Abstract
Objectives
Corynebacterium glutamicum that expresses the exogenous L-glutamate decarboxylase (GAD) gene can synthesize γ-aminobutyric acid (GABA). To prevent GABA decomposition in the recombinant C. glutamicum GAD strain, GABA uptake and the GABA shunt pathway were blocked.
Results
GABA uptake is catalyzed by GABA permease encoded by gabP. The first reaction of the GABA shunt pathway is catalyzed by the GABA transaminase encoded by gabT. Initially, the effects of pH on GABA decomposition in recombinant C. glutamicum co-expressing two GAD genes (gadB1 and gadB2) were analyzed, demonstrating that GABA could be decomposed under neutral pH. Next, the gabP and gabT were individually deleted, and the GABA production of the related GAD strains was investigated by controlling the pH of the final fermentation stage at a neutral state. During this stage, the GABA concentration of the gabT-deleted GAD strain decreased from 23.9 ± 1.8 to 17.7 ± 0.7 g/l. However, the GABA concentration of the gabP-deleted GAD strain remained at 18.6–19.4 g/l.
Conclusion
This study demonstrated that GABA was decomposed under neutral pH and that the deletion of gabP could effectively alleviate GABA decomposition in C. glutamicum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gamma-aminobutyric acid (GABA) is widely distributed in nature, from microorganisms to plants and mammals, in which GABA biosynthesis and GABA catabolism, along with its export and uptake system, constitute GABA metabolism. The biosynthesis of GABA is fulfilled by the irreversible decarboxylation of L-glutamate by L-glutamate decarboxylase (GAD) in some species. Additionally, some species can take up extracellular GABA from the environment by a GABA-specific permease. The synthesized or obtained GABA can be decomposed via the GABA shunt pathway (Feehily and Karatzas 2013), which is a pathway involving the conversion of GABA to succinic semialdehyde (SSA) by GABA transaminase (GABA-T) followed by the conversion of SSA to succinate by SSA dehydrogenase (SSADH). Corynebacterium glutamicum is the main producer of L-glutamate. Although no gene encoding GAD was identified in C. glutamicum, a GABA-T gene (gabT), two SSADH genes (gabD) and a GABA permease gene (gabP) were annotated and has indicated that C. glutamicum could metabolize GABA (Zhao et al. 2012).
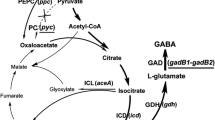
By the expression of exogenous GAD gene(s), C. glutamicum can synthesize GABA from its own accumulated L-glutamate (Shi and Li 2011; Shi et al. 2013). However, GABA may sometimes be degraded, as indicated by the sharply decreased GABA concentration after fermentation for 72 h in C. glutamicum expressing gadB of Escherichia coli W3110 (Takahashi et al. 2012). The metabolic pathway of GABA in recombinant C. glutamicum is shown in Fig. 1. The GABA permease (GabP) identified in C. glutamicum is considered to be the only GABA-specific transport system in C. glutamicum (Zhao et al. 2012). It plays a major role in GABA uptake and is essential to C. glutamicum growing in the presence of GABA. GABA-T is the first enzyme of the GABA shunt pathway. It can use GABA as an amino donor to form SSA and 2-oxoglutarate as an amino acceptor to form L-glutamate. To avoid the degradation of GABA in recombinant C. glutamicum, the GABA shunt pathway and GABA uptake system should be blocked.
The metabolic pathway of GABA in recombinant C. glutamicum that consists of the GABA shunt pathway and GABA uptake system. The GABA shunt pathway consists of two enzymatic steps that are catalyzed by GABA transaminase (GABA-T) and succinic semialdehyde dehydrogenase (SSADH). The GABA uptake system is carried out by the GABA permease (GabP) that transports GABA into the cytoplasm. TCA tricarboxylic acid cycle
In the present study, the gabT and gabP genes were individually deleted in C. glutamicum ATCC13032 to prevent GABA decomposition and GABA uptake. Next, a plasmid expressing two GAD genes (gadB1 and gadB2) was transformed into the deletion strains and wild-type strain. The effect of pH on GABA decomposition was investigated in the wild-type GAD strain. GABA fermentation in the gabT-deleted GAD strain and the gabP-deleted GAD strain was then investigated.
Materials and methods
Strains, media and growth conditions
The strains and plasmids used in this study are listed in Table 1. E. coli JM 109 was used as the host for constructing and propagating the plasmids. E. coli was grown in lysogeny broth (LB) at 37 °C and 200 rpm. C. glutamicum ATCC13032, a L-glutamate-producing strain, was used for gene deletion, expressing gadB1 and gadB2 genes and producing GABA. C. glutamicum was grown in LBG medium (LB supplemented with 5 g glucose/l) at 200 rpm and 30 °C. Epo medium (LB supplemented with 30 g glycine/l and 0.1 % Tween 80) and LBHIS medium (5 g Tryptone/l, 5 g NaCl/l, 2.5 g yeast extract/l, 18.5 g brain heart infusion powder/l and 91 g sorbitol/l) were used for C. glutamicum transformation (Wang et al. 2015). When necessary, 30 μg kanamycin/ml or 15 μg chloramphenicol/ml was added to the media.
Gene deletion and construction of recombinant C. glutamicum ATCC13032
Based on the homologous recombination and site-specific recombination system, gene disruption in C. glutamicum was performed according to a previous study (Hu et al. 2013). First, the deletion plasmid was constructed. Fragment gabTU, located upstream of the gene gabT, and fragment gabTD, located downstream of the gene gabT, were amplified from the genomic DNA of C. glutamicum ATCC13032. Fragment loxL-kan-loxR, which contains two recognition sites by Cre, was amplified from plasmid pDTW202. The three DNA fragments were ligated into plasmid pBluscriptII SK(+), resulting in plasmid pSYN1 (Fig. 2a). Similarly, plasmid pSYN2, in which fragments gabTU and gabTD were replaced by gabPU and gabPD, respectively, was constructed and is shown in Fig. 2a. Second, plasmid pSYN1 or pSYN2 was transformed into C. glutamicum ATCC13032 to allow for homologous recombination, as illustrated previously (Wang et al. 2015). The recombinant strains were selected by growth on LBHIS media supplemented with 30 μg kanamycin/ml. Plasmid pDTW-109 was transformed into the recombinant strain to remove the kan gene from the chromosome and was selected on LBHIS agar containing 15 μg chloramphenicol/ml. Finally, cells were cultured at 37 °C to remove the pDTW-109 plasmid. The cells that grew on the plate without antibiotics but not on the plate with kanamycin or chloramphenicol were chosen as the mutant strain ATCC13032 ΔgabT, renamed as SYN101, and the mutant strain ATCC13032 ΔgabP, renamed as SYN102. All of the strains were verified by restriction enzyme digestion or target gene amplification.
The gadB1 and gadB2 coexpression plasmid (pJYW-4-gadB1-gadB2) constructed previously (Wang et al. 2015, Fig. 2b) was finally transformed into the deletion mutants SYN101 and SYN102 and wild-type ATCC13032, generating three new recombinant C. glutamicum strains, SYN101/pJYW-4-gadB1-gadB2, SYN102/pJYW-4-gadB1-gadB2 and ATCC13032/pJYW-4-gadB1-gadB2, renamed as SYN201, SYN202 and SYN200, respectively.
Fermentation of different C. glutamicum strains in a fermentor
L-Glutamate and GABA production in the wild-type C. glutamicum strain ATCC13032, wild-type GAD strain SYN200, gabT-deleted GAD strain SYN201 and gabP-deleted GAD strain SYN202 were evaluated by fermentation in a fermentor. First, 100 ml seed medium (25 g glucose/l, 30 g corn steep liquor/l, 8 g urea/l, 1 g K2HPO4·3H2O/l, 0.2 g MgSO4/l, pH 7.0–7.2) was prepared in flasks at 30 °C and 110 rpm for 7 h and was then transferred to a 3 l fermentor (BioFlo 110; New Brunswick Scientific, USA) containing 1.2 l fermentation medium (100 g glucose/l, 2 g corn steep liquor/l, 2 g K2HPO4·3H2O/l, 0.4 g MgSO4/l, 0.2 g MnSO4·H2O/l and 0.29 g FeSO4·7H2O/l). Four g urea/l was added at the start of fermentation.
During fermentation, the temperature and aeration rates were kept at 30 °C and 1.5 vvm, respectively. The dissolved O2 level was controlled at 30 % by coupling with the agitation speed. From 12 h of fermentation, glucose was fed by a peristaltic pump when the residual glucose in the medium decreased to below 20 g/l. The pH was controlled differently during fermentation. For the fermentation of SYN200, the pH was controlled at 7–7.5 by adding 10 % (w/v) urea at 0–36 h and at pH 5–5.5 by adding 2 M HCl at 36–60 h. However, at 60–72 h, the pH was controlled at 7.5–8 by adding 2 M NaOH (neutral sample) and at 5–5.5 by adding 2 M HCl (acidic sample). For the fermentation of ATCC13032, SYN201 and SYN202, the pH was controlled at 7–7.5 for 0–36 h, at 5–5.5 for 36–72 h and at 7.5–8 for 72–84 h. Samples were taken approx. every 12 h to determine the OD562 value and residual glucose. GABA and L-glutamate concentrations were assayed using reversed-phase HPLC.
Cultivation of wild-type C. glutamicum with GABA as the sole carbon and nitrogen source
Cells of wild-type C. glutamicum ATCC13032 were precultured in seed medium at 30 °C and 110 rpm for 7 h. The precultured broth was inoculated into 20 ml fermentation medium in a 500 ml Erlenmeyer flask to a final OD562 of 1.8 and was shaken by a reciprocating shaker at 30 °C and 110 rpm for 36 h. Next, 4 g urea/l was added at the start of fermentation and 2 g urea/l was added to the culture every 3.5 h for 10–24 h of fermentation to maintain the neutral condition. The total cells were harvested at 36 h, transferred into 20 ml GABA medium (20 g GABA/l, 2 g K2HPO4·3H2O/l, 0.4 g MgSO4/l, 0.2 g MnSO4·H2O/l and 0.29 g FeSO4·7H2O/l, pH 7.5) with GABA as the sole carbon and nitrogen source and cultivated at 30 °C and 110 rpm for 36 h. During cultivation, the pH was adjusted to 7.5–8 every 12 h by adding 2 M NaOH. Samples were taken approx. every 12 h to determine the L-glutamate and GABA concentrations.
Results
To investigate their functions, the gabT and gabP genes of ATCC13032 were individually deleted, yielding the strains SYN101 and SYN102, respectively. Next, plasmid pJYW-4-gadB1-gadB2 was transformed into SYN101, SYN102 and ATCC13032, generating three new recombinant strains, SYN201, SYN202 and SYN200, respectively.
Effect of pH on GABA decomposition in recombinant C. glutamicum SYN200
Normally, there are two stages—i.e., the L-glutamate fermentation stage and the GABA fermentation stage—during the fermentation of GABA in recombinant C. glutamicum. At 0–36 h (the L-glutamate fermentation stage), the pH was controlled at 7–7.5 to ensure cell growth and L-glutamate biosynthesis. After 36 h (the GABA fermentation stage), the pH was adjusted to 5–5.5 to ensure GABA synthesis because GAD exhibits optimum activity at pH 4.5–5 and significantly loses activity at near-neutral pH (Shi et al. 2014). Accompanied by the conversion of L-glutamate to GABA by GAD, a proton was consumed that might increase the pH if the pH latter was not controlled. However, under neutral to slightly alkaline pH conditions (pH 7.5–8), GabP and GABA-T of C. glutamicum may be active (Zhao et al. 2012; Liu et al. 2005). GabP can take up extracellular GABA into cells. GABA-T, as the first enzyme of the GABA shunt pathway, can catalyze the conversion of GABA to SSA with the concomitant conversion of 2-oxoglutarate to L-glutamate. Both the active GABA uptake and GABA decomposition system can make GABA production decrease in recombinant C. glutamicum. To understand whether pH influences GABA decomposition, the pH was adjusted to 7.5–8 (neutral) and 5–5.5 (acidic) at the third stage (final stage, 60–72 h) of SYN200 fermentation, and GABA production under the two conditions was compared.
During fermentation, cells grew quickly during the first 12 h and the cell concentration became stable after 24 h (Fig. 3a). Glucose was consumed quickly during the first 24 h (Fig. 3b) and was supplemented when the residual glucose in the medium was lower than 20 g/l. L-Glutamate accumulated quickly during the first 36 h, decreased sharply during 36–60 h and was almost converted to GABA at 60 h (Fig. 3c). Accordingly, the GABA concentration increased quickly from 36 to 60 h (Fig. 3c). From 60 to 72 h, glutamate decreased from 2.8 ± 2.4 to 1.2 ± 0.3 g/l, and GABA further increased from 14.9 ± 0 to 17.4 ± 1.2 g/l under acidic conditions (Fig. 3c). However, under neutral conditions, glutamate increased from 4 ± 0.3 to 7 ± 0.8 g/l, and GABA decreased from 16.5 ± 0.7 to 12.6 ± 0.8 g/l from 60 to 72 h, indicating that GABA might be decomposed under neutral conditions. Additionally, the total amounts of L-glutamate and GABA of SYN200 under the two conditions were almost invariable from 60 to 72 h (Fig. 3d).
Time course of GABA fermentation by recombinant C. glutamicum SYN200 under controlled pH. a Cell growth; b glucose consumption (dotted lines) and residual glucose (solid lines); c production of L-glutamate (dotted lines) and GABA (solid lines); d total amount of L-glutamate and GABA. Filled squares, pH was controlled at 7.5–8.0 from 60 to 72 h; Open squares, pH was controlled at 5.0–5.5 from 60 to 72 h
GABA decomposition in wild-type C. glutamicum under neutral conditions
To confirm the decomposition of GABA under neutral conditions in C. glutamicum, GABA and L-glutamate concentrations were examined during the cultivation of wild-type C. glutamicum ATCC13032 with GABA as the sole carbon and nitrogen source. After cultivation for 36 h at pH 7.5–8, GABA decreased from 20 to 16.5 ± 0.5 g/l and L-glutamate increased from 0 to 1.6 ± 0.2 g/l (Fig. 4), demonstrating that GABA was decomposed under neutral conditions.
Effect of the deletion of gabT and gabP on GABA decomposition in recombinant C. glutamicum
To understand whether the deletion of gabT and gabP alleviates GABA decomposition, the pH was adjusted to 7.5–8 (neutral) at the final stage (72–84 h) of ATCC13032, SYN201 and SYN202 fermentation, and GABA production was investigated.
During the fermentation of the wild-type C. glutamicum strain ATCC13032, gabT-deleted GAD strain SYN201 and gabP-deleted GAD strain SYN202, cells grew quickly during the first 12 h and the cell concentration became stable after 24 h (Fig. 5a). Glucose was consumed quickly during the first 24 h (Fig. 5b) and was supplemented when the residual glucose in the medium was lower than 20 g/l. L-Glutamate accumulated quickly during the first 36 h and decreased from 36 to 60 h in both the SYN201 and SYN202 strains; however, at 60 h, a large amount of L-glutamate remained, and only at 72 h was L-glutamate transformed nearly completely to GABA (Fig. 5c). In the strain SYN202, GABA increased continuously from 36 to 72 h and then slightly decreased from 19.4 ± 1.7 to 18.6 ± 0.6 g/l from 72 to 84 h (Fig. 5c). The GABA decrement at the final stage of SYN202 fermentation was significantly lower than that of SYN200. In the strain SYN201, GABA increased continuously from 36 to 72 h, but decreased from 23.9 ± 1.8 to 17.7 ± 0.7 g/l from 72 to 84 h; meanwhile, the L-glutamate concentration increased from 2.9 ± 0.5 to 13.7 ± 1.2 g/l during 72–84 h (Fig. 5c). The total amount of L-glutamate and GABA in SYN201 and SYN202 was almost invariable from 72 to 84 h (Fig. 5d). In the wild-type strain ATCC13032, L-glutamate accumulated during the first 36 h and was maintained at that level thereafter, while no GABA was produced during all the time.
Time course of GABA fermentation by recombinant C. glutamicum SYN201 and SYN202 and wild-type ATCC13032 under controlled pH. The pH was controlled at 7.5–8.0 from 72 to 84 h. a Cell growth; b glucose consumption (dotted lines) and residual glucose (solid lines); c production of L-glutamate (dotted lines) and GABA (solid lines); d total amount of L-glutamate and GABA. Circles, SYN201; triangles, SYN202; squares, ATCC13032
Discussion
In this study, the effect of pH on GABA decomposition was first investigated by adjusting the pH of the final fermentation stage (60–72 h) to 7.5 to 8 and 5 to 5.5 in recombinant C. glutamicum ATCC13032 co-expressing two GAD genes (SYN200). GABA decreased by 3.9 ± 0.1 g/l from 60 to 72 h at pH 7.5–8.0, a level that was drastically reduced compared with the level at pH 5–5.5, which was increased by 2.6 ± 1.2 g/l (Fig. 3c), indicating that GABA could be decomposed at pH 7.5–8.
Wild-type C. glutamicum cannot synthesize GABA by itself, but can take up and utilize GABA as a carbon and/or nitrogen source by GABA permease and the GABA shunt pathway. GABA-T, which is the first enzyme of the GABA shunt pathway (Fig. 1), exhibits the highest activity at pH 7.8 (Liu et al. 2005). GABA permease GabP is active under neutral to slightly alkaline conditions and uptakes extracellular GABA into cells in which GABA can be further decomposed by the GABA shunt pathway (Zhao et al. 2012). In the recombinant C. glutamicum strain SYN200, the GABA concentration decreased when the pH was adjusted to 7.5–8 from 60 to 72 h (Fig. 3c). Meanwhile, the total amount of L-glutamate and GABA was almost constant (Fig. 3d) but the L-glutamate concentration was increased at this stage (Fig. 3c). The decrease in the GABA concentration in SYN200 may be due to GABA decomposition by the GABA shunt pathway and GABA uptake by GabP under pH 7.5–8.0. The decomposition of GABA was further confirmed in the wild-type C. glutamicum strain ATCC13032 under neutral pH (Fig. 4).
gabP encodes the only permease (GabP) of the GABA uptake system in C. glutamicum ATCC13032. GabP can take up extracellular GABA into cells. GabP of C. glutamicum was more active under neutral to slightly alkaline conditions and exhibited both a high affinity and activity on GABA at pH 6.5–8.0 (Zhao et al. 2012). After the gabP gene was deleted, GABA concentration did not decrease when the pH was controlled at 7.5–8 (Fig. 5c), indicating that the deletion of gabP can effectively prevent the uptake of GABA and its entry into the GABA shunt pathway.
GABA-T and SSADH are encoded by gabT and gabD in E. coli. However, GABA-T and SSADH activities persist even in the gabT- and gabD-deleted strains and were induced when putrescine in the media acted as a nitrogen source (Schneider et al. 2002). PuuE and YneI, identified as the second GABA-T and second SSADH, constitute a putrescine-inducible pathway that degrades GABA to succinate (Kurihara et al. 2010). A similar result has also been confirmed in Listeria monocytogenes EGD-e strain with a GABA-T-encoding gene being deleted (Feehily et al. 2013). As GABA-T activity can still be detected, this suggests that an alternative GABA-T activity is present in the cells. gabT encodes GABA-T in C. glutamicum ATCC13032, which catalyzes the conversion of GABA to SSA. It is possibly the only gene that encodes GABA transaminase in C. glutamicum ATCC13032. However, after the gabT gene was deleted, the GABA concentration of SYN201 still decreased significantly when pH was controlled at 7.5–8 (Fig. 5c), indicating that the deletion of the gabT gene could not prevent GABA from decomposing. It can be inferred that other transaminases acting on GABA might exist in C. glutamicum ATCC13032. However, this remains to be studied in detail.
The decrease in the GABA concentration in C. glutamicum at pH 7.5–8 was effectively alleviated by deleting the gabP gene. However, the decrease in GABA was not prevented in C. glutamicum by deleting the gabT gene. To further prevent GABA from decomposing, other transaminases acting on GABA will be weakened or deleted. Meanwhile, the double deletion mutant of gabT and gabP can be constructed, and its GABA concentrations during fermentation will be determined in the future. In a recent study, a C. glutamicum strain expressing an E. coli GAD mutant produced 5.9 ± 0.35 g GABA/l in flask cultivation at pH 7.0, a level that was 17-fold higher than the strain expressing wild-type GAD (Choi et al. 2015). Accordingly, the GAD mutant active in the expanded pH range could be further employed in the gabP-deleted strain SYN102 to provide a balanced condition for cell growth, GABA synthesis and GABA decomposition.
References
Choi JW, Yim SS, Lee SH, Kang TJ, Park SJ, Jeong KJ (2015) Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase activce in expanded pH range. Microb Cell Fact 14:21
Feehily C, Karatzas KA (2013) Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol 114:11–24
Feehily C, O’Byrne CP, Karatzas KA (2013) Functional γ-aminobutyrate shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl Environ Microbiol 79:74–80
Hu JY, Tan YZ, Li YY, Hu XQ, Xu DQ, Wang XY (2013) Construction and application of an efficient multiple-gene-deletion system in Corynebacterium glutamicum. Plasmid 70:303–313
Kurihara S, Kato K, Asada K, Kumagai H, Suzuki H (2010) A putrescine-inducible pathway comprising PuuE-YneI in which gamma-aminobutyrate is degraded into succinate in Escherichia coli K-12. J Bacteriol 192:4582–4591
Liu WS, Peterson PE, Langston JA, Jin XG, Zhou XZ, Fisher AJ, Toney MD (2005) Kinetic and crystallographic analysis of active site mutant of Escherichia coli gamma-aminobutyrate aminotransferase. Biochemistry 44:2982–2992
Schneider BL, Ruback S, Kiupakis AK, Kasbarian H, Pybus C, Reitzer L (2002) The Escherichia coli gabDTPC operon: specific gamma-aminobutyrate catabolism and nonspecific induction. J Bacteriol 184:6976–6986
Shi F, Li YX (2011) Synthesis of γ-aminobutyric acid by expressing Lactobacillus brevis-derived glutamate decarboxylase in the Corynebacterium glutamicum strain ATCC 13032. Biotechnol Lett 33:2469–2474
Shi F, Jiang JJ, Li YF, Li YX, Xie YL (2013) Enhancement of γ-aminobutyric acid production in recombinant Corynebacterium glutamicum by co-expressing two glutamate decarboxylase genes from Lactobacillus brevis. J Ind Microbiol Biotechnol 40:1285–1296
Shi F, Xie YL, Jiang JJ, Wang NN, Li YF, Wang XY (2014) Directed evolution and mutagenesis of glutamate decarboxylase from Lactobacillus brevis Lb85 to broaden the range of its activity toward a near-neutral pH. Enzyme Microb Technol 61–62:35–43
Takahashi C, Shirakawa J, Tsuchidate T, Okai N, Hatada K, Nakayama H, Tateno T, Ogino C, Kondo A (2012) Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxylase from Escherichia coli. Enzyme Microb Technol 51:171–176
Wang N, Ni Y, Shi F (2015) Deletion of odhA or pyc improves production of γ-aminobutyric acid and its precursor L-glutamate in recombinant Corynebacterium glutamicum. Biotechnol Lett 37:1473–1481
Zhao Z, Ding JY, Ma WH, Zhou NY, Liu SJ (2012) Identification of characterization of γ-aminobutyric acid uptake system GabPCg (NCgl0464) in Corynebacterium glutamicum. Appl Environ Microbiol 78:2596–2601
Acknowledgments
The authors thank the ‘‘Program of State Key Laboratory of Food Science and Technology” (contract no. SKLF-ZZB-201405) and ‘‘Fundamental Research Funds for the Central Universities” (contract no. JUSRP51303A) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ni, Y., Shi, F. & Wang, N. Specific γ-aminobutyric acid decomposition by gabP and gabT under neutral pH in recombinant Corynebacterium glutamicum . Biotechnol Lett 37, 2219–2227 (2015). https://doi.org/10.1007/s10529-015-1897-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1897-y