Abstract
The ability of most opportunistic bacteria to form biofilms, coupled with antimicrobial resistance, hinder the efforts to control widespread infections, resulting in high risks of negative outcomes and economic costs. Endolysins are promising compounds that efficiently combat bacteria, including multidrug-resistant strains and biofilms, without a low probability of subsequent emergence of stable endolysin-resistant phenotypes. However, the details of antibiofilm effects of these enzymes are poorly understood. To elucidate the interactions of bacteriophage endolysins LysAm24, LysAp22, LysECD7, and LysSi3 with bacterial films formed by Gram-negative species, we estimated their composition and assessed the endolysins’ effects on the most abundant exopolymers in vitro. The obtained data suggests a pronounced efficiency of these lysins against biofilms with high (Klebsiella pneumoniae) and low (Acinetobacter baumannii) matrix contents, or dual-species biofilms, resulting in at least a twofold loss of the biomass. These peptidoglycan hydrolases interacted diversely with protective compounds of biofilms such as extracellular DNA and polyanionic carbohydrates, indicating a spectrum of biofilm-disrupting effects for bacteriolytic phage enzymes. Specifically, we detected disruption of acid exopolysaccharides by LysAp22, strong DNA-binding capacity of LysAm24, both of these interactions for LysECD7, and neither of them for LysSi3.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial biofilm (BF) is a community of immobilized and phenotypically altered microorganisms embedded in self-produced exogenous polymeric substances (EPSs) (Donlan and Costerton 2002; Karygianni et al. 2020). In particular, biofilm-related phenotype is formed by various matrix structures and chemical compositions, such as exopolysaccharides, extracellular DNA (eDNA), proteins, lipids and membrane vesicles, low molecular compounds (for example, “quorum sensing” ligands, cyclic di-GMP, accumulated water, and polyvalent ions). These heterogeneous constituents greatly impact biofilm formation, sustainability, availability of nutrients, and tolerance toward antiseptics, antibiotics and bacteriophages. Being one of the most abundant states of bacterial existence both in natural and human-made environments, BFs have great biological significance owing to efficient expansion of the community and increased survival rates under severe conditions (Donlan and Costerton 2002; Flemming et al. 2016).
Specifically, there are challenges in the therapy of diseases associated with Gram-negative opportunistic bacteria, such as Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Escherichia coli, associated with rapid emergence of multidrug-resistance traits and hardly removable biofilms formed within the host organisms and on surfaces of hospital equipment and prosthetic devices. These features together with advanced natural antibiotic resistance led to a notable increase in the probability of negative outcomes, formation of persistent infection and spread of nosocomial infections (Omar et al. 2017; Stewart 2015). Despite actively developed restrictive measures to control Gram-negative pathogens, there are limited innovative treatment approaches (Giacobbe et al. 2018; Prasad et al. 2022), while common chemotherapy is often considered as non-effective or excessively deleterious (Eriksson et al. 2022). At the same time, the search for reliable antibacterial strategies continues, thus an application of nanoparticles, antibodies, bacteriophages, bacteriophage-derived antimicrobial proteins (also called enzybiotics) and peptides (AMPs) are expected to be highly advanced options as a supplementation to antibiotic therapy or as an alternative treatment strategy (Heselpoth et al. 2021; Koo et al. 2017).
Endolysins are promising peptidoglycan-degrading enzybiotics that are already investigated for clinical relevance due to their pronounced activity and safety (Karau et al. 2023). Except for observation of physiological resistance to endolysins, likewise undergoing of bacteria to cell-wall deficient state or peptidoglycan modifications, no strict evidence of acquired resistance to these proteins has been reported, that is explained particularly by the high fitness costs of most changes in the peptidoglycan, especially across Gram-negative species. Thus, L-form conversion of bacterial cells under phage or endolysins action, chemical modifications of peptidoglycan, production of specific inhibitors, and some unspecific mechanisms could contribute to the maintenance of bacterial resistance in both Gram-positive and Gram-negative species (Grishin et al. 2020; Wohlfarth et al. 2023). Although, during the in vitro experiments, different species do not easily develop the resistance to phage endolysins (Grishin et al. 2020), in vivo the abovementioned aspects may affect the susceptibility status of bacteria.
Several studies reveal an ability of endolysins not only to eliminate multidrug resistant bacteria, but also to efficiently disrupt their biofilms (Arroyo-Moreno et al. 2022; Gutiérrez et al. 2014; Karau et al. 2023). However, the key mechanisms of endolysin-biofilm interactions are uncovered, and the investigation is mostly limited to the fact of BFs disruption and prevention of their formation. Whilst interactions between endolysins and EPSs and their effects on the enzymes’ activity are poorly described.
Our previous research was focused on recombinant endolysins, muramidases LysAm24, LysAp22, LysSi3 and endopeptidase LysECD7, derived from different bacteriophages infected Gram-negative hosts. Recently, promising safety properties and antibacterial activity of the endolysins against a broad spectrum of Gram-negative representatives were described (Antonova et al. 2019), furthermore, highlighted the substantial disruption of 24 h BFs, affected by these enzymes (Vasina et al. 2021). However, the catalytic mechanism of endolysins, associated with the hydrolysis of CW (cell-wall) peptidoglycan bonds, is not enough to explain their influence on preformed BFs, degrading complex three-dimensional structures of the matrix, down to individual bacterial cells. In the present study we investigated the composition of biofilms formed by polyresistant strains of A. baumannii and K. pneumoniae, revealing interactions between either of the endolysins and the abundant defensive compounds of preformed BFs.
Materials and methods
Bacterial strains and culture conditions
Acinetobacter baumannii Ts 50-16 and Klebsiella pneumoniae F 104-14 clinical isolates (collection of the N.F. Gamaleya Federal Research Center for Epidemiology and Microbiology, Ministry of Health of the Russian Federation), were implicated in the present study as biofilm-forming Gram-negative pathogens of the ESKAPE group. A. baumannii Ts 50-16 was isolated from patient’s sputum, intensive care unit, and K. pneumoniae F 104-14 was obtained from patient’s sputum, outpatient hospital. Both strains were stored at − 80 °C and cultivated in pre-autoclaved Tryptic Soy Broth (TSB; SIFIN, Berlin, Germany) at 37 °C, at 250 rpm overnight before performing further tests. K. pneumoniae F 104-14 genome sequencing and phenotyping showed that it is resistant to a number of antibiotics (including the resistance to third-generation cephalosporins) and refers to capsular system type K2 (contains KL2 locus), which is hypervirulent-associated (Paczosa and Mecsas 2016).
Recombinant expression and purification of proteins
Recombinant endolysins LysAm24, LysAp22, LysECD7, and LysSi3 were obtained as described previously (Vasina et al. 2021). In brief, proteins modified with histidine tag (8 His) were recombinantly expressed in Escherichia coli BL21(DE3) pLysS strain using 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) induction at 37 °C for 3 h. The cells were harvested by centrifugation (6,000 × g for 10 min at 4 °C) and incubated with 100 μg/ml lysozyme in lysis buffer (20 mM Tris–HCl, 250 mM NaCl, and 0.1 mM EDTA, pH = 8.0), and disrupted by sonication. Soluble proteins were purified on an NGC Discovery™ 10 FPLC system (Bio-Rad, Hercules, CA, U.S.) with a HisTrap FF column (GE Healthcare, Munich, Germany) pre-charged with Ni2+ ions. The filtered lysate was mixed with 30 mM imidazole and 1 mM MgCl2 and loaded on the column pre-equilibrated with binding buffer (20 mM Tris–HCl, 250 mM NaCl, and 30 mM imidazole, pH = 8.0). The fractions were eluted using a linear gradient to 100% elution buffer (20 mM Tris–HCl, 250 mM NaCl, and 500 mM imidazole, pH = 8.0). Resulting protein fractions were dialyzed against 20 mM Tris–HCl buffer (pH = 7.5).
The purity of endolysins was determined by 16% SDS-PAGE, and protein concentrations were measured using a spectrophotometer (Implen NanoPhotometer; Implen, München, Germany) at 280 nm and calculated using the predicted extinction coefficients [0.840, 0.831, 1.460, and 1.029 (mg/mL)−1 cm−1 for LysAm24, LysAp22, LysECD7, and LysSi3, respectively]. The isoelectric points of endolysins LysAm24, LysAp22, LysECD7, and LysSi3 were predicted using the internet-source http://isoelectric.org.
Biofilm composition and morphology
Macrocolony assays were conducted on solid culture medium with application of three different staining techniques and without any dye as an intact control. For the formation of BFs, cells from overnight cultures were harvested by centrifugation (6,000 × g, 10 min, RT), resuspended in PBS (pH = 7.4), and diluted to achieve a cell density of approximately 3 × 106 CFU/ml. To obtain dual-species biofilms, the appropriate bacterial suspensions were mixed in a 1:1 (v/v) ratio. Thereafter, 7 μl of the suspension was inoculated onto solid TSB agar medium on 90 mm Petri dishes and BFs were grown at 37 °C for 48 h without agitation. All images of colonies were acquired using HD automatic colony counter Scan 300 and provided software (Interscience, Saint-Nom-la-Bretèche, France).
Alcian blue 8GX 0.3% stain (PanReac AppliChem, Darmstadt, Germany) in a 4% acetic acid aqueous solution (pH = 2.5) was prepared to visualize polysaccharides with abundant carboxyl groups. Five ml of pre-filtered Alcian blue stain was carefully dripped on plates containing macrocolonies and then incubated for 15 min at RT without agitation. Subsequently, the plates were rinsed with 5 ml of deionised distilled water.
The results were confirmed by the Congo red stain absorption test (Freeman et al. 1989). For this, TSB agar was supplemented with pre-filtered 0.08% Congo red stain (Sigma, U.S.) to prepare agar plates. The 48-h old biofilms were cultured on the medium at 37 °C without agitation. The intensity of red staining was considered proportional to the quantity of extracellular polysaccharides and proteins (Ahmad et al. 2020).
The morphotyping assay was conducted according to (Römling et al. 1998) with modifications. In brief, TSB agar was supplemented with a CRCB aqueous mixture, made up of pre-filtered 0.04% Congo red stain and 0.02% Coomassie blue G-250 stain (VWR (Avantor), Radnor, U.S.) to prepare agar-containing Petri dishes. Biofilms were grown on these CRCB agar plates for 24, 48, and 144 h.
Microtiter biofilm formation assay (CV Mtp)
Overnight bacterial cultures in TSB were harvested (6,000 × g, 10 min, RT), then suspended in PBS (pH = 7.4), diluted in the buffer to achieve a cell density of approximately 3 × 106 CFU/ml. Next, 100 μl of the suspension was added to sterile wells of a 96-well polystyrene cell culture plate, and incubated for 48 h at 37 °C and 200 rpm. To obtain dual-species biofilms suspensions of both bacteria were mixed in a 1:1 (v/v) ratio. The wells’ content with planktonic cells was discarded, and the plate was washed three times with 200 μl of PBS (pH = 7.4), then air-dried for approximately 20 min. The dried BFs were stained with a 0.1% aqueous solution of Crystal violet (CV) for 15 min, at RT, followed by triple rinsing with water. The stained content was resolubilized in 200 μl of 33% acetic acid, and OD590 of the obtained solutions was measured using SPECTROstar NANO spectrophotometer (BMG LABTECH, Ortenberg, Germany). All experiments were performed with quadruplicate technical replicates and repeated at least in two independent assays. The values were normalized by dividing them by the OD590 of biofilm treated with the blank buffer. The level of biofilm formation was interpreted according to (Stepanović et al. 2007).
Quantification of eDNA in the biofilm
A measurement of eDNA was carried out with DNAse I (PanReac AppliChem, Darmstadt, Germany) treatment of biofilms, grown for 24 and 48 h. BFs were grown as described in the subsection “Microtiter biofilm formation assay” and treated with 100 μl of DNAse I solution (20 μg/ml of DNAse I in 20 mM Tris–HCl buffer, 1 mM CaCl2 (pH = 7.5)) or the same volume of the buffer at 37 °C and 200 rpm, for 2 h. Thereafter, biofilms were rinsed with water, air-dried, and stained with 0.1% CV, following the OD590 measurement. The eDNA content was calculated as the difference in BF staining with and w/o of DNAse I treatment (BF biomass reduction).
Quantification of proteins content in the biofilm
A measurement of proteins’ quantity was conducted for 24 and 48 h old biofilms. Proteinase K (PanReac Applichem, Darmstadt, Germany) in 20 mM Tris–HCl buffer supplemented with 100 mM NaCl, 1 mM CaCl2 (pH = 7.5), at 50 and 800 μg/ml concentration was added to A. baumannii biofilms; and at 50, 800 and 1600 μg/ml concentration was added to K. pneumoniae biofilms. Preformed biofilms were treated with 100 μl of Proteinase K solutions or blank buffer at 37 °C and 200 rpm, for 2 h. Thereafter, treated biofilms were rinsed, air-dried, stained with CV and analyzed as described in the subsection “Quantification of eDNA in the biofilm”.
Antibiofilm activity assessment
Mono-species and dual-species biofilms grown for 48 h and prepared for microtiter assay, were treated with either 100 μl of endolysin solutions at concentrations of 100 or 1000 μg/ml, or equal volume of 20 mM Tris–HCl (pH = 7.5) buffer as a negative control, for 2 h at 37 °C and 200 rpm. After incubation, biofilms were rinsed twice, air-dried, stained with 0.1% CV, rinsed again, dissolved and analyzed as was described in the subsection “Microtiter biofilm formation assay”.
Microscopy of biofilms
Sterile glass coverslips (Hampton Research, Aliso Viejo, CA, U.S.) were plunged into the overnight cultures, diluted in fresh TSB medium on Petri dishes and incubated at 37 °C for 48 h without shaking. The slides were then carefully washed three times with sterile distilled water and air-dried. Two slides were treated with 300 μl of 20 mM Tris–HCl (pH = 7.5) control buffer, other pairs of slides were exposed to 300 μl of 100 μg/ml LysAm24, LysAp22, LysECD7, or LysSi3 solutions for 2 h at RT. Afterward, all slides were again washed with water two times. Air-dried slides were stained with a 0.1% aqueous solution of CV for 15 min at RT. All stained samples were rinsed once with water. Then, half of the slides were immediately rinsed twice and air-dried for microscopy. Another half of the slides were negatively stained to identify acid polysaccharides, which are crucial for antibacterial tolerance in most bacterial capsules and biofilms. A negative stain was performed using the Anthony method (Hughes and Smith 2013) with modifications. Following the method, CV-stained slides were submerged into 20% aqueous CuSO4 for 10 s, rinsed thoroughly with water, and then dried. All slides were imaged using Axiostar Plus Transmitted Light Microscope (Zeiss AG, Jena, Germany) at × 630 magnification.
DNA-binding effect of the endolysins
To detect interaction between eDNA and the endolysins we obtained genomic DNA of A. baumannii using a modified CTAB method (Minas et al. 2011). Briefly, cells from an overnight bacterial culture were harvested, once washed, and solubilized in a CTAB solution (2% w/v cetrimonium bromide (Helicon, Moscow, Russia), 100 mM Tris–HCl, 20 mM EDTA and 1.4 M NaCl, pH = 8.0), ex tempore supplemented with 0.2% β-mercaptoethanol, at 65 °C for 40 min. Then, the suspensions were mixed with an equal volume of chloroform-isoamyl alcohol (24:1), and the aqueous phase was collected after centrifugation at 12,000 × g for 10 min, RT. The DNA precipitation proceeded in 0.6 volume of isopropanol overnight at – 25 °C. The precipitates were harvested at 8,000 × g for 15 min and washed three times with 80% ethanol. After ethanol was discarded, the pellets were suspended in 20 mM Tris–HCl (pH = 7.5) buffer with 1 μg/ml RNAse A (PanReac Applichem, Darmstadt, Germany), incubated for 15 min at 37 °C, afterwards RNAse A was inactivated at 65 °C for 15 min. Quality of the DNA samples was estimated spectrophotometrically, analyzing A260/230 and A260/280, thereafter a concentration of DNA was measured using Qubit DNA HS Assay Kit and Qubit 3.0 fluorometer (Thermo Fisher Scientific Eugene, Oregon, U.S.).
Endolysins LysAm24, LysAp22, LysECD7, and LysSi3 solutions in 20 mM Tris–HCl buffer at 10 or 50 ng/μl concentration were mixed with DNA solutions (at a concentration of 20 ng/μl) at a protein:DNA (P:DNA) mass ratio of 2:1 or 10:1 (final volume of the mixtures was 7.5 μl) and incubated for 30 min at RT. Solutions containing either DNA or investigated endolysin in 20 mM Tris–HCl buffer (pH = 7.5) were used as controls. The influence of electrostatic interactions was assessed by adding NaCl to final concentrations of 150 or 300 mM to the mixtures (P:DNA w/w ratio of 10:1) during their incubation. Post-incubated solutions were mixed with the 6X DNA Loading Dye (Thermo Fisher Scientific Eugene, Oregon, U.S.) and loaded in 1% agarose gel, containing 0.2 μg/ml ethidium bromide (Helicon, Moscow, Russia). An electrophoretic separation of all samples was performed in the Tris–borate buffer (50 mM Tris-base, 50 mM boric acid, 2 mM EDTA, pH = 8.3) at 80 V for an hour. Imaging was performed using the Gel Doc EZ gel documentation system (Bio-Rad, Hercules, CA, U.S.).
To correlate the binding assay with the inhibitory effect of eDNA, an antibacterial activity test was performed. An overnight bacterial culture of A. baumannii was diluted 30-fold in LB broth and grown to the exponential phase (OD600 = 0.6). Subsequently, the cells were harvested by centrifugation (6,000 × g, 10 min) and resuspended in the same volume of PBS (pH = 7.4). Each suspension was diluted in 20 mM Tris–HCl (pH = 7.5) to a final density of approximately 106 cells/mL. Afterward, 100 μl of the bacterial suspensions and 100 μL of the pre-incubated protein-DNA solutions (a final protein concentration was 10 µg/ml, a final DNA concentration was 1 or 5 µg/ml) were mixed in 96-well plates. Buffers with or without DNA were used as controls. The mixtures were incubated at 37 °C for 30 min with shaking at 200 rpm and then were diluted tenfold in PBS (pH = 7.4). Then, 100 μl of each dilution was plated onto LB agar, and the number of bacterial colonies was counted after an overnight incubation at 37 °C. All experiments were performed in quadruplicate, and the antibacterial activity was expressed as follows: Antibacterial activity (%) = 100% – (CFUexp/CFUcont) × 100%, where CFUexp is the number of bacterial colonies in the experimental culture plates, and CFUcont is the mean number of bacterial colonies in the control culture plates.
Statistical analysis
The data were analyzed and illustrated using GraphPad Prism 9.0 software. According to the results of normality tests (Kolmogorov–Smirnov’s method), datasets were compared using appropriate statistical tests with corrections for multiple comparisons (detailed information on the chosen analysis methods is provided in captions and results).
Results
Exopolysaccharides content
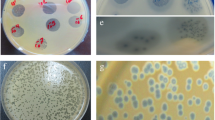
To visualize different EPSs, various staining procedures of bacterial BF-macrocolonies on nutritious media with glucose are often implemented. Cultivation of bacterial macrocolonies on solid medium, containing Congo red (CR) dye is an express test system of biofilm formation properties. CR interacts with most β-folded proteins, glucans with β (1- > 4) and β (1- > 3) bonds, and with various polycationic polysaccharides (Puchtler et al. 1962). During the 48-h incubation on CR agar, the investigated bacterial colonies accumulated small amounts of the stain and slightly decolorized the medium (Fig. 1a, the second row). The K. pneumoniae originated macrocolonies with a crystalline red center and sharp, clear edges, while A. baumannii formed homogeneous brown and rough colonies indicating production of moderate quantity of polysaccharides and amyloid proteins. Dual-species colonies resembled those of K. pneumoniae, with a rougher structure and partially colored periphery.
Macrocolony staining of K. pneumoniae and A. baumannii biofilm components. a Extracellular polymers of colonies, grown on solid media for 48 h: TS, TSB agar without staining; CR, TSB agar stained with Congo red, indicating amyloids and spectrum of carbohydrates; AlcB, TSB agar stained with Alcian blue, indicating polysaccharides in colonies’ exterior, but not proteins. b Formation of EPS-containing structures within colonies, grown on CRCB-TSB agar media for 24 and 48 h
A different matrix architecture was observed by staining with polycationic dye Alcian blue, revealing carboxyl-rich polysaccharides (Scott and Dorling 1965), located on the air-contacting exterior of formed macrocolonies (Fig. 1a, the third row). The colonies of K. pneumoniae had a concentric staining, significantly more intense compared to A. baumannii, while the dual-species colonies possessed well resolved dark-blue strands radiating from the center, revealing the conglomerated polycationic and polyanionic polysaccharides’ plots.
To investigate the coexistence of bacteria in mixed biofilms the morphotyping assay with culture media containing Coomassie brilliant blue dye (CBB) was done. CBB stains biofilm-associated proteins, and Congo red dye, binding with both proteins and polysaccharides (Nesse et al. 2020). The significant heterogeneity of biofilm composition was observed for dual-species colonies (Fig. 1b and Online Resource Fig. 1) combining phenotypic traits of both species and characterized by increased cell motility.
Quantification of eDNA
The biomass density of either K. pneumoniae or A. baumannii 24 h old biofilms pretreated with DNAse I, was reduced by 20% on average (Fig. 2), while the enzymatic treatment of 48 h biofilms led to a decline in the biofilm biomass by 30.9 and 25.9% respectively. Neither the increase in DNAse I concentration (up to tenfold) nor the time of biofilm formation led to a significant reduction in the BFs’ biomass (p > 0.05, two-way ANOVA with Tukey’s correction).
Biomass reduction of 24 and 48 h biofilms of A. baumannii a and K. pneumoniae b after treatment with 20 µg/ml DNAse I (DNAse + , white columns). Each dataset contains 4 technical and 2 biological replicates and is presented as a normalized mean value ± standard deviation (SD). Only statistically relevant differences from controls and between cohorts of distinct aged biofilms are marked as asterisks: *, p < 0,05; **, p < 0,01 (one-way ANOVA with Tukey’s correction)
Quantification of proteins within biofilms
The presence of fibrillar proteins, pili or pilus-like structures within examined biofilms of 24 and 48 h macrocolonies of A. baumannii (brown staining) and K. pneumoniae (red staining) was demonstrated in CRCB-TSB assay (Fig. 1b). To quantify the approximate content of proteins within the matrix of biofilms, treatment with Proteinase K (PK) was used. It was shown that biofilms of investigated strains, particularly of A. baumannii, were significantly resistant to proteolysis (Fig. 3). No effective concentration of PK was found to disrupt biofilms of A. baumannii (Fig. 3a), where the addition of 800 µg/ml of PK resulted in a negligible loss of biomass, while the further increase in the proteinase concentration did not lead to any additional reduction. Thereby, proteins make up a relatively small fraction of the A. baumannii biofilm matrix (less than 10–15%), although some EPS proteins might evade proteolysis, for instance, due to aggregation in fibers, O-glycosylation, or their covering by other compounds of the matrix (Iwashkiw et al. 2012).
Content of proteins in 24 (grey) and 48 h (black) preformed biofilms of A. baumannii (a) and K. pneumoniae (b). Each dataset contains 4 technical and 2 biological replicates and represents a normalized mean value ± SD. Only statistically relevant differences from controls and between cohorts of 24 and 48 h biofilms are marked as asterisks: *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Mixed-effects analysis with Tukey’s correction)
Twenty-four hours old biofilms formed by K. pneumoniae also remained recalcitrant to the activity of PK (Fig. 3b), that apparently correlated with the results of morphotyping, where 24 h old macrocolonies of K. pneumoniae had no amyloid content (Fig. 1b, the first row). On the contrary, biofilms of K. pneumoniae, grown for 48 h, were susceptible to the action of Proteinase K, in a dose-dependent manner in concentrations range 50 to 1600 µg/ml of PK, resulting in the loss of up to 45% of biomass at 1600 µg/ml. Notably, simultaneous treatment with DNAse and PK did not lead to a synergistic decline in biomass of the investigated biofilms (Online Resource Fig. 2), which indicates tough stability of these BFs, provided by multiple contacts between various EPSs.
Activity of the endolysins against 48-h biofilms
A pronounced reduction in biomass of 48 h old BFs was observed after 2 h incubation with endolysins as shown by the CV Mtp assay for A. baumannii (Fig. 4a), K. pneumoniae (Fig. 4b) or mixed biofilms (Fig. 4c), which all formed moderate or strong BFs on the polystyrene. Endolysins in many cases acted regardless the dose, especially versus mono-species Klebsiella BFs. Each protein led to at least a twofold loss in the biomass of preformed biofilms, in particular, LysAp22 possessed the strongest disruption of BFs, whilst antibiofilm activity on EPS-rich biofilms by LysSi3 was less evident than that of other endolysins.
The activity of endolysins LysAm24, LysAp22, LysECD7, and LysSi3 versus the 48-h biofilms. a A. baumannii biofilms; b K. pneumoniae biofilms; c dual-species (mixed) biofilms. Tris–HCl buffer (pH = 7.5) was used as control. Each dataset includes 4 technical and 3 biological replicates was conducted and is demonstrated as normalized mean value ± SD. Only differences between data related to distinct concentrations of enzyme interpreted as follows: ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001. All differences between the biofilm growth control (buffer-treated BFs) and endolysin-treated groups were supposed highly reliable, p < 0.0001 (one per row Mann–Whitney tests with Holm-Šídák’s correction)
Exposure to a 1000 µg/ml of LysAp22 solution decreased the biofilms’ biomass up to 33, 25 and 27% from initial (Fig. 4). Two endolysins, LysECD7 and LysAm24, acted similarly versus mono-species BFs, albeit the impact of LysECD7 on dual-species BFs was comparable to that of LysAp22. The 2 h treatment with LysAm24 or LysECD7 at a concentration of 1000 µg/ml led to a decrease up to approximately 41, 35 and 27% of A. baumannii, K. pneumoniae or mixed biofilms’ biomass respectively, compared to the controls treated with the buffer. At 100 µg/ml, these proteins exhibited a lower activity. Except for LysSi3, endolysins at a higher concentration removed dual-species biofilms better than low-EPS containing A. baumannii BFs (p < 0.01, Mann–Whitney tests with Holm-Šídák’s correction). LysSi3 acted on mixed biofilms worse than other endolysins, disrupting the biomass by approximately 54%, independently on dose.
Exopolysaccharides and biomass staining of biofilms pretreated with endolysins
As demonstrated, three-dimensional biofilms were formed after a 48-h incubation of A. baumannii cells in TSB media on glass slides (Fig. 5). These aggregates consisted of dense and tightly connected structures (Fig. 5a), which were disrupted by endolysins to incoherent monolayers (BFs treated with LysAm24, LysAp22, LysSi3), or microcolonies (LysECD7). After destaining with CuSO4, light-blue color of the biofilm’s structures indicated the presence of acid, mostly capsular carbohydrates. Otherwise, if there is a lack of these EPSs, the biomass was colored violet. Thus, it allows to distinguish a significant number of cells covered with capsular polysaccharides and also the heterogeneous arrangement of different EPSs, especially on the BF edges (Fig. 5a’). Biofilms treated with LysAp22 or LysECD7 were poorly stained by CV (Fig. 5c, d), except for a few areas with dark violet peaks, and did not differ from the cells, additionally rinsed with CuSO4, suggesting interactions with carbohydrates, subsequent dissociation, and destabilization of biofilms’ matrix. The BF-disrupting effect of LysSi3 (Fig. 5e, e’) appeared to be similar to that of LysECD7, however, it was less pronounced, and did not cause evident destabilization of exopolysaccharides, but also efficiently passed through EPSs. A different manner of endolysin-biofilm interactions was observed for LysAm24 (Fig. 5b, b’). Preparations treated with this endolysin and stained with CV possessed an intense violet color, whereas double-stained biofilms apparently lacked any purple areas that demonstrate the interactions between LysAm24 and EPS, other than acid carbohydrates, providing additional sites for CV staining.
Effects of bacterial DNA on endolysins
We estimated that in abundance of endolysins (10:1 ratio), the DNA samples incubated with three out of 4 investigated endolysins displayed a notable decline in the electrophoretic mobility of nucleic acids (Fig. 6a), suggesting the complexes’ formation of endolysin with genomic DNA, which remained in the loading well or formed significant smears. Among the examined proteins, LysAm24 and LysECD7 exhibited the most pronounced DNA-binding activity (Fig. 6a). However, with the exception of LysAm24, endolysin-eDNA complexes were not detected when the ratio of P:DNA was 2:1. Only LysSi3 showed no interactions with DNA samples in this test.
a Binding of endolysins to A. baumannii genomic DNA. The Protein:DNA mass ratios are shown; Cont means only DNA-containing buffer solution; Mr, molecular length in base pairs (bp) of the DNA standards. b Changes in the antibacterial activity of 10 μg/ml lysins in the presence of DNA at the same ratios, versus free-living cells of A. baumannii (only statistically significant differences between datasets are shown, *, p < 0.05; **, p < 0.01 (Kruskal–Wallis test with Dunn’s correction). Two technical and 2 biological replicates were conducted
Exogenous nucleic acids led to the inhibition of muramidases’ activity versus free-living A. baumannii cells. At the lower concentration (10:1 ratio), DNA did not impact antibacterial properties of these proteins, whereas it dramatically inhibited them at the ratio of 2:1 (Fig. 6b). At the same time, endopeptidase LysECD7 remained effective against the bacteria even in the excess of DNA.
Discussion
Clinically relevant biofilms, formed by A. baumannii and K. pneumoniae species or their combination, are associated with a variety of skin and soft tissue infections, as well as nosocomial infections of the respiratory (Said et al. 2022) and urinary tracts (Paczosa and Mecsas 2016). Because of producing a heterogeneous and thick matrix, these causative agents possess an extreme tolerance towards various antimicrobials (Stewart 2015). Herein, we characterized the content of biofilms formed by two bacterial isolates and estimated that 48 h A. baumannii BFs contained a moderate amount of highly deacetylated glycosaminoglycans, acid polysaccharides, eDNA, and a relatively large fraction of the cellular biomass (Online Resource Table 1). For K. pneumoniae, the BF matrix makes up at least half of the dry biofilm’s volume, with a pronounced content of negatively charged polysaccharides, DNA, and non-amyloid proteins.
The enzymes contain phage lysozyme-like domain (family GH24), except for LysECD7 – a putative zinc-binding endopeptidase, which belongs to peptidase M15C family and contains a conserved LAS-type motif. LysAm24 and LysAp22 are naturally encoded by Acinetobacter phages, however, unlikely most Gram-negative endolysins, LysAm24 is made up two functional domains, the previously mentioned glucoside hydrolase domain and N-terminal cell-wall binding domain (PGBD1 family). LysSi3 and LysECD7 are produced by phages infecting S. typhi and E. coli correspondingly. Although, three out of the four endolysins referred as muramidases, the two most close (LysAp22 and LysAm24) possess about 62% amino acid sequence identity in catalytic domains. All the enzymes have no predicted signal protein (SP)- or signal-arrest-release (SAR)-domains within their structures (Gontijo et al. 2021).
Despite the difference in composition, we have shown that investigated enzymes disrupt heterogeneous biofilms, interacting directly with different BF compounds. All of the endolysins at a concentration of 100 µg/ml led to a decrease in biomass of mono- and dual-species films of Gram-negative bacteria by at least 2–3 times. Moreover, treatment with LysAm24, LysAp22, or LysECD7 at a concentration of 1000 µg/ml was linked to the pronounced destruction of the biofilms. Application of standard methods, such as CV-staining, to assessment of biofilm disruption, revealed a staining reduction up to 30% from the initial. As noted, this method of biofilm detection is not highly specific, thus, the decrease in staining may occur not due to degradation of the matrix as such, but due to matrix destabilization effects under the lysins’ action followed by the release of cells, BF components during the experimental procedures. However, the low correlation of endolysins action with the applied dose and the higher activity against mixed films suggests that lysins act relatively selective on biofilm components.
Although the investigated endolysins are considered highly specific hydrolases, disrupting the peptidoglycan at specific sites, and which no supposed to employ in direct enzymatic degradation of exopolysaccharides or eDNA in regard of other research on similar lysins, a side ability to influence compounds of the biofilm matrix could be recognized. Thus, we detected weak interactions between endolysins and exopolysaccharides and eDNA. These interactions, however, were associated with a significant destabilization of the treated biofilm and could be beneficial in terms of bacteriophage ecology. Endolysin-EPS interactions may include effects on adhesion of competitive viruses, availability of phages’ receptors, and formation of cell-wall deficient cells to sustain the host population (Wohlfarth et al. 2023). Furthermore, lysins can be engaged in BF-specific signaling pathways due to the products of peptidoglycan hydrolysis (Irazoki et al. 2019). Extracellular carbohydrates are a significant component involved in biofilm formation, crucial for stable cell adhesion on various surfaces, and establish multiple contacts with other exogenous molecules (Ostapska et al. 2018). They include enterobacterial cellulose (Nesse et al. 2020), and poly-β-1,6-N-acetyl-D-glucosamine (PNAG), found in numerous species of bacteria (Choi et al. 2009). Capsular polysaccharides, frequently consisting of various negatively and neutrally charged subunits, serve as a physical shield against environmental factors (Paczosa and Mecsas 2016; Singh et al. 2019), and within BFs modulate surface adhesion (Mann and Wozniak 2012; Pompilio et al. 2021). Therefore, massive detachment of acid exopolysaccharides by LysAp22 and LysECD7 may impair BF stability, without relevant loss of the antibacterial activity, and subsequently seems to be an intriguing feature for further development of enzybiotics. The exact mechanism of this change is not fully understood. However, it is unlikely to be related to capsule digestion by specific phage depolymerases. In particular, their active sites differ from those of endolysins and structures of the investigated lysins possess no significant similarity with any of this wide group of enzymes. It is worth noting, that exopolysaccharides can trigger changes in the catalytic activity of other enzymes, more related to endolysins. For instance, a polyanionic exopolysaccharide of Xantamonas spp. inhibits egg white lysozyme and lysostaphin, but activates two bacterial muramidases (Stepnaya et al. 2001).
The results of the eDNA-endolysin binding assay partly correlated with the isoelectric points (pIs) of the lysins: LysAp22 (pI 9.19), LysAm24 (pI 8.89), LysECD7 (pI 8.83) and LysSi3 (pI 8.52), proposing a higher probability of binding with an increase of pI. Hence, unspecified electrostatic forces may determine the interactions between the endolysins and eDNA. However, the differences in pIs between the studied proteins are small; thereby an actual net charge distribution of molecules may be distinct, affecting the binding capacity. Anyway, as shown in Online Resource Fig. 3, the strength of the endolysin-DNA association was significantly reduced by the addition of NaCl. Thus, LysECD7 partially ceased to bind with DNA in the mixtures, containing 150 and 300 mM NaCl, LysAm24 showed a similar tendency at the lower salt’s concentration, whereas the DNA-binding effect of LysAp22 was evident in the mixture without NaCl. Therefore, the interplay observed between eDNA and the investigated enzymes should be considered nonspecific, and primarily driven by electrostatic binding. Strong electrostatic interactions are suggested to destabilize eDNA in the cases of LysECD7 and possibly LysAm24, serving as a mechanism for the antibiofilm effect of these enzymes within BF areas depleted in free cations, formed by eDNA. LysSi3 is believed to have a reduced antibiofilm activity due to the indirect influence of exogenous nucleic acids, while eDNA affected LysAp22 ambivalently. We suggest that DNA mixed with a bacterial suspension can bind positively charged compounds on the outer membrane (OM) of bacteria and enhance their surface hydrophobicity (Das et al. 2014), impairing CW accessibility for either of hydrolases. It is interesting to note, that the investigated endolysins contain positively charged terminal structures putatively involved in permeabilization of membranes, except for LysECD7. The mechanism of passing through OM for LysECD7 is probably based on other, not only electrostatic interactions with a cell’s surface (Antonova et al. 2019).
There is plenty of findings noted eDNA can be a component of biofilm tolerance exhibited by different species against charged antimicrobial peptides (Batoni et al. 2016) and proteins, as well as aminoglycosides (Chiang et al. 2013), fluoroquinolones (Tetz et al. 2009), and non-phage derived lysozymes. In this regard, eDNA interaction with positively charged structures often found in endolysins’ molecules may also affect their catalytic activity and access to target sites, hampering endolysin-based therapy. On the other hand, enzybiotics may destabilize exposed biofilms through unspecific interactions with extracellular nucleic acids, allowing biofilm control. For example, side antibacterial effects have been reported for egg lysozyme and antistaphylococcal chimeric peptidoglycan hydrolase (Fernández et al. 2017; Liu et al. 2023). It has been proposed to be based on strong endolysin-DNA binding, that influence the expression of bacterial genes. Another interesting mechanism of biofilm removal by targeting defensive BF compounds has been reviewed for a few AMPs, which possess similarity with semi-conservative structures of the studied proteins (Batoni et al. 2016).
Therefore, our study revealed contrasting aspects in the antibiofilm activity of the phage endolysins, which deviated from bacterial cell lysis caused by catalytic digestion of the CW peptidoglycan. Specifically, we detected the interaction with the carbohydrate component of the matrix (LysAp22 and LysECD7), as well as the electrostatic binding of polycationic sites of endolysins to eDNA (LysAm24 and LysECD7). These interactions can result in the biofilm removal by endolysins, despite a decrease in their initial antibacterial activity. The plate staining results together with microscopy of biofilm samples enable to clearly see the matrix destabilization effects under the lysins’ action followed by the release of cells and BF components. Thereafter, a direct impact of lytic activity (bacterial membrane permeabilization and cleavage of peptidoglycan) of the endolysins is thought to occur. However, this research did not evaluate the influence of abnormal lysin-induced release of various bacterial compounds on the stability of BFs, which could also be compelling, as well as probable engagement of endolysins in degradation of eDNA and exopolysaccharides. Additional experiments should be conducted to clarify the endolysin-exoproteome interactions (De Gregorio et al. 2015), the specificity and intensity of endolysin-exopolysaccharides and endolysin-DNA interactions. This could be investigated through ELISA assays, mass-spectrometric structural analysis involving isolated EPS and its components, accompanied by a design of possible binding in silico. Treatment of biofilms with the cell-free lysate or products of peptidoglycan hydrolysis could also provide additional information. Thus, the present and further findings are expected to highlight a complex mechanism underlying the endolysins’ antibiofilm activity, allowing them to successfully evade a biofilm’s defense, provided by its matrix.
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information. Additionally, the raw data are available from the authors upon reasonable request.
References
Ahmad I, Nygren E, Khalid F, Myint SL, Uhlin BE (2020) A Cyclic-di-GMP signaling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci Rep 10:1–11. https://doi.org/10.1038/s41598-020-58522-5
Antonova NP, Vasina DV, Lendel AM, Usachev EV, Makarov VV, Gintsburg AL, Tkachuk AP, Gushchin VA (2019) Broad bactericidal activity of the myoviridae bacteriophage lysins LysAm 24, LysECD7, and LysSi3 against gram-negative ESKAPE pathogens. Viruses 11:284. https://doi.org/10.3390/v11030284
Arroyo-Moreno S, Cummings M, Corcoran DB, Coffey A, McCarthy RR (2022) Identification and characterization of novel endolysins targeting Gardnerella vaginalis biofilms to treat bacterial vaginosis. npj Biofilms Microbiomes 8:29. https://doi.org/10.1038/s41522-022-00285-0
Batoni G, Maisetta G, Esin S (2016) Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta, Biomembr 1858:1044–1060. https://doi.org/10.1016/j.bbamem.2015.10.013
Chiang WC, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T (2013) Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa Biofilms. Antimicrob Agents Chemother 57:2352–2361. https://doi.org/10.1128/AAC.00001-13
Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litrán T (2009) The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191:5953–5963. https://doi.org/10.1128/JB.00647-09
Das T, Sehar S, Koop L, Wong YK, Ahmed S, Siddiqui KS, Manefield M (2014) Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS ONE 9:e91935. https://doi.org/10.1371/journal.pone.0091935
De Gregorio E, Del Franco M, Martinucci M, Roscetto E, Zarrilli R, Di Nocera PP (2015) Biofilm-associated proteins: news from Acinetobacter. BMC Genomics 16:1–14. https://doi.org/10.1186/s12864-015-2136-6
Donlan RM, Costerton JW (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. https://doi.org/10.3201/eid0809.020063
Eriksson E, Liu PY, Schultz GS, Martins-Green MM, Tanaka R, Weir D, Gould LJ, Armstrong DG, Gibbons GW, Wolcott R (2022) Chronic wounds: treatment consensus. Wound Rep Reg 30:156–171. https://doi.org/10.1111/wrr.12994
Fernández L, González S, Campelo AB, Martínez B, Rodríguez A, García P (2017) Downregulation of autolysin-encoding genes by phage-derived lytic proteins inhibits biofilm formation in Staphylococcus aureus. Antimicrob Agents Chemother 61:e02724-e2816. https://doi.org/10.1128/AAC.02724-16
Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S (2016) Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. https://doi.org/10.1038/nrmicro.2016.94
Freeman DJ, Falkiner FR, Keane CT (1989) New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol 42:872–874. https://doi.org/10.1136/jcp.42.8.872
Giacobbe DR, Mikulska M, Viscoli C (2018) Recent advances in the pharmacological management of infections due to multidrug-resistant gram-negative bacteria. Expert Rev Clin Pharmacol 11:1219–1236. https://doi.org/10.1080/17512433.2018.1549487
Gontijo MTP, Vidigal PMP, Lopez MES, Brocchi M (2021) Bacteriophages that infect gram-negative bacteria as source of signal-arrest-release motif lysins. Res Microbiol 172:103794. https://doi.org/10.1016/j.resmic.2020.103794
Grishin AV, Karyagina AS, Vasina DV, Vasina IV, Gushchin VA, Lunin VG (2020) Resistance to peptidoglycan-degrading enzymes. Crit Rev Microbiol 46:703–726. https://doi.org/10.1080/1040841X.2020.1825333
Gutiérrez D, Ruas-Madiedo P, Martínez B, Rodríguez A, García P (2014) Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 9:e107307. https://doi.org/10.1371/journal.pone.0107307
Heselpoth RD, Swift SM, Linden SB, Mitchell MS, Nelson DC (2021) Enzybiotics: endolysins and bacteriocins. In: Harper DR, Abedon ST, Burrowes BH, McConville ML (eds) Bacteriophages: biology, technology, therapy. Springer International Publishing, Cham, pp 989–1030
Hughes RB, Smith AC (2013) Capsule stain protocols. Am J Microbiol 13:1–12
Irazoki O, Hernandez SB, Cava F (2019) Peptidoglycan muropeptides: release, perception, and functions as signaling molecules. Front Microbiol 10:500. https://doi.org/10.3389/fmicb.2019.00500
Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, Feldman MF (2012) Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog 8:e1002758. https://doi.org/10.1371/journal.ppat.1002758
Karau MJ, Mandrekar J, Lehoux D, Schuch R, Cassino C, Patel R (2023) In vitro activity of exebacase against methicillin-resistant Staphylococcus aureus biofilms on orthopedic Kirschner wires. BMC Res Notes 16:209. https://doi.org/10.1186/s13104-023-06468-y
Karygianni L, Ren Z, Koo H, Thurnheer T (2020) Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol 28:668–681. https://doi.org/10.1016/j.tim.2020.03.016
Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L (2017) Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 15:740–755. https://doi.org/10.1038/nrmicro.2017.99
Liu L, Jia X, Zhao X, Li T, Luo Z, Deng R, Peng B, Mao D, Liu H, Zheng Q (2023) In vitro PCR verification that lysozyme inhibits nucleic acid replication and transcription. Sci Rep 13:6383. https://doi.org/10.1038/s41598-023-33228-6
Mann EE, Wozniak DJ (2012) Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. https://doi.org/10.1111/j.1574-6976.2011.00322.x
Minas K, McEwan NR, Newbold CJ, Scott KP (2011) Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett 325:162–169. https://doi.org/10.1111/j.1574-6968.2011.02424.x
Nesse LL, Osland AM, Mo SS, Sekse C, Slettemeås JS, Bruvoll AEE, Urdahl AM, Vestby LK (2020) Biofilm forming properties of quinolone resistant Escherichia coli from the broiler production chain and their dynamics in mixed biofilms. BMC Microbiol 20:46. https://doi.org/10.1186/s12866-020-01730-w
Omar A, Wright JB, Schultz G, Burrell R, Nadworny P (2017) Microbial biofilms and chronic wounds. Microorganisms 5:1–15. https://doi.org/10.3390/microorganisms5010009
Ostapska H, Howell PL, Sheppard DC (2018) Deacetylated microbial biofilm exopolysaccharides: it pays to be positive. PLoS Pathog 14:e1007411. https://doi.org/10.1371/journal.ppat.1007411
Paczosa MK, Mecsas J (2016) Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. https://doi.org/10.1128/mmbr.00078-15
Pompilio A, Scribano D, Sarshar M, Di Bonaventura G, Palamara AT, Ambrosi C (2021) Gram-negative bacteria holding together in a biofilm: the Acinetobacter baumannii way. Microorganisms 9:1353. https://doi.org/10.3390/microorganisms9071353
Prasad NK, Seiple IB, Cirz RT, Rosenberg OS (2022) Leaks in the pipeline: a failure analysis of gram-negative antibiotic development from 2010 to 2020. Antimicrob Agents Chemother 66:e00054-e122. https://doi.org/10.1128/aac.00054-22
Puchtler H, Sweat F, Levine M (1962) On the binding of Congo red by amyloid. J Histochem Cytochem 10:355–364. https://doi.org/10.1177/10.3.355
Römling U, Sierralta WD, Eriksson K, Normark S (1998) Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28:249–264. https://doi.org/10.1046/j.1365-2958.1998.00791.x
Said KB, Alsolami A, Alshammari F, Alreshidi FS, Fathuldeen A, Alrashid F, Bashir AI, Osman S, Aboras R, Alshammari A, Alshammari T, Alharbi SF (2022) Selective COVID-19 coinfections in diabetic patients with concomitant cardiovascular comorbidities are associated with increased mortality. Pathogens 11:508. https://doi.org/10.3390/pathogens11050508
Scott JE, Dorling J (1965) Differential staining of acid glycosaminoglycans (mucopolysaccharides) by Alcian blue in salt solutions. Histochemie 5:221–233. https://doi.org/10.1007/BF00306130
Singh JK, Adams FG, Brown MH (2019) Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front Microbiol 9:3301. https://doi.org/10.3389/fmicb.2018.03301
Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Ćirković I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115:891–899. https://doi.org/10.1111/j.1600-0463.2007.apm_630.x
Stepnaya OA, Ryazanova LP, Krupyanko VI, Kulaev IS (2001) Influence of acidic exopolysaccharide of Xanthomonas campestris IBPM 124 on the kinetic parameters of extracellular bacteriolytic enzymes. Biochemistry 66:662–666. https://doi.org/10.1023/A:1010263432155
Stewart PS (2015) Antimicrobial tolerance in biofilms. Microbiol Spectr 3:269–285. https://doi.org/10.1128/microbiolspec.mb-0010-2014
Tetz GV, Artemenko NK, Tetz VV (2009) Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother 53:1204–1209. https://doi.org/10.1128/AAC.00471-08
Vasina DV, Antonova NP, Grigoriev IV, Yakimakha VS, Lendel AM, Nikiforova MA, Pochtovyi AA, Remizov TA, Usachev EV, Shevlyagina NV, Zhukhovitsky VG, Fursov MV, Potapov VD, Vorobev AM, Aleshkin AV, Laishevtsev AI, Makarov VV, Yudin SM, Tkachuk AP, Gushchin VA (2021) Discovering the potentials of four phage endolysins to combat gram-negative infections. Front Microbiol 12:748718. https://doi.org/10.3389/fmicb.2021.748718
Wohlfarth JC, Feldmüller M, Schneller A, Kilcher S, Burkolter M, Meile S, Pilhofer M, Schuppler M, Loessner MJ (2023) L-form conversion in gram-positive bacteria enables escape from phage infection. Nat Microbiol 8:387–399. https://doi.org/10.1038/s41564-022-01317-3
Acknowledgements
Not applicable.
Funding
The study was supported by the Russian Science Foundation (RSF), grant No. 23–74-10027, https://rscf.ru/project/23-74-10027/.
Author information
Authors and Affiliations
Contributions
Conceptualization: Daria V. Vasina; Methodology: Anastasiya M. Lendel; Formal analysis and investigation: Anastasiya M. Lendel, Nataliia P. Antonova, Igor V. Grigoriev, Evgeny V. Usachev; Writing—original draft preparation: Anastasiya M. Lendel; Writing—review and editing: Nataliia P. Antonova, Igor V. Grigoriev, Daria V. Vasina; Funding acquisition: Daria V. Vasina; Resources and project administration: Vladimir A. Gushchin; Supervision: Daria V. Vasina. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no financial or proprietary interests in any material discussed in this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Authors information
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lendel, A.M., Antonova, N.P., Grigoriev, I.V. et al. Biofilm-disrupting effects of phage endolysins LysAm24, LysAp22, LysECD7, and LysSi3: breakdown the matrix. World J Microbiol Biotechnol 40, 186 (2024). https://doi.org/10.1007/s11274-024-03999-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-024-03999-9