Abstract
The present study aimed at characterizing lactic acid bacteria (LAB) strains isolated from traditional sourdoughs collected in different regions of Morocco. Isolated strains were firstly identified using Gram staining and catalase reaction test. Presumptive LAB strains were then checked for various phenotypical properties including growth at 45 °C, resistance to NaCl, enzyme production, acidification capacity, diacetyl and exopolysaccharide (EPS) production, and antifungal activity. Finally, selected LAB strains were identified using 16S rDNA sequencing. Results showed that 32.1% of the isolates were thermophilic (45 °C) and 83.9% were resistant to NaCl (6.5%). Moreover, 51.7 and 37.5% were able to produce diacetyl and EPS, respectively. Regarding enzyme production, 55.3 and 7.1% of the isolates showed lipolytic and proteolytic activities, respectively. Low pH values (3.37–3.76) were obtained after 24 h of incubation of LAB strains in de Man, Rogosa and Sharpe (MRS) broth. Antifungal activity test against Aspergillus flavus, Aspergillus niger and Penicillium spp. showed an inhibition rate up to 50%. Bacterial DNA sequencing showed that LAB isolates belong to seven species, chiefly Levilactobacillus brevis, Lentilactobacillus parabuchneri, Lactiplantibacillus plantarum, Pediococcus pentosaceus, Enterococcus hirae, Bifidobacterium pseudocatenulatum, and Companilactobacillus paralimentarius. These findings, for the first time in Moroccan sourdoughs, indicate that the isolated LAB strains have good multifunctional properties and could be suitable as good starters for sourdough bread production under controlled conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The origin of bread, as one of the earliest food technologies used by humans being, is not precisely known and the use of sourdough for bread dough leavening is one of the oldest biotechnological processes known by humankind (Reale et al. 2019; Novotni et al. 2021). Sourdough can be defined as an acidic mixture of water flour with pronounced taste, obtained after fermentation and used for making bread and other bakery products (Bartkiene et al. 2020). This mixture forms a sponge-like product which is then kept at room temperature and refreshed on a daily basis, for several days, to develop into a sourdough (Moujabbir et al. 2023). Since the beginning of the twenty-first century, traditional sourdough or sourdough-based products have been increasingly used as a fermentation step or as an ingredient in wheat and rye baking. The increased use of sourdough or sourdough products is linked to the possibility of producing "clean label" products and the superior nutritional and sensory quality of bread (Novotni et al. 2021). Sourdough bread is also increasingly popular as a natural, healthy and nutritious food having a characteristic flavor and taste and an extended shelf-life with considerable resistance to spoilage (Brandt 2019; Novotni et al. 2021). Many breads and many other baked goods, such as pizzas and various sweet baked goods, are made in European countries using sourdough fermentation. For example, maize has been used for centuries to produce an ethnic bread called Broa in Portugal (Rocha et al. 2023; Rocha and Malcata 2016), and over 200 different types of traditional/typical Italian breads are made with sourdough as a natural starter (Reale et al. 2019).
During sourdough fermentation, complex activities and metabolic interactions occur and a complex and unique microbial ecosystem is established (Gobbetti et al. 1995). This microbial community mainly made up of LAB and yeasts is responsible for this fermentation process (Carbonetto et al. 2020). LAB dominate the sourdough microbiota and along with acid-tolerant yeasts, and being generally present in a ratio of 100:1, respectively (Fraberger et al. 2020). The synergistic interaction between LAB, and yeasts (e.g. Saccharomyces cerevisiae, Saccharomyces exiguus, Candida krusei, and Candida guilliermondii, etc.), during the fermentation phase, allows the production of aromatic precursors, increases the volume of dough and decreases the firmness of bread (Moujabbir et al. 2023).
LAB strains including Lactobacillus species (Lactiplantibacillus plantarum, Levilactobacillus brevis, Lactobacillus paralimentarius, Limosilactobacillus fermentum, Limosilactobacillus pontis, Companilactobacillus crustorum, and Lentilactobacillus parabuchneri, etc.) have been isolated from sourdoughs (Rocha et al. 2023; Rocha and Malcata 2012). LAB species belonging to Leuconostoc, Lactococcus, Pediococcus, Enterococcus, or Weissella, and streptococci genera have also been isolated from sourdoughs (Rocha and Malcata 1999). In sourdoughs present over the world with, Lactobacillus is the common and the most present genera while Enterococcus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, and Weissella species are less frequently exist (Kaya et al. 2022).
In Morocco, bread is a staple food for the population and its consumption represents the bulk of the country's diet. Preliminary surveys in the country expectably reported a significant role of LAB strains in sourdough breadmaking (Chaoui et al. 2006; Faid et al. 1993). Based on the literature, occurrence of LAB strains in sourdough ferments depends on their origin, raw-material and type of sourdough fermentation used in their preparation, among other endogenous factors in cereals and processing parameters (i.e., exogeneous factors). Several basic ingredients of various origin (Lben, date nuts, green tea leaves, garlic, carob flour, lemon and figs, etc.) are generally integrated into wheat or barley flours during traditional Moroccan breadmaking, to accelerate the dough fermentation and create special flavors (Moujabbir et al. 2023). These ingredients generally may constitute a new source of desirable with functional (biological activity or technological functionality) LAB strains (Faid et al. 1994). The major LAB strains involved in the acidification of sourdough using “lben”, a traditional fermented milk, were identified as P. pentosaceus, Ln. mesenteroides, L. delbrueckii, Len. parabuchneri, Lp. Plantarum, and Fructolactobacillus sanfranciscensis, while traditional sourdough ferments made with garlic and lemon showed the presence of Lp. plantarum, Limosilactobacillus fermentum, L. delbrueckii, P. pentosaceus, and Ln. mesenteroides (Faid et al. 1994). It should be highlighted that all these LAB strains were identified by traditional phenotypic methods. Despite the availability of scientific information on the biodiversity of LAB in various foods available in Morocco including olive table (Rokni et al. 2021), cow's and camel milk ( Khedid et al. 2009), cheeses (Bennani et al. 2017), the biodiversity and biotechnological properties of LAB isolated from in Moroccan sourdoughs have never been investigated.
Thus, the purposes of the current study were to identify and characterize for the first time LAB strains isolated from various Moroccan traditional sourdoughs collected in different geographical regions of the country by using molecular methods, as well as to evaluate their potential multifunctional properties for further applications. It is expected that this study will contribute to a better and in-depth understanding of sourdough breads from Morocco.
Material and methods
Sourdough sampling
A total of seventeen traditional sourdough samples were collected in four geographical areas of Morocco, chiefly Fès-Meknès, El Jadida, Rabat, and Taza. All selected sourdoughs (liquid and semi-solid samples) are maintained in households for daily baking. Collected sourdoughs were transported in plastic bottles (at 4 °C) to the laboratory and stored at 4 °C until analysis.
pH and dornic acidity (DA) of sourdoughs
The pH of sourdough samples was measured using a pH meter (Adwa, AD1000 pH, Hungary) previously calibrated using buffer standards of 4.0 and 7.0. For liquid sourdough samples, pH was directly measured by emerging the sensor in 100 g of sourdough. For semi-solid samples, pH measurement was performed after mixing 100 g of sourdough with 20 mL of distilled water. DA was determined by titration with sodium hydroxide (0.1 N) using phenolphthalein as color indicator. Results were expressed as degree Dornic (°D).
LAB isolation
LAB strains were isolated from each sourdough sample after a pre-enrichment step in MRS broth (Biokar, France). The pre-enrichment step consisted of incubating 1 mg of sourdough sample in 9 mL (dilution of 1/10) of sterile MRS broth at 30 °CC for 24 h under aerobiosis. LAB strains were isolated using the spread plate method on MRS agar plates (Biokar, France). Serial dilutions from 10–1 to 10–7 were prepared and 0.1 mL of overnight incubated cultures were spread on MRS agar plate and incubated at 30 °C for 24–48 h under aerobic conditions. Different colony shapes were then purified on MRS agar medium in the same incubation conditions. Strains with Gram-positive staining and tested catalase negative were stored at 4 °C on MRS agar slant tubes for further analysis.
Properties of LAB isolates
Overnight cultures of LAB isolates were first incubated on MRS broth at different temperatures (30 and 45 °C) for 48 h under aerobiosis to assess their thermophilic or mesophilic character. For the acidifying aptitude, a 24 h microbial culture of each LAB strain was incubated in a 1% (v/v) level of fermented wheat flour (10% w/v), already sterilized and divided in a volume of 10 mL into sterile tubes. The pH was measured at 0, 6 and 24 h of incubation at 30 °C under aerobiosis. The tolerance of LAB strains to different NaCl concentrations was tested by their growth assays at 1, 4 and 6.5% of NaCl. For this, modified MRS broth containing bromocresol purple as indicator was prepared and inoculated with 50 µL of 1% LAB culture (incubated overnight) in each tube, and then incubated at 30 °C for 24–48 h under aerobic conditions. Color change from purple to yellow was considered as positive indication of their growth (Boyaci Gunduz et al. 2022).
The homo- or heterofermentative traits of LAB isolates were studied based on their ability, or not, to produce CO2 on a sample tube with MRS semi-agar enriched with sugar (5% glucose) after incubation at 30 °C for 48 h under aerobiosis. Homofermentative LAB (Streptococcus, Lactococcus, and Enterococcus) grow well in the culture medium using sugar but do not produce gas. Conversely, heterofermentative LAB (Betabacterium group of the genus Lactobacillus and Leuconostoc) are able to produce CO2 in the middle of the sample tube (Ismaili Alaoui et al. 2017).
The ability of LAB isolates to produce diacetyl, a flavoring compound responsible for aroma in fermented food products, was determined in duplicate by inoculating each LAB strain at 1% (v/v) in 10 mL of UHT milk and incubated at 30 °C for 24 h under aerobiosis, as previously described by Boyaci Gunduz et al. (2022). For this purpose, 1 mL of each cell suspension was mixed with 0.5 mL of α-naphthol (1% w/v) and KOH solution (16% w/v) and incubated at 30 °C for 10 min under aerobic conditions. The formation of a red ring at the top of the tubes indicates diacetyl production.
The EPS production by LAB isolates was evaluated as described by Pelin et al. (2020), with slight modifications. LAB isolates were streaked in MRS agar medium supplemented with 5% of glucose and incubated aerobically at 37 °C for 48 h. The EPS production was considered positive when colonies showed mucoid or viscous shapes on the agar medium.
Proteolytic activity of the LAB strains was evaluated by depositing 20 μL of LAB cultures on sterile Whatman paper disks, previously placed on the surface of an MRS agar supplemented with 10% skimmed milk. Proteolytic activity was determined as a clear zone (halo) around the discs after incubation at 37 °C for 24 h under aerobiosis. Moreover, lipolytic activity was tested by inoculating LAB isolates on agar spot (MRS supplemented by 1% of tributyrin). After aerobic incubation at 37 °C for 72 h, positive lipolytic activity was elucidated as an opaque zone (halo) around the disc due to the formation of fatty acids and esters released by calcium.
Antifungal activity of LAB isolates
The antifungal activity was tested against the growth of fungal strains of Aspergillus flavus, Aspergillus niger, and Penicillium spp. previously isolated from contaminated foodstuffs and being part of the fungal collection of BIOMARE Laboratory. Mycelium growth inhibition by the cell-free supernatants (CFS) of LAB strains was determined on Dichloran Rose-Bengal Chloramphenicol (DRBC) Agar (Biokar, France). Plates containing DRBC medium, added at 10% (v/v) with sterile CFS of each LAB strain, were inoculated in the center with disc (5 mm) of A. flavus, A. niger, and Penicillium spp. After aerobic incubation at 25 °C for 5 days, the diameters of fungal colonies were measured. A negative control was used by adding MRS broth to DRBC medium. The percentage of mycelium growth inhibition (MI, %) was calculated using the Eq. 1, as follows (Abouloifa et al. 2021):
where MI (%) Percentage of mycelium growth inhibition, Tc Total fungal colony diameter (mm) used as control, TT Diameter (mm) of total fungal colony obtained with CFS.
Molecular identification of LAB strains
LAB strains were selected according to their metabolic characteristics and their multifunctional properties such as diacetyl and EPS production, acidification, NaCl tolerance, lipase and protease production and antifungal activity, and identified by DNA sequencing. DNA was extracted from LAB isolates according to the method of Wang et al. (2016) with slight modifications. For each LAB strain, 10 colonies suspended in 500 µL of molecular biology grade water (Sigma-Aldrich), were vortexed for 2 min, and centrifuged at 12,000×g for 10 min. Bacterial pellet was then suspended in 100 µL of 25 mg/mL lysozyme and 100 µL of lysis buffer (10 mMTris-Cl, 1 mM EDTA, pH 8), mixed and incubated for 5 min at ambient temperature. The suspension was added to 100 µL of 20 mg/mL proteinase K and incubated for 20 min at 42 °C. 50 µL of sodium dodecyl sulfate (SDS, 20%) were added and the suspension homogenized and incubated at ambient temperature for 5 min, then at 42 °C for 10 min. In order to remove cell wall debris, 400 µL of cetyltrimethylammonium bromide (CTAB)/NaCl solution (10% Tris 1M pH 8, 28% NaCl 5M, 4% EDTA, 20 g CTAB) were added to the suspension, homogenized and incubated for 10 min at 65 °C. Then, 700 µL of chloroform-isoamyl-alcohol (25:24:1, v/v/v) were added and the tubes were rapidly inverted 10 times, and centrifuged at 12,000×g for 15 min. Aqueous supernatant was recovered in a micro-centrifuge tube and subjected to the previous step (× 2). 600 µL of chloroform-isoamyl-alcohol (24:1, v/v) were added to the supernatant and centrifuged for 10 min at 12,000×g, then 30 µL of 100% isopropanol and one volume of sodium acetate (3M, pH 5) were added in micro-centrifuge tubes. The solution was maintained at − 20 °C for 1 h and centrifuged for 30 min at 12,000×g. After elimination of the supernatant, DNA was washed with 500 µL ethanol (70%) and centrifugated for 5 min at 12,000×g. DNA pellet was dried overnight in hood, re-dissolved in sterile distilled water (100 µL) and stored at 4 °C. After running on agarose gel electrophoresis (0.8%, w/v) and staining with a GelRed® Nucleic Acid Gel Stain, 10,000X (Biotium, USA), bacterial DNA was visualized using an UV transilluminator (318 nm). Sample of DNA extract (5 µL) were mixed with 3 µL of 1 Kb DNA ladder (Promega, USA) and 2 µL blue/orange loading dye 6X (Promega, USA) and run in TAE 1X buffer (Tris–Acetate-EDTA pH 8.3, Euromedex, France) at 100 V for 30 min.
LAB isolates were identified by 16S rDNA gene sequence analysis. Bacterial DNA was amplified by Polymerase Chain Reaction (PCR) using the primer pair 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1495r (5′-CTACGGCTACCTTGTTACGA-3′) as previously reported (Ho et al. 2018). The PCR reaction mixtures (50 µL) contained 5 µL of extracted DNA, 13.5 µL sterile water, 10 µL of each primer (10 µL/mL), 1 µL dNTP (Promega, USA), 0.25 µL Taq polymerase (Promega, USA) and 10 µL PCR buffer for Taq polymerase (Promega, USA). DNA amplification was performed using thermocycler (Mastercycler X50s) with initial denaturation of 2 min at 94 °C, followed by 36 cycles of 30 s at 94 °C, 30 s at 55 °C, 1 min at 72 °C, and an extension step at 72 °C for 5 min. DNA amplicons (approximately 340 pb) were visualized on 2% (w/v) agarose gel electrophoresis using 100 pb DNA leader (Promega, USA), and stored at 4 °C until DNA sequencing.
The sequencing of amplified DNA fragments was realized at the GPTR GenSeq UMR (Montpellier, France). Obtained sequences were submitted to database of National Centre of Biotechnology Information (GenBank NCBI) for LAB identification using Basic Local Alignment Search Tool (BLAST).
Statistical analysis
All data were statistically reported from three replicates (n = 3). Means and standard deviations were calculated in Microsoft Excel software (Microsoft Corporation, Seattle, WA, USA).
Results
Physico-chemical characterization of sourdoughs
Dornic acidity and pH of analyzed sourdough samples are summarized in Table 1. The pH measurements of sourdough samples varied depending on the sampling area from 3.00 to 4.46 for a sourdough collected in Taza and Rabat-Salé area, respectively. The acidity reached high values ranging from 21.05 to 50°D for a sourdough from Taza and Rabat-Salé area, respectively. These results are similar to those obtained by Fekri et al. (2020) and are higher than those described by Felicia (2015).
LAB isolation
A total of 56 LAB strains were isolated on MRS agar from the collected household sourdough samples. All purified strains were checked for negative catalase reaction and Gram-positive staining. The cocci species (64%) dominated, followed by the rod-shaped ones (34%). Repartition of the isolated strains depending on the sampling geographic area (Table 1).
Properties of LAB isolates
The technological characteristics of LAB isolates are presented in Table 2. Results of this study showed that all tested LAB strains were capable to grow under mesophilic conditions (30 and 37 °C), whereas 32.1% of isolates LAB strains showed thermophilic ability by growing at 45 °C. Acidifying ability of LAB strains is an important criterion used for the best selection of an appropriate starter culture for sourdough fermentation. As shown, 10.7, 80.3 and 9% of LAB isolates decreased the pH after 24 h incubation to values of 3, 4 and 5, respectively (Table 2). A pH decrease of MRS broth was obtained at time 0, 6 and 24 h. LAB strains S18 and S22 were able to decrease the initial pH value of 6.67, after 6 h, to 5.15 and, after 24 h, to 4.74. However, a drastic reduction to pH values of 3.37, 3.43, 3.53, 3.63 and 3.76 was obtained by the LAB strains S9, S28, S16, S27, and S4 after 24 h, respectively.
Regarding others properties, most of isolated LAB strains (83.9%) tolerated high NaCl concentrations (6.5%). Salt is an important contributor to the shelf-life, taste and structure of bread. However, salt addition in bread has been gradually reduced worldwide to prevent cardiovascular diseases and hypertension caused by an excessive intake of sodium (Novotni et al. 2021). Such regulations on salt content in bread brings new challenges to the bakery industry.
On the other hand, 51.7% of LAB isolates were capable of producing diacetyl. Regarding EPS production, 37.5% of tested strains were able to produce these compounds. Concerning the enzymatic capacities of LAB, a total of 4 out 56 isolates (7.1%) possessed proteolytic activity with the presence of a transparent halo around discs, with a diameter zone ranging up 17.5 ± 0.07 mm for the LAB strain S3 (Tables 2 and 3).
Antifungal activity of LAB strains
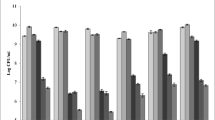
The antifungal activity of LAB strains was performed against spoilage fungi, particularly A. flavus, A. niger and Penicillium spp. Initial screening for the antifungal activity of LAB isolates against the spoilage fungi demonstrated that out of 56 isolates, only 13 LAB (23.2%) have shown growth inhibition against the studied fungi. Further screening with four LAB (S4, S17, S21, and S23) revealed that MI only went up to around 50% of inhibition. The mean values of inhibition rates (MI%) of each LAB against the tested fungal strains was calculated from 3 replicates inhibition assays (Fig. 1).
LAB identification
In this current study, among all isolates, thirteen LAB isolated from Moroccan sourdough samples were identified. The 16S rDNA sequencing was considered reliable for identifying individual species (> 99%). LAB strains were identified as belonging to the seven species, chiefly Lev. brevis, Len. parabuchneri, Lp. plantarum, P. pentosaceus, E. hirae, B. pseudocatenulatum, and Co. paralimentarius (Table 4). Regarding the profiles of LAB strains isolated from Moroccan sourdoughs, it should be emphasized that, to the authors knowledge, no published data is currently available in the literature on the molecular identification of LAB strains originating from Moroccan sourdoughs. Previous studies showed the among 41 LAB strains isolated from durum wheat sourdoughs, six LAB species were identified as Lactobacillus plantarum, Leuconostoc mesenteroides, Lactobacillus curvatus, Lactobacillus paraplantarum, Weissella cibaria, and Lactobacillus pentosus (Zotta et al. 2008). Recently, LAB strains isolated from “El Hammoum”, an Algerian fermented wheat, were identified as Enterococcus faecium and E. durans (Badji et al. 2023).
Discussion
Physico-chemical properties of sourdough showed low pH and high acidity values of all collected samples. This could be explained by the raw material and the method used for the household traditional sourdough preparation in the areas which may affect the final microbiological composition of samples. Indeed, earlier investigations reported that LAB strains are always involved in association with some yeasts species in Moroccan traditional sourdoughs (Boraam et al. 1993). According to Faid et al. (1994b), physico-chemical characteristics of the sourdoughs are very useful to know whether the sourdough preparation was successful or not before using them in traditional sourdough bread fermentation. The low pH and high acidity values obtained in some samples are due to the acidifying microbiota (LAB) present in the dough starter. Bom et al. (2018) reported that differences in pH values might be related to the LAB strain capacity to produce varying amounts of organic acids, mainly lactic and acetic acids. Indeed, low pH values (less than 5) is one of the important parameters for controlling the growth of hazardous bacteria and preventing microbial contamination especially undesirable microorganisms and outcompeting yeasts. For this reason, the spontaneous fermentation of baking doughs using mother-doughs/sourdoughs constitutes an important tool towards food safety of baking goods. It was reported that dough acidification through the production of lactate is a key contributor to lactic metabolism in sourdough fermentation as it influences all aspects of bread quality, directly or indirectly through the modulation of cereal enzymes (Novotni et al. 2021). These findings suggest that LAB strains isolated from Moroccan sourdough produce high amounts of organic acids preventing the growth of pathogenic and foodborne bacteria and molds (Fujimoto et al. 2019) and playing an utmost role in the texture and organoleptic properties of the final sourdough bread (Rocha and Malcata 2012).
Regarding thermophilic ability, 32.1% of LAB isolates were able to grow at 45 °C (Table 2). Thermophilic LAB strains were reported to have several advantages and play important role in sourdough breadmaking at the industrial scale, referred to as type II sourdoughs that are dominated by thermophilic and acid-tolerant heterofermentative lactobacilli. Indeed, it was reported that the application of thermophilic sourdough increased bread porosity, elasticity, crumbliness and moisture content. Furthermore, the fungal spoilage on the bread crust surface was proved to be suppressed using sourdough prepared with thermophilic LAB (Cizeikiene et al. 2020).
Concerning enzyme activity, moderate lipolytic activity was observed for eight LAB strains including Lev. brevis S3, Co. paralimentarius S4, E. hirae S5, P. pentosaceus (S12, S15, S27, and S28), and Len. parabuchneri S17. While a strong proteolytic activity (17.5 ± 0.7) was obtained by Levilactobacillus brevis S3. Because of the higher proteolytic activity of some LAB strains, sourdough can contain many peptides and amino acids. Indeed, it was reported that good proteolytic activity improves the taste and flavor profiles of the leavened baked goods, and, nutritionally, it leads to improve the protein digestibility (Nionelli et al. 2018). Even though sourdough microbiota have not been shown to have relevant lipolytic activity and most od isolated strains did not show extracellular protease activities, some sourdough LAB would increase the amount of free amino acids which are important for flavor formation via Maillard reactions during baking, as well in Amadori reaction responsible for the condensation of amino acids with reducing sugars (Novotni et al. 2021). However, it was reported that proteolytic breakdown is enhanced at low values of pH during sourdough fermentation of wheat dough, and main effects relate to changes in pH rather than to microbial proteolytic activity of sourdough (Rocha 2011).
Regarding aroma and EPS production by LAB isolates, diacetyl is an important component of the food industry providing a specific desirable flavor and have inhibitory activity against hazardous strains, especially pathogenic and foodborne bacteria. Formation of diacetyl from pyruvate during sourdough fermentation has been particularly attributed to homofermentative lactobacilli (Damiani et al. 1996). While EPS production by LAB strains is one of the most sought after and desired biotechnological properties, in particular for traditional sourdough bread fermentation since this property plays a critical role on the texture of fermented sourdough breads. Some sourdough microbial strains for instance belonging to genera Leuconostoc, Weissella, and Pediococcus are able to synthesize and excrete to the medium EPS from sucrose by extracellular glucansucrases or fructansucrases (İspirli et al. 2020). According to previous studies, the major qualities required for sourdoughs are the production of EPS and organic acids, as well as the release of carbon dioxide, and the presentation of good dough growth profiles (Bom et al. 2018; Păcularu-Burada et al. 2020). Indeed, EPS produced by LAB in sourdough improve bread rheological proprieties and quality by increasing bread volume and lightness and by reducing its firmness.
Most of the LAB isolates (84%) were homofermentative. Homofermentative bacteria metabolize sugars (hexoses) through glycolysis pathway, where fermentation of 1 mol of hexose can yield 2 mol of lactic acid and 2 mol of ATP. In comparison, heterofermentative bacteria use another pathway, and hexoses could be converted to equimolar amounts of lactic acid, acetate or ethanol, and carbon dioxide, yielding 1 mol of ATP per mol of hexose fermented (Rocha et al. 2023). It was reported that heterofermentative LAB produces strong spicy, acidic, aldehydic, fruity, caramel-like, and butter-like flavors, whereas homofermentative LAB produces softer, sweeter and more “flower-like” flavors (Rocha et al. 2023; Rocha 2011).
The composition of sourdough microbiota depends on the technological purpose of the use of sourdough in breadmaking. Thus, sourdoughs are fermented to produce a dough leavening without the addition of baker's yeast or to obtain a sourdough or a sourdough product as a breadmaking improver (Brandt 2019). It has been reported that sourdoughs that are maintained in households for baking every few weeks harbored a combination of Lp. plantarum and Lev. brevis group organisms (Gänzle and Zheng 2019). In the present study, seven species, chiefly Lev. brevis, Len. parabuchneri, Lp. plantarum, P. pentosaceus, E. hirae, B. pseudocatenulatum, and Co. paralimentarius were isolated and identified for the first time from Moroccan sourdoughs. Indeed, Lactobacillus species (Len. parabuchneri, Lev. brevis, and Lp. plantarum, etc.) have been found to play a key role in sourdough fermentation and showed better ability to increase the dough volume during sourdough bread fermentation. Indeed, Faid et al. (1993) studied LAB interactions in Moroccan sourdough bread fermentation and reported a correlation between the physico-chemical properties (pH, acidity, and CO2) and LAB viable growth. The authors reported that the fermentation of sourdough bread is composed of two steps. First, yeast strains begin to grow, resulting in dough rising activity, and when the dough is saturated with CO2, LAB strains start to grow, leading to acidification in the second step of sourdough bread fermentation. For example, homofermentative (Lp. plantarum) and heterofermentative (Lev. brevis) LAB strains gave best results (pH decrease and dough rising capacity) in combination with the yeasts Sacchoromyces cerevisiae and Candida milleri in wheat flour sourdough, respectively. On the other hand, Lev. brevis play a role in phytate reduction (compared to baker’s yeasts), and is capable to improve the nutritional properties of Barbari bread (Hadaegh et al. 2019). With this LAB strain, 30% of sourdough in bread formulation gave the highest phytate reduction with fermentation time being an important factor.
Regarding P. pentosaceus, it was reported that this strain can improve the quality of gluten-free cereals. According to Olojede et al. (2022), sourdough made with this species contain high crude protein, ash and dietary fiber contents, with significant high levels of total phenol and tannin contents in breads. Moreover, P. pentosaceus showed in vitro antifungal activity against A. flavus and the addition of this strain with S. cerevisiae into sourdough starters positively impacted the volatile compound profiles and contributed to more complex aroma through dough fermentation, which enhances the quality and safety of sourdough bread with high scores for taste, aroma, texture and overall acceptability (Jin et al. 2021). On the other hand, sourdough fermented with Len. parabuchneri was used in bread production and its effect on fungal has been illustrated (Zhang et al. 2010). Thus, the use of starter cultures of this LAB strain allowed the accumulation of substantial amounts of propionic and acetic acids in sourdough, supporting the use of these sourdoughs as biopreservative.
Strains of E. hirae may be considered as starter cultures for baking sourdough. These strains can acidify dough and inhibits undesired microorganisms, such as molds and pathogenic bacteria. It may also play an important role for proteolysis during sourdough fermentation. Free amino acids generation can positively influence the growth of yeasts during dough fermentation and the growth of flavor compounds, in addition to hydrolysis of gluten may influence the rheology of the dough and the texture of the final baked goods (Sana et al. 2012). During bread and dough fermentation, the application of Bifidobacterium strains may contribute positively to the fermentation process by increasing phytate hydrolysis during fermentation owing to the activation of endogenous cereal phytase and its own phytase, resulting in bread with significantly lower levels of the antinutrient phytate (Sanz-Penella et al. 2011). B. pseudocatenulatum is considered a probiotic bacterium and associated with the human host (Sanz-Penella et al. 2011). It was reported that ingestion of B. pseudocatenulatum strain CECT 7765 by high-fat diet-fed obese mice caused positively the reduction in serum cholesterol, triacylglycerols, glucose levels, insulin resistance, and also improved glucose tolerance. Finally, Co. paralimentarius (formerly L. paralimentarius) is one of many genera of bacteria that can be present in sourdoughs. Co. paralimentarius in combination with Lp. plantarum have been frequently reported in many sourdoughs as the sourdough-specific LAB species that predominate in sourdough fermentations (Boyaci Gunduz et al. 2022). Co. paralimentarius favorably contributes to the baked product by providing numerous benefits arising mainly from metabolic processes such as catabolism of amino acids that can generate flavor and aromatic compounds and improving the sensory profile, production of EPS improving bread texture by enzymatic reactions, and extending the shelf-life by releasing antimicrobial compounds (Marco et al. 2022).
Cereal products are a common medium for fungal growth due to their high-water activity and nutrients availability (Mota-Gutierrez et al. 2021). In this study, four isolated LAB strains (Co. paralimentarius S4, Len. parabuchneri S17, Lp plantarum S21, and Len. parabuchneri S23) showed inhibitory effect against A. flavus, while a moderate activity was obtained with the strain Pediococcus pentosaceus S12 against A. flavus and Penicillium spp. Similar results reported that LAB strains isolated from spontaneously fermented hull-less barley sourdough, especially Lactobacillus paralimentarius, Lactobacillus brevis, Pediococcus acidilactici, and Lactobacillus plantarum showed antifungal effects against three mold species of Penicillium carneum, Aspergillus flavus, and A. niger (Çakır et al. 2020). On the other hand, LAB belonging to Lp. plantarum and Lacticaseibacillus casei isolated from spontaneous sourdough fermentation showed a strain-dependent antifungal activity, with a strong antifungal activity obtained by Lp. plantarum (Mota-Gutierrez et al. 2021). These results are also in agreement with those already reported by Muhialdin et al. (2018), Abouloifa et al., (2020) and Ben Salah-Abbès et al. (2021) on the ability of LAB strains to inhibit the growth of various fungal species. Indeed, LAB strains are able to produce multiple antimicrobial compounds, including organic acids (lactic, acetic, citric, formic, and succinic acids, among others), bacteriocins, phenyllactic acid, hydroxy-fatty acids, proteinaceous compounds, reuterin, and many other compounds with antimicrobial effects (Bartkiene et al. 2019; Matevosyan et al. 2020). Thus, LAB strains isolated from sourdough has shown great potential in controlling the growth of spoilage fungi, guarantee quality and prolong the shelf life of fermented bread. Isolated strains presented good multifunctional properties (acidifying capacity; CO2, diacetyl, and EPS production).
Conclusion
The present study showed that Moroccan traditional sourdoughs can be considered as a good biotope for LAB strains. Seven LAB species (Lev. brevis, Len. parabuchneri, Lp. plantarum, P. pentosaceus, E. hirae, B. pseudocatenulatum, and Co. paralimentarius) were identified for the first time in Moroccan sourdoughs. These strains presented good biotechnological properties (acidifying capacity; CO2, diacetyl, and EPS production, and antifungal activity) that can boost microbial safety, offer many features such as organoleptic, technological, nutritional or health advantages. Based on the results of this study, the isolated LAB will be combined as starter cultures (in association with baker’s yeast and some local aromatic plants) for the manufacture of enriched fermented bread at the laboratory scale. The microbiological, physico-chemical, nutritional and sensorial properties of the novel produced bread will then be completely assessed.
Data availability
The authors declare data transparency.
References
Abouloifa H, Gaamouche S, Rokni Y et al (2021) Antifungal activity of probiotic Lactobacillus strains isolated from natural fermented green olives and their application as food bio-preservative. Biol Control 152:104450. https://doi.org/10.1016/j.biocontrol.2020.104450
Abouloifa H, Rokni Y, Bellaouchi R et al (2020) Characterization of probiotic properties of antifungal Lactobacillus strains isolated from traditional fermenting green olives. Probiotics Antimicrob Proteins 12:683–696. https://doi.org/10.1007/s12602-019-09543-8
Badji T, Durand N, Bendali F et al (2023) In vitro detoxification of aflatoxin B1 and ochratoxin A by lactic acid bacteria isolated from Algerian fermented foods. Biol Control 179:105181. https://doi.org/10.1016/j.biocontrol.2023.105181
Bartkiene E, Lele V, Sakiene V et al (2019) Improvement of the antimicrobial activity of lactic acid bacteria in combination with berries/fruits and dairy industry by-products. J Sci Food Agric 99:3992–4002. https://doi.org/10.1002/jsfa.9625
Bartkiene E, Lele V, Ruzauskas M et al (2020) Lactic acid bacteria isolation from spontaneous sourdough and their characterization including antimicrobial and antifungal properties evaluation. Microorganisms 8:64. https://doi.org/10.3390/microorganisms8010064
Ben Salah-Abbès J, Mannai M, Belgacem H et al (2021) Efficacy of lactic acid bacteria supplementation against Fusarium graminearum growth in vitro and inhibition of zearalenone causing inflammation and oxidative stress in vivo. Toxicon 202:115–122. https://doi.org/10.1016/j.toxicon.2021.09.010
Bennani S, Mchiouer K, Rokni Y et al (2017) Characterisation and identification of lactic acid bacteria isolated from Moroccan raw cow’s milk. J Mater Environ Sci 8:4934–4944
Bom S, Jagan L, Rao M et al (2018) Isolation of lactic acid bacteria starters from jeung - pyun for sourdough fermentation. Food Sci Biotechnol 27:73–78. https://doi.org/10.1007/s10068-017-0274-0
Boraam F, Faid M, Larpent J et al (1993) Lactic acid bacteria and yeasts associated with traditional Moroccan sour-dough bread fermentation. Sci Aliment 13:501–509
Boyaci Gunduz CP, Agirman B, Gaglio R et al (2022) Evaluation of the variations in chemical and microbiological properties of the sourdoughs produced with selected lactic acid bacteria strains during fermentation. Food Chem X 14:100357. https://doi.org/10.1016/j.fochx.2022.100357
Brandt MJ (2019) Industrial production of sourdoughs for the baking branch: an overview. Int J Food Microbiol 302:3–7. https://doi.org/10.1016/j.ijfoodmicro.2018.09.008
Çakır E, Arıcı M, Durak MZ (2020) Biodiversity and techno-functional properties of lactic acid bacteria in fermented hull-less barley sourdough. J Biosci Bioeng 130:450–456. https://doi.org/10.1016/j.jbiosc.2020.05.002
Carbonetto B, Nidelet T, Guezenec S et al (2020) Interactions between kazachstania humilis yeast species and lactic acid bacteria in sourdough. Microorganisms 8:240. https://doi.org/10.3390/microorganisms8020240
Chaoui A, Faid M, Belahsen R (2006) Making bread with sourdough improves iron bioavailability from reconstituted fortified wheat flour in mice. J Trace Elem Med Biol 20:217–220. https://doi.org/10.1016/j.jtemb.2006.04.002
Cizeikiene D, Jagelaviciute J, Stankevicius M et al (2020) Thermophilic lactic acid bacteria affect the characteristics of sourdough and whole-grain wheat bread. Food Biosci 38:100791. https://doi.org/10.1016/j.fbio.2020.100791
Damiani P, Gobbetti M, Cossignani L et al (1996) The sourdough microflora. characterization of hetero- and homofermentative lactic acid bacteria, yeasts and their interactions on the basis of the volatile compounds produced. LWT - Food Sci Technol 29:63–70. https://doi.org/10.1006/fstl.1996.0009
Faid M, Boraam F, Achbab A et al (1993) Yeast-lactic acid bacteria interactions in Moroccan Sour-dough Bread Fermentation. LWT - Food Sci Technol 26:443–446. https://doi.org/10.1006/fstl.1993.1087
Faid M, Boraam F, Zyani I et al (1994) Characterization of sourdough bread ferments made in the laboratory by traditional methods. Z Lebensm Unters Forsch 198:287–291. https://doi.org/10.1007/BF01193176
Fekri A, Torbati M, Yari Khosrowshahi A et al (2020) Functional effects of phytate-degrading, probiotic lactic acid bacteria and yeast strains isolated from iranian traditional sourdough on the technological and nutritional properties of whole wheat bread. Food Chem 306:125620. https://doi.org/10.1016/j.foodchem.2019.125620
Felicia C (2015) The technological evaluation of sourdoughs prepared in different conditions. Manag Sustain Dev 7:33–36. https://doi.org/10.1515/msd-2015-0019
Fraberger V, Ammer C, Domig KJ (2020) Functional properties and sustainability improvement of sourdough bread by lactic acid bacteria. Microorganisms 8:1895
Fujimoto A, Ito K, Narushima N et al (2019) Identification of lactic acid bacteria and yeasts, and characterization of food components of sourdoughs used in Japanese bakeries. J Biosci Bioeng 127:575–581. https://doi.org/10.1016/j.jbiosc.2018.10.014
Gänzle MG, Zheng J (2019) Lifestyles of sourdough lactobacilli: do they matter for microbial ecology and bread quality? Int J Food Microbiol 302:15–23. https://doi.org/10.1016/j.ijfoodmicro.2018.08.019
Gobbetti M, Corsetti A, Rossi J (1995) Interaction between lactic acid bacteria and yeasts in sour-dough using a rheofermentometer. World J Microbiol Biotechnol 11:625–630. https://doi.org/10.1007/BF00361004
Hadaegh H, Ardabili SMS, Ebrahimi MT et al (2019) Effect of using Lactobacillus Brevis IBRC- M10790 and Baker’s yeast on Phytate degradation of sourdough and Barbari bread. J Food Biosc Technol 9:11–18
Ho VTT, Fleet GH, Zhao J (2018) Unravelling the contribution of lactic acid bacteria and acetic acid bacteria to cocoa fermentation using inoculated organisms. Int J Food Microbiol 279:43–56. https://doi.org/10.1016/j.ijfoodmicro.2018.04.040
Ismaili Alaoui M, Hamama A, Saidi B et al (2017) Chemical Composition, microbial profile and identification of lactic acid bacteria of moroccan fermented camel milk “Lfrik.” Curr Res Nutr Food Sci 5:383–390. https://doi.org/10.12944/CRNFSJ.5.3.24
İspirli H, Özmen D, Yılmaz MT et al (2020) Impact of glucan type exopolysaccharide (EPS) production on technological characteristics of sourdough bread. Food Control 107:106812. https://doi.org/10.1016/j.foodcont.2019.106812
Jin J, Nguyen TTH, Humayun S et al (2021) Characteristics of sourdough bread fermented with Pediococcus pentosaceus and Saccharomyces cerevisiae and its bio-preservative effect against Aspergillus flavus. Food Chem 345:128787. https://doi.org/10.1016/j.foodchem.2020.128787
Kaya Y, Erten T, Vurmaz M et al (2022) Comparison of the probiotic characteristics of lactic acid bacteria isolated from sourdough and infant feces. Food Biosci 47:101722. https://doi.org/10.1016/j.fbio.2022.101722
Khedid K, Faid M, Mokhtari A et al (2009) Characterization of lactic acid bacteria isolated from the one humped camel milk produced in Morocco. Microbiol Res 164:81–91. https://doi.org/10.1016/j.micres.2006.10.008
Marco I, Silva C, Moraes M et al (2022) A systematic review of drying methods and their impact on technological characteristics of sourdough type III. Biotechnol Res Innov 6:1–17. https://doi.org/10.4322/biori.202203
Matevosyan LA, Bazukyan IL, Trchounian AH (2020) Antifungal activity of lactic acid bacteria isolates and their associations: The effects of Ca and Mg divalent cations. Curr Microbiol 77:959–966. https://doi.org/10.1007/s00284-020-01897-5
Mota-Gutierrez J, Franciosa I, Ruggirello M, Dolci P (2021) Technological, functional and safety properties of lactobacilli isolates from soft wheat sourdough and their potential use as antimould cultures. World J Microbiol Biotechnol 37:146. https://doi.org/10.1007/s11274-021-03114-2
Moujabbir S, Aboukhalaf A, Kalili A et al (2023) Sourdoughs used in the preparation of traditional bread in the province of figuig in eastern Morocco. Int J Food Stud 12:71–83. https://doi.org/10.7455/ijfs/12.1.2023.a5
Muhialdin BJ, Hassan Z, Saari N (2018) In vitro antifungal activity of lactic acid bacteria low molecular peptides against spoilage fungi of bakery products. Ann Microbiol 68:557–567. https://doi.org/10.1007/s13213-018-1363-x
Nionelli L, Montemurro M, Pontonio E et al (2018) Pro-technological and functional characterization of lactic acid bacteria to be used as starters for hemp (Cannabis sativa L.) sourdough fermentation and wheat bread fortification. Int J Food Microbiol 279:14–25. https://doi.org/10.1016/j.ijfoodmicro.2018.04.036
Novotni D, Gänzle M, Rocha JM (2021) Composition and activity of microbiota in sourdough and their effect on bread quality and safety. In: Galanakis CM (ed) Trends in wheat and bread making. Elsevier, Amsterdam, pp 129–172. https://doi.org/10.1016/B978-0-12-821048-2.00005-2
Olojede AO, Sanni AI, Banwo K (2022) Improvement of texture nutritional qualities and consumers perceptions of sorghum-based sourdough bread made with Pediococcus pentosaceus and Weissella confusa Strains. Fermentation 8:32. https://doi.org/10.3390/fermentation8010032
Păcularu-Burada B, Georgescu LA, Vasile MA et al (2020) Selection of wild lactic acid bacteria strains as promoters of postbiotics in gluten-free sourdoughs. Microorganisms 8:643. https://doi.org/10.3390/microorganisms8050643
Pelin C, Gunduz B, Gaglio R et al (2020) Molecular analysis of the dominant lactic acid bacteria of chickpea liquid starters and doughs and propagation of chickpea sourdoughs with selected Weissella confusa. J Food Microbiol 91:103490. https://doi.org/10.1016/j.fm.2020.103490
Reale A, Di Renzo T, Boscaino F et al (2019) Lactic acid bacteria biota and aroma profile of italian traditional sourdoughs from the irpinian area in Italy. Front Microbiol. https://doi.org/10.3389/fmicb.2019.01621
Rocha JM, Brás A, Miranda J, Malcata FX (2023) Broa: a Portuguese traditional sourdough bread, made of maize and rye flours. In: Garcia-Vaquero M, Pastor K, Orhun GE, McElhatton A, Rocha JMF (eds) Traditional European Breads. Springer, Cham, pp 251–293. https://doi.org/10.1007/978-3-031-23352-4_13
Rocha JM (2011) Microbiological and lipid profiles of broa: contributions for the characterization of a traditional portuguese bread. Thesis Doctoral, University of Lisbon
Rocha JM, Malcata FX (1999) On the microbiological profile of traditional portuguese sourdough. J Food Prot 62:1416–1429. https://doi.org/10.4315/0362-028X-62.12.1416
Rocha JM, Malcata FX (2016) Microbial ecology dynamics in portuguese broa sourdough. J Food Qual 39:634–648. https://doi.org/10.1111/jfq.12244
Rocha JM, Malcata FX (2012) Microbiological profile of maize and rye flours, and sourdough used for the manufacture of traditional portuguese bread. Food Microbiol 31:72–88. https://doi.org/10.1016/j.fm.2012.01.008
Rokni Y, Abouloifa H, Bellaouchi R et al (2021) Characterization of β-glucosidase of Lactobacillus plantarum FSO1 and Candida pelliculosa L18 isolated from traditional fermented green olive. J Genet Eng Biotechnol 19:117. https://doi.org/10.1186/s43141-021-00213-3
Sana M, Minervini F, Cagno R (2012) Technological, functional and safety aspects of enterococci in fermented vegetable products : a mini-review. Ann Microbiol 62:469–481. https://doi.org/10.1007/s13213-011-0363-x
Sanz-Penella JM, Tamayo-Ramos JA, Haros M (2011) Application of bifidobacteria as starter culture in whole wheat sourdough breadmaking. Food Bioprocess Technol 5:2370–2380. https://doi.org/10.1007/s11947-011-0547-1
Wang D, Liu W, Ren Y et al (2016) Isolation and identification of lactic acid bacteria from traditional dairy products in Baotou and Bayannur of Midwestern inner Mongolia and q-PCR analysis of predominant species. Korean J Food Sci Anim Resour 36:499–507. https://doi.org/10.5851/kosfa.2016.36.4.499
Zhang C, Brandt MJ, Schwab C, Gänzle MG (2010) Propionic acid production by cofermentation of Lactobacillus buchneri and Lactobacillus diolivorans in sourdough. Food Microbiol 27:390–395. https://doi.org/10.1016/j.fm.2009.11.019
Zotta T, Piraino P, Parente E et al (2008) Characterization of lactic acid bacteria isolated from sourdoughs for Cornetto, a traditional bread produced in Basilicata (Southern Italy). World J Microbiol Biotechnol 24:1785–1795. https://doi.org/10.1007/s11274-008-9671-0
Acknowledgements
This research was funded by the project “PHC Maghreb (09MAG20)”. The authors would like to acknowledge the CNRST and the Ministry MESRSI (Morocco) for the grant given and the CIRAD of Montpellier (France) for the technical facilities. The authors would like also to acknowledge the COST Action 18101 SOURDOMICS-Sourdough biotechnology network towards novel, healthier and sustainable food and bioprocesses (https://sourdomics.com/; https://www.cost.eu/actions/CA18101/, accessed on 31st July 2023), where the author A. Zinedine is member of the working groups 2, 3, 4, 5, 7, and 8, and the author J.M. Rocha is the Chair and Grant Holder Scientific Representative and is supported by COST (European Co-operation in Science and Technology) (https://www.cost.eu/, accessed on 31st July 2023). COST is a funding agency for research and innovation networks. Author J.M. Rocha also acknowledges the Universidade Católica Portuguesa, CBQF—Centro de Biotecnologia e Química Fina—Laboratório Associado, Escola Superior de Biotecnologia, Porto, Portugal, as well as the support made by LA/P/0045/2020 (ALiCE) and UIDB/00511/2020-UIDP/00511/2020 (LEPABE) funded by national funds through FCT/MCTES (PIDDAC).
Author information
Authors and Affiliations
Contributions
Material preparation, data collection and analysis were performed by MB, AA, and NM. The first draft of the manuscript was written by MB and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals experiments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
EL Boujamaai, M., Mannani, N., Aloui, A. et al. Biodiversity and biotechnological properties of lactic acid bacteria isolated from traditional Moroccan sourdoughs. World J Microbiol Biotechnol 39, 331 (2023). https://doi.org/10.1007/s11274-023-03784-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03784-0