Abstract
l-Glutamate decarboxylase (GAD) transforms l-glutamate into γ-aminobutyric acid (GABA). Corynebacterium glutamicum that expresses exogenous GAD gene(s) can synthesize GABA from its own produced l-glutamate. To enhance GABA production in recombinant C. glutamicum strain SH, metabolic engineering strategies were used to improve the supply of the GABA precursor, l-glutamate. Five new strains were constructed here. First, the ppc gene was coexpressed with two GAD genes (gadB1 and gadB2). Then, the mdh gene was deleted in C. glutamicum SH. Next, gadB1-gadB2 and gadB1-gadB2-ppc co-expression plasmids were transformed into C. glutamicum strains SH and Δmdh, resulting in four recombinant GAD strains SE1, SE2, SDE1, and SDE2, respectively. Finally, the mdh gene was overexpressed in mdh-deleted SDE1, generating the mdh-complemented GAD strain SDE3. After fermenting for 72 h, GABA production increased to 26.3 ± 3.4, 24.8 ± 0.7, and 25.5 ± 3.3 g/L in ppc-overexpressed SE2, mdh-deleted SDE1, and mdh-deleted ppc-overexpressed SDE2, respectively, which was higher than that in the control GAD strain SE1 (22.7 ± 0.5 g/L). While in the mdh-complemented SDE3, GABA production decreased to 20.0 ± 0.6 g/L. This study demonstrates that the recombinant strains SE2, SDE1, and SDE2 can be used as candidates for GABA production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gamma-aminobutyric acid (GABA), a non-protein amino acid, is a well-characterized inhibitory neurotransmitter in mammals and has been applied as a bioactive component in functional foods, feeds, and pharmaceuticals due to its various physiological functions, including hypotensive, anti-anxiety, tranquilising, analgesic, and diuretic effects and regulation of hormones and cells (Mohler 2012; Diana et al. 2014; Shi et al. 2016). In addition to its bioactivities beneficial to mammals, GABA also involves in acid-resistance in some bacteria such as Listeria monocytogenes (Karatzas et al. 2010) and environmental stress-resistance in plants (Bouche and Fromm 2004). Furthermore, GABA has been used as a major building block for the synthesis of 2-pyrrolidone and biodegradable polyamide nylon 4 (Park et al. 2013).
Naturally, GABA is synthesized from l-glutamate by l-glutamate decarboxylase (GAD) in certain species (Li and Cao 2010; Shi et al. 2016). GAD genes (gad) are present in Escherichia coli (De Biase et al. 1996), Lactobacillus brevis (Hiraga et al. 2008), and several other species of Lactobacillus as well as Enterobacteria. However, during the production of GABA by strains of these species, l-glutamate must be added as the precursor (Zhao et al. 2016). Such bioprocess is not cost-effective, thus it is necessary to establish a new approach for sustainable industrial production of GABA. Corynebacterium glutamicum is a gram-positive actinobacterium widely used for the industrial production of l-glutamate and other amino acids (Hermann 2003; Leuchtenberger et al. 2005). By expressing exogenous GAD gene(s), C. glutamicum could be engineered to synthesize GABA from its own accumulated l-glutamate (Shi and Li 2011; Takahashi et al. 2012). Recently, through co-expressing two GAD genes (gadB1 and gadB2) derived from L. brevis in C. glutamicum ATCC13032 and optimizing urea supplementation, GABA production increased to 18.7 ± 2.1 g/L (Shi et al. 2013). To improve GABA production, a more robust l-glutamate-producing strain shall be used as the host. Furthermore, l-glutamate biosynthesis must be enhanced because l-glutamate is the direct precursor of GABA.
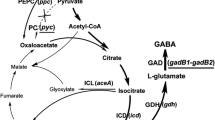
The biosynthetic pathway of l-glutamate and GABA in recombinant C. glutamicum is shown in Fig. 1. l-Glutamate is synthesized from glucose via glycolysis pathway, anaplerotic pathway, first half of tricarboxylic acid (TCA) cycle, and l-glutamate dehydrogenase (GDH); among them phosphoenolpyruvate (PEP)-pyruvate-oxaloacetate (OAA) node works as the switch point for carbon flux distribution (Sauer and Eikmanns 2005). OAA supply determines the flux flowing into the TCA cycle. In addition, OAA is continuously withdrawn from TCA cycle during l-glutamate and GABA production. But OAA cannot be regenerated efficiently via the TCA cycle because GDH activity increases and 2-oxoglutarate dehydrogenase complex specific activity decreases during l-glutamate production (Hirasawa and Wachi 2016). Therefore, the anaplerotic pathway must be enhanced for efficient OAA supply and l-glutamate and GABA overproduction. In C. glutamicum, two anaplerotic enzymes, phosphoenolpyruvate carboxylase (PEPC) and pyruvate carboxylase (PC), mainly generate OAA, from PEP and pyruvate, respectively (Peters-Wendisch et al. 1998). Meanwhile, CO2 released via the TCA cycle can be assimilated by these two anaplerotic reactions. However, PC, a biotin-requiring enzyme, may not function well during l-glutamate fermentation under biotin limitation (Shirai et al. 2007). Thus, PEPC may be crucial for OAA supply. In addition to OAA generation, OAA can be reduced to malate by malate dehydrogenase (MDH), the last enzyme of TCA cycle. C. glutamicum possesses two types of MDH, a membrane-associated malate:quinone oxidoreductase (MQO) and a cytoplasmic MDH (Molenaar et al. 2000). MQO catalyzes the oxidation of malate to OAA, whereas MDH catalyzes the opposite reaction.
In the present study, a more robust l-glutamate-producing industrial strain, C. glutamicum SH, was used for expressing gadB1-gadB2 and producing GABA. To enhance OAA supply in recombinant C. glutamicum SH strains, ppc gene which encodes the PEPC was co-overexpressed with gadB1-gadB2 and mdh gene which encodes the MDH was deleted. Their influence on GABA fermentation was then researched.
Materials and methods
Strains, media, and growth conditions
The strains and plasmids used in this study are listed in Table 1. E. coli JM 109 was used as the host for constructing and propagating the plasmids. E. coli was grown in Luria–Bertani (LB) medium at 37 °C and 200 rpm. Corynebacterium glutamicum SH, an l-glutamate-producing strain, was used for mdh gene deletion, expressing gadB1-gadB2 and ppc genes and producing GABA. SH was deposited in the China General Microbiological Culture Collection (CGMCC) center with accession number CGMCC 1.581. C. glutamicum was grown in LBG medium (LB supplemented with 5 g/L glucose) at 200 rpm and 30 °C. When necessary, 30 μg/mL kanamycin or 15 μg/mL chloramphenicol was added to the media.
Construction of gadB1-gadB2 and gadB1-gadB2-ppc co-expression strains
The primers used in this study are listed in Table 2. Firstly, gadB1-gadB2 genes were amplified via PCR from the plasmid pDXW-10-gadB1-gadB2 (Shi et al. 2013) using the primer pair of B1F and B2R. The PCR product was digested with AflII and BamHI, and ligated into pJYW-4 (Hu et al. 2014) that was similarly digested, resulting in the plasmid pJYW-4-gadB1-gadB2. Secondly, ppc gene was PCR amplified using the primer pair of ppc-F and ppc-R from genomic DNA of C. glutamicum SH. The PCR product was digested with NotI and SalI, and ligated into the pJYW-4 that was similarly digested, resulting in the plasmid pJYW-4-ppc. Next, the DNA fragment containing the tac-M promoter and ppc gene was PCR amplified from pJYW-4-ppc using primer pair of ppc-tacM-F and ppc-R, digested with SalI, and ligated into pJYW-4-gadB1-gadB2 that was similarly digested, resulting in the plasmid pJYW-4-gadB1-gadB2-ppc. Finally, the two plasmids were transformed into C. glutamicum SH by the method described previously (Wang et al. 2015), generating two recombinant strains, C. glutamicum SH/pJYW-4-gadB1-gadB2 and C. glutamicum SH/pJYW-4-gadB1-gadB2-ppc, renamed as SE1 and SE2, respectively.
Deletion of mdh gene in C. glutamicum
Based on the homologous recombination and site-specific recombination system, gene deletion in C. glutamicum SH was performed according to a previous study (Hu et al. 2013). First, deletion plasmid was constructed as follows. Fragment mdhU, located upstream of the gene mdh, and fragment mdhD, located downstream of the gene mdh, were amplified from the genomic DNA of C. glutamicum SH using the primer pairs mdhU-F/mdhU-R and mdhD-F/mdhD-R, respectively; Fragment loxL-kan-loxR, which contained two recognition sites (loxL and loxR) by Cre, was amplified from plasmid pDTW202 using the primer pair kan-F/kan-R. The three DNA fragments were ligated into the XbaI and XhoI restriction sites of plasmid pBluscriptII SK(+), resulting in plasmid pSD. Second, plasmid pSD was transformed into C. glutamicum SH to allow for homologous recombination. The recombinant strains were selected by growth on LBHIS medium (Wang et al. 2015) supplemented with 30 μg/mL kanamycin. Third, plasmid pDTW-109 was transformed into the recombinant strain to remove the kan gene out of the chromosome, and selected on LBHIS agar containing 15 μg/mL chloramphenicol. Finally, cells were cultured at 37 °C to remove the pDTW-109 plasmid. The cells that grew on the plate without antibiotics but not on the plate with kanamycin or chloramphenicol were chosen as the mutant strain SH Δmdh, renamed SD. The strains were verified by restriction enzyme digestion or target gene amplification.
The mdh complemented plasmid was constructed as follow. The mdh gene was PCR amplified using the primer pair of mdh-F and mdh-R from genomic DNA of C. glutamicum SH, then digested with BamHI and SalI, and ligated into pJYW-4-gadB1-gadB2 that was similarly digested, resulting in the plasmid pJYW-4-gadB1-gadB2-mdh. The three plasmids, pJYW-4-gadB1-gadB2, pJYW-4-gadB1-gadB2-ppc, and pJYW-4-gadB1-gadB2-mdh were finally transformed into the mdh deletion mutant, generating the new recombinant C. glutamicum strains, Δmdh/pJYW-4-gadB1-gadB2, Δmdh/pJYW-4-gadB1-gadB2-ppc, and Δmdh/pJYW-4-gadB1-gadB2-mdh, renamed as SDE1, SDE2, and SDE3, respectively.
GABA fermentation of recombinant C. glutamicum strains in a fermentor
l-Glutamate and GABA production in recombinant C. glutamicum strains were evaluated in a fermentor (BioFlo 110; New Brunswick Scientific, USA) as described previously (Wang et al. 2015) with a little modification. The pH was controlled at 7.0–7.5 by adding concentrated ammonium hydroxide at 0–33 h, declined spontaneously to 5.5 at 33–36 h, and then controlled at 5.0–5.5 by adding 2 M HCl at 36–72 h. After 12 h of fermentation, glucose was fed by a peristaltic pump when the residual glucose in the medium decreased below 20 g/L. Samples were taken every 12 h to determine the optical density at 562 nm (OD562), residual glucose level and amino acids concentrations. Residual glucose concentration was determined using a biosensor (Institute of Biology, Shandong Academy of Science, China). GABA and l-glutamate concentrations were assayed using reversed-phase high-performance liquid chromatography (HPLC, Agilent 1200, USA) by the method described previously (Shi et al. 2013).
Extraction of crude enzyme and assay of GAD, MDH, and PEPC activity
At 24 and 42 h of fermentation, the fermenting cells of C. glutamicum strains were collected, washed, and the crude enzyme was extracted as described previously by Shi et al. (2013). GAD activity was determined at 42 h by measuring the formation of GABA in a GAD reaction (Shi et al. 2013). MDH activity was determined at 24 h by monitoring the oxidation of NADH to NAD+ accompanied by the reduction of OAA to malate in a MDH reaction (Mansouri et al. 2017). PEPC activity was determined at 24 h by monitoring the oxidation of NADH to NAD+ in a PEPC–MDH coupling reaction (Cheng et al. 2016). The formation of GABA was analyzed using reversed-phase HPLC. The oxidation rate of NADH to NAD+ was measured spectrophotometrically at 340 nm with a Shimadzu UV-1800 spectrophotometer. One unit (U) of enzymatic activity is defined as 1.0 µmol GABA produced (for GAD) or NADH consumed (for MDH and PEPC) in 1 min at 30 °C in the initial reaction mixture. The specific activity is expressed as U/mg or U/g of protein.
Results
To improve OAA supply and GABA production in recombinant C. glutamicum SH, the ppc gene was overexpressed and mdh gene was deleted. First, the mdh deletion plasmid pSD was constructed and then the mdh deletion strain SD was generated. Second, gadB1-gadB2, gadB1-gadB2-ppc, and gadB1-gadB2-mdh co-expressing plasmids were constructed and then transformed into wild type SH and mdh-deleted SD stains, generating five new recombinant GAD strains, the control SE1, ppc-overexpressed SE2, mdh-deleted SDE1, mdh-complemented SDE3, and mdh-deleted ppc-overexpressed SDE2. The five new recombinant C. glutamicum strains were fermented and production of l-glutamate and GABA were investigated.
The fermentation process of recombinant C. glutamicum in a fermentor
The optimum pH for cell growth and GDH activity of C. glutamicum is 7.0–7.5, whereas that for GAD activity is 4.5–5.0. Meanwhile, more than half of GDH activity is lost at pHs lower than 6.5 (Shiio and Ozaki 1970) and nearly all of the GAD activity is lost at pHs higher than 6.0 (Shi et al. 2014). For effective production of GABA in recombinant C. glutamicum SH strains, pH was controlled at 7.0–7.5 during the first stage of fermentation by adding concentrated ammonium hydroxide to ensure cell growth and l-glutamate biosynthesis and later adjusted to 5.0–5.5 during the last stage of fermentation by adding 2 M HCl to ensure GAD activity and conversion of l-glutamate to GABA (Wang et al. 2015). To control the optimum pH of these two stages, recombinant C. glutamicum SH strains were fermented in fermentor.
During fermentation, all five strains grew fast in the first 12 h, slowly thereafter and did not grow after 36 h (Fig. 2a). Before 36 h, the growth rate of the control GAD strain SE1 and mdh-complemented GAD strain SDE3 was a little lower than those of ppc-overexpressed SE2, mdh-deleted SDE1, and mdh-deleted ppc-overexpressed SDE2; while after 36 h, cell concentration of SE1 became stable, whereas those of SE2, SDE1, SDE2, and SDE3 decreased slightly. Finally, cell concentrations of the five strains were nearly the same. So overexpression of ppc and deletion or complement of mdh did not influence cell growth significantly. In addition, glucose consumption patterns of all the five strains were similar. Glucose was consumed quickly and nearly completely in the first 24 h (Fig. 2b), in accordance with the fast cell growth (Fig. 2a) and quick l-glutamate biosynthesis at this stage (Fig. 3a). After 24 h, additional glucose was added when the residual glucose in the medium was lower than 20 g/L and glucose was consumed continuously and slowly thereafter, resulting in the continuous and relatively slower l-glutamate biosynthesis during 24–36 h (Fig. 3a).
Effect of ppc overexpression and mdh deletion on l-glutamate and GABA biosynthesis in recombinant C. glutamicum
During the fermentation of five recombinant C. glutamicum SH strains, the accumulation trends of l-glutamate and GABA were similar (Fig. 3). l-Glutamate accumulated quickly during 12–36 h when pH was controlled at 7.0–7.5 and then decreased quickly and converted nearly completely to GABA during 36–60 h when pH was controlled at 5.0–5.5. But their production was different. In the control GAD strain SE1, the l-glutamate concentration increased to the highest value 45.5 ± 0.7 g/L at 36 h, decreased sharply to 2.4 ± 0.8 g/L until 60 h and increased slightly during the final 12 h. Accordingly, GABA concentration increased continuously from 36 to 60 h and maintained thereafter. Finally, 22.7 ± 0.5 g/L GABA accumulated at 72 h.
In the ppc overexpressed GAD strain SE2, the PEPC activity increased by 5.4-fold and the GAD activity decreased slightly by 4.2% (Table 3). The l-glutamate concentration increased to the highest value 46.4 ± 5.1 g/L at 36 h, which was slightly higher than that in SE1, and decreased continuously to 9.0 ± 2.6 g/L thereafter. In accordance, GABA concentration increased continuously from 36 to 72 h and 26.3 ± 3.4 g/L GABA accumulated at 72 h, 16% higher than that in SE1, indicating that overexpression of ppc was benefit for GABA biosynthesis. Moreover, no significant amounts of other amino acids were detected in the final fermentation product, indicating that SE2 can be a good candidate for industrial production of GABA because of the easy downstream separation and purification of GABA.
In the mdh deleted GAD strain SDE1, the MDH activity decreased completely and the GAD activity decreased slightly by 7.1% (Table 3). The l-glutamate concentration increased to the highest value 47.0 ± 1.6 g/L at 36 h, which was slightly higher than that in SE1, and decreased sharply and converted completely to GABA thereafter. Accordingly, the GABA concentration increased continuously to 24.8 ± 0.7 g/L during 36–72 h, 9% higher than that in SE1, indicating that deletion of mdh was also benefit for GABA biosynthesis.
In the mdh complemented GAD strain SDE3, the MDH activity increased by 14.1-fold and the GAD activity decreased by 9.9% (Table 3). The l-glutamate concentration increased to the highest value 36.8 ± 1.1 g/L at 36 h, which was obviously lower than that in SE1, and then decreased sharply and converted nearly completely to GABA during 36–60 h and maintained thereafter. The GABA concentration increased to 20.0 ± 0.6 g/L during 36–48 h and fluctuated thereafter, 12% lower than that in SE1, indicating that complement of mdh was not benefit for GABA biosynthesis.
In the mdh deleted and ppc overexpressed GAD strain SDE2, the MDH activity decreased completely, the PEPC activity increased by 5.4-fold and the GAD activity decreased by 11.1% (Table 3). The l-glutamate concentration increased to the highest value 46.3 ± 0.8 g/L at 36 h, which was slightly higher than that in SE1 and was comparable to those in ppc overexpressed SE2 and mdh deleted SDE1, then decreased sharply and converted nearly completely to GABA during 36–60 h and maintained thereafter. Meanwhile, GABA concentration increased much quickly to 25.7 ± 0.4 g/L during 36–48 h and maintained at 25.7–25.5 g/L thereafter, 12% higher than that in SE1 but nearly same with those in ppc overexpressed SE2 and mdh deleted SDE1, indicating that the beneficial effects of ppc overexpression and mdh deletion on GABA biosynthesis were not synergistic.
Discussion
In this study, ppc was overexpressed and mdh was deleted in recombinant C. glutamicum SH expressing gadB1-gadB2 genes. The l-glutamate concentration increased to 46.4 ± 5.1 and 47.0 ± 1.6 g/L at 36 h in the ppc-overexpressed GAD strain SE2 and mdh-deleted GAD strain SDE1, respectively, slightly higher than that in the control GAD strain SE1 (45.5 ± 0.7 g/L), whereas in mdh-complemented GAD strain SDE3, l-glutamate concentration decreased to 36.8 ± 1.1 g/L at 36 h (Fig. 3a), indicating that l-glutamate accumulation was improved slightly after PEPC activity increased or MDH activity lost, but weakened obviously after MDH activity increased. Meanwhile, GABA concentration increased to 26.3 ± 3.4 and 24.8 ± 0.7 g/L at 72 h in ppc-overexpressed SE2 and mdh-deleted SDE1, respectively, 16 and 9% higher than that in SE1 (22.7 ± 0.5 g/L), whereas in mdh-complemented SDE3, GABA concentration decreased to 20.0 ± 0.6 g/L at 72 h (Fig. 3b), indicating that GABA biosynthesis was improved obviously after ppc was overexpressed or mdh was deleted, but weakened obviously after mdh was complemented. In addition, there are not significant amounts of other amino acids being detected in the final fermentation product of SE2 and SDE1, indicating them as the good candidates for industrial production of GABA. However, the l-glutamate concentration at 36 h (46.3 ± 0.8 g/L) and GABA concentration at 72 h (25.5 ± 3.3 g/L) in the mdh-deleted and ppc-overexpressed GAD strain SDE2 did not increase further, suggesting that ppc overexpression and mdh deletion did not improve GABA biosynthesis synergistically.
ppc gene overexpression and mdh gene deletion enhanced l-glutamate production at 24 h (Fig. 3a), in accordance with the increased PEPC activity and the lost MDH activity; however, such an effect was not observed at 36 h, perhaps due to the improper condition for these enzymes and cellular metabolism. C. glutamicum is a moderate alkaliphile (Barriuso-Iglesias et al. 2008). Meanwhile, the optimum pH of PEPC, MDH, and GDH is 7.5, 9.5, and 7.5, respectively. The pH of fermentation broth decreased spontaneously from 7.3 to 5.5 during 33–36 h of fermentation, therefore might make these enzymes inactive and cellular metabolism slow. At the last stage of fermentation when pH was controlled at 5.0–5.5, ppc gene overexpression and mdh gene deletion enhanced GABA production at 48 h; however, such an effect was not observed at 60 or 72 h (Fig. 3b), likely due to the more serious cellular damage in SE2 and SDE1, as the cell concentration of SE2 and SDE1 decreased obviously after 48 h. Therefore, GABA should be produced as quickly as possible.
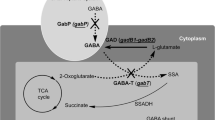
OAA is the precursor metabolite for l-glutamate and GABA biosynthesis and it is mainly supplied by anaplerotic pathway. It has been demonstrated that between the two anaplerotic enzymes, the activity of PC is diminished under biotin-limited condition, and deletion of pyc which encodes PC do not affect l-glutamate production under biotin limited condition (Sato et al. 2008). In the present study, l-glutamate and GABA was produced under corn steep liquor limited condition and the active component of corn steep liquor is biotin. Our recent research showed that deletion of pyc improved l-glutamate and GABA production under corn steep liquor limitation, partially due to the increased transcription level of ppc (Wang et al. 2015). Here, after ppc was overexpressed and PEPC activity was enhanced, l-glutamate production improved slightly and finally GABA production increased by 16%, demonstrating again the beneficial effect of ppc overexpression on l-glutamate and GABA biosynthesis.
On the other hand, to prevent OAA reduction and provide more OAA precursor for GABA formation, the cytosolic reductive pathway of OAA was blocked, as predicted by the analysis of a genome-scale metabolic network of C. glutamicum S9114 (Mei et al. 2016). After mdh was deleted and MDH activity lost, l-glutamate production improved a little and finally GABA production increased by 9%, demonstrating the contribution of mdh deletion on l-glutamate and GABA biosynthesis. But the increment of GABA production was not as high as that in ppc overexpressed GAD strain SE2, likely due to the fact that CO2 released by the decarboxylation of l-glutamate to GABA as well as via the TCA cycle can be assimilated and fixed more efficiently by PEPC when ppc was overexpressed, but can not when mdh was deleted.
After simultaneous overexpression of ppc and deletion of mdh, GABA production increased only by 12% and this production was not higher than that of ppc-overexpressed GAD strain SE2, suggesting that the two strategies both focused on OAA supply can not act synergistically. Perhaps only the overexpression of ppc or deletion of mdh is enough to provide appropriate OAA for l-glutamate and GABA production. However, GABA produced more quickly in ppc overexpressed and mdh deleted SDE2 (Fig. 3b).
Although the production of GABA increased in SE2, SDE1, and SDE2, it was not high enough. Fermentation conditions shall also be optimized in the future. In addition, other metabolic strategies focused on the α-ketoglutarate node of l-glutamate biosynthetic pathway including deletion of pknG (Okai et al. 2014) and deletion of odhA (Wang et al. 2015) were demonstrated recently to be able to improve GABA biosynthesis in recombinant C. glutamicum. Furthermore, a new metabolic route for the production of GABA via putrescine was established in C. glutamicum (Jorge et al. 2016a, b). All these researches suggest recombinant C. glutamicum as the good species for the production of GABA from glucose.
References
Barriuso-Iglesias M, Schluesener D, Barreiro C, Poetsch A, Martín JF (2008) Response of the cytoplasmic and membrane proteome of Corynebacterium glutamicum ATCC 13032 to pH changes. BMC Microbiol 8:225
Bouche N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9(3):110–115
Cheng G, Wang L, Lan H (2016) Cloning of PEPC-1 from a C4 halophyte Suaeda aralocaspica without Kranz anatomy and its recombinant enzymatic activity in responses to abiotic stresses. Enzyme Microb Technol 83:57–67
De Biase D, Tramonti A, John RA, Bossa F (1996) Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr Purif 8(4):430–438
Diana M, Quilez J, Rafecas M (2014) Gamma-aminobutyric acid as a bioactive compound in foods: a review. J Funct Foods 10:407–420
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104(1–3):155–172
Hiraga K, Ueno Y, Oda K (2008) Glutamate decarboxylase from Lactobacillus brevis: activation by ammonium sulfate. Biosci Biotechnol Biochem 72(5):1299–1306
Hirasawa T, Wachi M (2016) Glutamate fermentation-2: mechanism of l-glutamate overproduction in Corynebacterium glutamicum. Adv Biochem Eng Biotechnol. doi:10.1007/10_2016_26
Hu JY, Tan YZ, Li YY, Hu XQ, Xu D, Wang XY (2013) Construction and application of an efficient multiple-gene-deletion system in Corynebacterium glutamicum. Plasmid 70(3):303–313
Hu JY, Li YY, Zhang HL, Tan YZ, Wang XY (2014) Construction of a novel expression system for use in Corynebacterium glutamicum. Plasmid 75:18–26
Jorge JM, Leggewie C, Wendisch VF (2016a) A new metabolic route for the production of gamma-aminobutyric acid by Corynebacterium glutamicum from glucose. Amino Acids 48(11):2519–2531
Jorge JM, Nguyen AQ, Perez-Garcia F, Kind S, Wendisch VF (2016b) Improved fermentative production of gamma-aminobutyric acid via the putrescine route: systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol Bioeng. doi:10.1002/bit.26211
Karatzas KA, Brennan O, Heavin S, Morrissey J, O’Byrne CP (2010) Intracellular accumulation of high levels of gamma-aminobutyrate by Listeria monocytogenes 10403 S in response to low pH: uncoupling of gamma-aminobutyrate synthesis from efflux in a chemically defined medium. Appl Environ Microbiol 76(11):3529–3537
Leuchtenberger W, Huthmacher K, Drauz K (2005) Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69(1):1–8
Li HX, Cao YS (2010) Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 39(5):1107–1116
Mansouri S, Shahriari A, Kalantar H, Moini Zanjani T, Haghi Karamallah M (2017) Role of malate dehydrogenase in facilitating lactate dehydrogenase to support the glycolysis pathway in tumors. Biomed Rep 6(4):463–467.
Mei J, Xu N, Ye C, Liu LM, Wu JR (2016) Reconstruction and analysis of a genome-scale metabolic network of Corynebacterium glutamicum S9114. Gene 575(2 Pt 3):615–622
Mohler H (2012) The GABA system in anxiety and depression and its therapeutic potential. Neuropharmacology 62(1):42–53
Molenaar D, van der Rest ME, Drysch A, Yucel R (2000) Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J Bacteriol 182(24):6884–6891
Okai N, Takahashi C, Hatada K, Ogino C, Kondo A (2014) Disruption of pknG enhances production of gamma-aminobutyric acid by Corynebacterium glutamicum expressing glutamate decarboxylase. AMB Express 4:20
Park SJ, Kim EY, Noh W, Oh YH, Kim HY, Song BK, Cho KM, Hong SH, Lee SH, Jegal J (2013) Synthesis of nylon 4 from gamma-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst Eng 36(7):885–892
Peters-Wendisch PG, Kreutzer C, Kalinowski J, Pátek M, Sahm H, Eikmanns BJ (1998) Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144(4):915–927
Sato H, Orishimo K, Shirai T, Hirasawa T, Nagahisa K, Shimizu H, Wachi M (2008) Distinct roles of two anaplerotic pathways in glutamate production induced by biotin limitation in Corynebacterium glutamicum. J Biosci Bioeng 106(1):51–58
Sauer U, Eikmanns BJ (2005) The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29(4):765–794
Shi F, Li YX (2011) Synthesis of γ-aminobutyric acid by expressing Lactobacillus brevis-derived glutamate decarboxylase in the Corynebacterium glutamicum strain ATCC 13032. Biotechnol Lett 33(12):2469–2474
Shi F, Jiang JJ, Li YF, Li YX, Xie YL (2013) Enhancement of γ-aminobutyric acid production in recombinant Corynebacterium glutamicum by co-expressing two glutamate decarboxylase genes from Lactobacillus brevis. J Ind Microbiol Biotechnol 40(11):1285–1296
Shi F, Xie YL, Jiang JJ, Wang NN, Li YF, Wang XY (2014) Directed evolution and mutagenesis of glutamate decarboxylase from Lactobacillus brevis Lb85 to broaden the range of its activity toward a near-neutral pH. Enzyme Microb Technol 61–62:35–43
Shi F, Ni YL, Wang NN (2016) Metabolism and biotechnological production of gamma-aminobutyric acid (GABA). In: Vandamme EJ, Revuelta JL (eds) Industrial biotechnology of vitamins, biopigments, and antioxidants. Wiley-VCH, Weinheim
Shiio I, Ozaki H (1970) Regulation of nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase from Brevibacterium flavum, a glutamate-producing bacterium. J Biochem 68:633–647
Shirai T, Fujimura K, Furusawa C, Nagahisa K, Shioya S, Shimizu H (2007) Study on roles of anaplerotic pathways in glutamate overproduction of Corynebacterium glutamicum by metabolic flux analysis. Microb Cell Fact 6(1):19
Takahashi C, Shirakawa J, Tsuchidate T, Okai N, Hatada K, Nakayama H, Tateno T, Ogino C, Kondo A (2012) Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxylase from Escherichia coli. Enzyme Microb Technol 51(3):171–176
Wang NN, Ni YL, Shi F (2015) Deletion of odhA or pyc improves production of γ-aminobutyric acid and its precursor l-glutamate in recombinant Corynebacterium glutamicum. Biotechnol Lett 37(7):1473–1481
Zhao AQ, Hu XQ, Li Y, Chen C, Wang XY (2016) Extracellular expression of glutamate decarboxylase B in Escherichia coli to improve gamma-aminobutyric acid production. AMB Express 6(1):55
Acknowledgements
The authors thank the “Program of State Key Laboratory of Food Science and Technology” (Contract No. SKLF-ZZB-201405) for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, F., Zhang, M. & Li, Y. Overexpression of ppc or deletion of mdh for improving production of γ-aminobutyric acid in recombinant Corynebacterium glutamicum . World J Microbiol Biotechnol 33, 122 (2017). https://doi.org/10.1007/s11274-017-2289-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-017-2289-3