds.S0022-0302(91)78453-1
Velazco JG, Jordan DR, Mace ES, Hunt CH, Malosetti M, Van Eeuwijk FA (2019) Genomic prediction of grain yield and drought-adaptation capacity in sorghum is enhanced by multi-trait analysis. Front Plant Sci 10:997. https://doi.org/10.3389/fpls.2019.00997
Wermuth N (1992) On block-recursive linear regression equations. Brazilian J Probab Stat 6:1–32
Zas R, Sampedro L, Solla A, Vivas M, Lombardero MJ, Alía R, Rozas V (2020) Dendroecology in common gardens: population differentiation and plasticity in resistance, recovery and resilience to extreme drought events in Pinus pinaster. Agric For Meteorol 291:108060. https://doi.org/10.1016/j.agrformet.2020.108060
Acknowledgements
The authors sincerely acknowledge to Robert Silvestre for help in assessing the trials and for assistance with field sampling. The contribution of Alexandra Simonov in processing of wood samples for the wood quality traits used in this study is also greatly acknowledged.
Funding
EPC’s research is partially supported by research grant 2019-PE-E6-I146-001 from the National Institute of Agricultural Technology (INTA). EJJ´s was supported by doctoral fellowships from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).
Author information
Authors and Affiliations
Contributions
Esteban J. Jurcic: Methodology, Software, Formal analysis, Data Curation, Writing - Original Draft, Writing - Review and Editing. Pamela V. Villalba: Resources, Writing - Review and Editing. Joaquín Dutour: Resources, Data Curation, Funding acquisition. Carmelo Centurión: Resources, Data Curation, Funding acquisition. Sebastián Munilla: Writing - Original Draft, Writing - Review and Editing. Eduardo P. Cappa: Conceptualization, Methodology, Software, Validation, Visualization, Writing - Original Draft, Writing - Review and Editing, Supervision, Project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Data archiving statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Communicated by C. Kulheim
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Contributions to the predicted breeding values of trees without records on a target trait from related trees under the single-trait and multiple-trait ABLUP and ssGBLUP models
This section describes the derivation of the equations corresponding to the contributions to the predicted breeding values of open-pollinated trees without phenotype on a target trait from related trees under the single-trait and multiple-trait pedigree-based ABLUP and combined pedigree-genomic ssGBLUP models.
Single-trait model
Let the mixed model equations (MME) for the single-trait pedigree-based ABLUP model for a single target trait (Eq. (1) in the manuscript) be:
All vectors of fixed and random effects and incidence matrices of Eq. (4) were described in the manuscript (Eq. (1)).
The equations for the tree´s predicted breeding values (\(\hat{\boldsymbol{u}}\)) from the fourth block of MME (4) are:
Operating we find out that,
For a row corresponding to a tree (i) without records on a target trait within this block, the corresponding row on the matrix \({\boldsymbol{Z}}_u^{\prime }\) is null, whereas the A−1 matrix has non-zero off-diagonals only for the individual’s mother (m) (Mrode 2005; page 46). Then, the breeding value solution for a tree i without records for any target scarcely recorded trait s (\({\hat{u}}_{i_s}\)) is:
where ami are the off-diagonal elements of the A−1 matrix corresponding to the tree i and its mother m, and aii is the diagonal element of the A−1 matrix corresponding to the tree i. Given that, the open-pollinated trees have only one known parent (the mother) and have no progeny, ami is equal to \({~}^{-2}\!\left/ \!{~}_{3}\right.\) and aii is equal to \({~}^{4}\!\left/ \!{~}_{3}\right.\) (Henderson 1976). Therefore,
For the single-trait ssGBLUP model, the inverse of A-matrix in the MME (4) (A−1) is replaced by the inverse of H-matrix (H−1) and the solutions for the predicted breeding values are:
For a row corresponding to genotyped tree i without records, the H−1 matrix has non-zero off-diagonals for the individual’s mother (m) and the remaining genotyped individuals (j). Therefore, the breeding value solution for a genotyped tree i without records for any target scarcely recorded trait s (\({\hat{u}}_{i_s}\)) may be written:
where hmi are the off-diagonal element of the H−1 matrix corresponding to the tree i and its mother m, and hii are the diagonal element of the H−1 matrix corresponding to the tree i. Since there are no genotyped mothers, the element hmi is equal to ami for all trees \(\left({h}^{mi}={a}^{mi}=-{~}^{2}\!\left/ \!{~}_{3}\right.\right)\), and the contribution of genomic information for the single-trait model [1], \({GI}_{{\textrm{ST}}_s}\), is:
Finally, the equation for the predicted breeding values of non-genotyped trees is equal to that obtained by ABLUP (Eq. (5)).
Multiple-trait models
Let the MME for the multiple-trait ABLUP model for one target trait (s) and one or more predictor (p) traits be:
We use the subscript l for all traits: l = 1, 2, … , t and the subscript p for the predictor trait/s: p = 1, 2, …, t-1. The incidence matrices \(\boldsymbol{X}=\genfrac{}{}{0pt}{}{t}{\genfrac{}{}{0pt}{}{\oplus }{l=1}}{\boldsymbol{X}}_l\), \({\boldsymbol{Z}}_r=\genfrac{}{}{0pt}{}{t}{\genfrac{}{}{0pt}{}{\oplus }{l=1}}{\boldsymbol{Z}}_{r_l}\), \({\boldsymbol{Z}}_b=\genfrac{}{}{0pt}{}{t}{\genfrac{}{}{0pt}{}{\oplus }{l=1}}{\boldsymbol{Z}}_{b_l}\), and \({\boldsymbol{Z}}_u=\genfrac{}{}{0pt}{}{t}{\genfrac{}{}{0pt}{}{\oplus }{l=1}}{\boldsymbol{Z}}_{u_l}\) relate the phenotype to the means of the genetic group fixed effects in \({\hat{\boldsymbol{\upbeta}}}^{\prime }=\left[{\hat{\boldsymbol{\upbeta}}}_1^{\prime },{\hat{\boldsymbol{\upbeta}}}_2^{\prime },\dots, {\hat{\boldsymbol{\upbeta}}}_t^{\prime}\right]\), of replicate random effects in \({\hat{\boldsymbol{r}}}^{\prime }=\left[{\hat{\boldsymbol{r}}}_1^{\prime },{\hat{\boldsymbol{r}}}_2^{\prime },\dots, {\hat{\boldsymbol{r}}}_t^{\prime}\right]\), incomplete block random effects in \({\hat{\boldsymbol{b}}}^{\prime }=\left[{\hat{\boldsymbol{b}}}_1^{\prime },{\hat{\boldsymbol{b}}}_2^{\prime },\dots, {\hat{\boldsymbol{b}}}_t^{\prime}\right]\), and additive genetic random effects in \({\hat{\boldsymbol{u}}}^{\prime }=\left[{\hat{\boldsymbol{u}}}_1^{\prime },{\hat{\boldsymbol{u}}}_2^{\prime },\dots, {\hat{\boldsymbol{u}}}_t^{\prime}\right]\), respectively. The vector of phenotypes y is equal to\(\left[{\hat{\boldsymbol{y}}}_1^{\prime },{\hat{\boldsymbol{y}}}_2^{\prime },\dots, {\hat{\boldsymbol{y}}}_t^{\prime}\right]\), and R−1 is the inverse of the error covariance matrix defined as \({\boldsymbol{R}}^{-1}={\boldsymbol{R}}_{\textbf{0}}^{-1}\bigotimes \boldsymbol{I}\) , and \({\boldsymbol{\varSigma}}_{\boldsymbol{r}}^{-\textbf{1}}\), \({\boldsymbol{\varSigma}}_{\boldsymbol{b}}^{-\textbf{1}}\) and \({\boldsymbol{\varSigma}}_u^{-1}\) are the covariance matrices of the replicate, incomplete block and additive genetic random effects, respectively, of order t x t. The symbols ⊕, ⊗and ′ indicate the direct sum of matrices notation, Kronecker product and the transpose operation, respectively.
The equations for the tree´s predicted breeding values (\(\hat{\boldsymbol{u}}\)), taken from the fourth block of MME (8), are:
For a row corresponding to tree i without records on the target trait s, each column of Zu has all elements equal to 0 except for 1´s in the row belonging the individual i with phenotype for the p predictor trait/s. In turn, the A−1 matrix has non-zero off-diagonals only for the individual’s mother (m) (Mrode 2005; page 46). Then, the breeding value solution for a tree i without records for the target trait s (\({\hat{u}}_{i_s}\)) is:
rearranging,
clearing \({\hat{u}}_{i_s}\),
or, more succinctly,
where \({\sigma}_u^{ps}\) are the off-diagonal entries of the inverse of the genetic covariance matrix \({\boldsymbol{\varSigma}}_u^{-1}\) between the predictor trait p and target trait s, and \({\sigma}_a^{ss}\) are the diagonal entries of the \({\boldsymbol{\varSigma}}_u^{-1}\) matrix corresponding to the target trait s. Given that the open-pollinated trees have only one known parent and have no progeny, ami is equal to \({~}^{-2}\!\left/ \!{~}_{3}\right.\) and aii is equal to \({~}^{4}\!\left/ \!{~}_{3}\right.\) (Henderson 1976). Therefore,
Let us write, now the partial regression coefficients of target trait s on predictor trait/s p (\({\beta}_{p_s}\)) as:
Then, the equation for the breeding value solution of a tree i without records on the target trait s from a multiple-trait ABLUP model is:
For the multiple-trait ssGBLUP model, the inverse of A-matrix in the MME (8) (A−1) is replaced by the inverse of H-matrix (H−1) and the solutions for the breeding values are:
Specifically, the breeding value solution for a genotyped tree i without records on the target trait s (\({\hat{\ u}}_{i_s}\)) is equal to:
or, more succinctly, is:
where the contribution of the genomic information under the multiple-trait ssGBLUP model, GIMT, is:
Again, the equation for the predicted breeding values of non-genotyped trees is equal to that obtained by ABLUP (Eq. (10)).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jurcic, E.J., Villalba, P.V., Dutour, J. et al. Breeding value predictive accuracy for scarcely recorded traits in a Eucalyptus grandis breeding population using genomic selection and data on predictor traits. Tree Genetics & Genomes 19, 35 (2023). https://doi.org/10.1007/s11295-023-01611-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-023-01611-z