Abstract
Mesophilic and thermophilic anaerobic digesters (MD and TD, respectively) utilizing Gracilaria and marine sediment as the substrate and inoculum, respectively, were compared by analyzing their performances and microbial community changes. During three successive transfers, the average cumulative methane yields in the MD and TD were 222.6 ± 17.3 mL CH4/g volatile solids (VS) and 246.1 ± 11 mL CH4/g VS, respectively. The higher hydrolysis rate and acidogenesis in the TD resulted in a several fold greater accumulation of volatile fatty acids (acetate, propionate, and butyrate) followed by a larger pH drop with a prolonged recovery than in the MD. However, the operational stability between both digesters remained comparable. Pyrosequencing analyses revealed that the MD had more complex microbial diversity indices and microbial community changes than the TD. Interestingly, Methanomassiliicoccales, the seventh methanogen order was the predominant archaeal order in the MD along with bacterial orders of Clostridiales, Bacteriodales, and Synergistales. Meanwhile, Coprothermobacter and Methanobacteriales dominated the bacterial and archaeal community in the TD, respectively. Although the methane yield is comparable, both MD and TD show a different profile of pH, VFA and the microbial communities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Anaerobic digestion (AD) is a complex microbiological process in which organic material is converted into biogas, mainly methane and carbon dioxide, by numerous different groups of microorganisms (Cantrell et al. 2008). This technology helps to provide alternative sources of renewable energy that have fewer environmental impacts compared with those from fossil fuel-derived energy (Adekunle and Okolie 2015; Chynoweth et al. 2001). A variety of different substrates, ranging from lignocellulosic substrates to municipal solid wastes, have been utilized as substrates for AD (Chynoweth et al. 2001; Wei et al. 2013).
The use of seaweed as a substrate for AD has gained increasing attention recently because it lacks lignin and, thus, does not require pretreatment. Some seaweeds contain high amounts of polysaccharides, which can be used as substrates for AD (Wei et al. 2013). A number of seaweed genera, such as Saccharina, Laminaria, and Ulva, which have been used as substrates have shown good hydrolysis efficiencies, methane yields, and process stabilities during AD (Hughes et al. 2012; Vanegas and Bartlett 2013). For AD that uses seaweed as the substrate, marine sediments, which are expected to contain large numbers of bacteria, are often used as inocula. Previously, it was reported that the bioconversion efficiency of green seaweed inoculated with marine sediment was higher than that of seaweed inoculated with non-marine origin sediment (Schramm and Lehnberg 1984). In other digestion processes that used Saccharina japonica as the substrate, the methane yields from various marine sediments that were used as inocula were significantly higher than that from a methanogenic granule (Miura et al. 2014). Improved degradation rates and methane yields have also been reported using seaweed as a substrate and anoxic lagoon sediment as an inoculum (Migliore et al. 2012). These results indicate that marine sediment is a suitable inoculum for an AD process that employs seaweed as the substrate.
Temperature is one of the significant factors that affect the kinetics and microbial compositions during AD. AD processes are regularly conducted under mesophilic (30–40 °C) and thermophilic (50–60 °C) conditions. Generally, high temperatures facilitate faster reaction rates, higher substrate degradation efficiencies, and a significant reduction of pathogens. However, thermophilic conditions might decrease the stability of the digestion process because of higher accumulations of volatile fatty acids (VFAs). In addition, more energy will be required to maintain thermophilic conditions (Li and Yu 2011). In contrast, mesophilic conditions, which are more commonly employed in AD, require less energy and are more stable than thermophilic conditions (Shi et al. 2013).
A successful AD performance depends on a dynamic balance among diverse microbes (Amani et al. 2010). Until now, most knowledge of the microbial communities of anaerobic digesters was generated from those that used terrestrial lignocellulose biomass as a substrate and non-marine waste as inocula (Mhuantong et al. 2015; Shi et al. 2013). Here, we report the investigation of the composition and succession of a microbial community from mesophilic (37 °C; MDs) and thermophilic digesters (55 °C; TDs), in which Gracilaria and marine sediment were used as the substrate and inoculum, respectively. The results of this study will improve our knowledge of the mesophilic and thermophilic microbes that are potentially important for the AD of seaweed. Additionally, we identified the physicochemical factors that shape the composition of the microbial community and correlated them with digester performance.
Materials and methods
Preparation of substrates and inocula
Three genera of fresh seaweeds (Ulva, Laminaria, and Gracilaria) were purchased from the local market in Gwangju, Korea in March 2013. All seaweeds were desalted by freshwater washing, sun-dried, and then milled into powder. The milled seaweeds were subsequently kept at −20 °C prior to use. Three inocula were collected from the wastewater treatment plant at Ansan, Korea, while manure was collected from an anaerobic digester at Paju, Korea, and anoxic marine sediment was collected from Oido Island, Korea. Prior to AD, inocula were pre-incubated at room temperature for 20 d to deplete the residual biodegradable organic materials, and gas was removed every second day.
Batch cultures in 150-mL digesters
Preliminary experiments were conducted using sets of three seaweeds and three inocula under mesophilic (37 °C) and thermophilic (55 °C) conditions. The basal medium was prepared in distilled water, and its composition was: NH4Cl (0.30 g/L), NaCl (0.30 g/L), MgCl2·6H2O (0.10 g/L), CaCl2·2H2O (0.11 g/L), KH2PO4 (0.41 g/L), Na2HPO4 (0.53 g/L), NaHCO3 (4.00 g/L), 0.1 % (w/v) resazurin solution, 1 % (v/v) trace elements, 1 % (v/v) filter sterilized of vitamin solution, cysteine HCl (0.50 g/L), and Na2S·9H2O (0.50 g/L) (Balch et al. 1979). The pH was adjusted to 7.4–7.6 in all cases with a 10 % NaOH solution.
To prepare the medium under strict anaerobic conditions, the medium was boiled using boiling flasks, while the gas phase was simultaneously exchanged with a mixture of gases (N2:CO2, 80:20 v/v). The inoculum/substrate (I/S) ratio was 1 on the basis of volatile solids (VS); equal amounts of each substrate (0.2 g VS) were transferred individually into separate serum bottles (150 mL) and mixed with each inoculum (0.2 g VS) in an anaerobic chamber. The autoclaved medium was added last, bringing the final volume to 50 mL, with the remaining 100 mL used for the headspace; then, the serum bottle was sealed with a rubber stopper and capped with aluminum crimps. Negative controls that contained only inoculum and medium were also prepared for all samples.
In total, three successive transfers were conducted in duplicate during 120 days of preliminary study. Methane production was measured twice per week. After reaching the stationary stage during each of the successive transfers, the serum bottle was vigorously shaken to homogenize the culture, and then the inocula were transferred into fresh medium.
Batch cultures in 2-L digesters
Gracilaria and marine sediment were used as the substrate and inoculum respectively for larger-scale digesters due to the higher methane production and digester stability under mesophilic and thermophilic conditions during preliminary experiment using 150 mL digesters. Triplicate samples were prepared for each MD (37 °C) and TD (55 °C), which used 2-L aspirator Duran glass bottles with a 1-L working volume, and which were equipped with a GL 45 threaded screw cap with an inserted rubber septa and a GL 32 outlet at the base. The I/S ratio was 1 on the basis of VS; Gracilaria (4 g VS) was transferred into 2-L bottles and mixed with marine sediment (4 g VS) in an anaerobic chamber. The medium was then added to a final working volume of 1 L with the remaining 1 L as headspace. Negative controls that contained only inoculum and medium were also prepared. All the experimental methods and preparations were the same as those used in the preliminary study, except the amounts of substrates, inocula, and media were increased by 20-fold. Three successive transfers were conducted in triplicate during 135 days as it is shown in Figs. 2 and 3.
Analytical methods
Total solids, VS, and pH were determined according to standard methods (APHA 1995). Methane and carbon dioxide that accumulated in the bottle headspace were measured by injecting a 100-µL sample volume, via a gas-tight syringe, into a gas chromatograph (YL 6100GC, Anyang, Korea) equipped with a flame ionization detector and a Porapack N, 80–100 mesh, 10 ft. ×8 in. column matrix. Argon was used as the carrier gas at a flow rate of 13 mL/min, and the temperatures of the column, injector, and detector were 40, 110 and 110 °C, respectively. The accumulated methane was measured twice per week and expressed as an average with standard errors. Methane gas production calculated from the headspace was converted into the volume of methane at standard temperature and pressure according to the ideal gas law. The methane production in the negative control was subtracted from the cumulative methane production. The methane production potential of seaweed was defined as the total volume of methane produced per amount of substrate initially added (i.e., mL CH4/gVS) during the digestion period.
The concentrations of VFAs (acetate, propionate, and butyrate) were determined by a gas chromatograph (GC-900C) with a flame ionization detector equipped with a fused silica column (30 m × 0.32 mm × 0.25 µm). The liquid samples from the anaerobic digesters were centrifuged at 10,397 g for 10 min, acidified to approximately pH 2 with formic acid, filtered through a 0.45-µm membrane, and finally measured by injecting 1 µL of the sample. The temperatures of the column and detector were 110 and 220 °C, respectively. Helium was used as the carrier gas at a flow rate of 5 mL/min.
Sampling, DNA extraction, and pyrosequencing
Over a 45-d digestion period, five samples were taken on d 0, 5, 15, 25, and 35 from the MD and TD (indicated as MD 0, MD 5, MD 15, MD 25, and MD 35, and TD 0, TD 5, TD 15, TD 25, and TD 35, respectively) on the basis of the methane and pH profiles. Marine sediment (indicated as MS) as the original inoculum was also subjected to pyrosequencing analysis. These samples were immediately frozen at −20 °C prior to DNA extraction, which was performed with the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA). Pyrosequencing was performed according to a protocol from Chunlab Inc. (Seoul, Korea), with some modifications. The extracted DNA was used as a template for fusion PCRs of the hypervariable regions (V1–V3) of the bacterial and archaeal 16S rRNA genes. The primers for the bacterial sequences were V1-27F (5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAG-AC-GAGTTTGATCMTGGCTCAG-3′) (gene-specific sequences are underlined) and V3-518R (5′-CCAT CTCATCCCTGCGTGTCTCCGAC-TCAG-X-AC-WTTACCGCGGCTGCTGG-3′); the X barcode was uniquely designed for each sample, followed by the common linker AC. The primers for the archaeal sequences were AV1-21F (5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAG-AG-TCCGGTTGATCCYGCCGG-3′) and AV3-519R (5′-CCTATCCCCTGTGTGCCTTGGCAGTC-TCAG-X-GA-GGTDTTACCGCGGCKGCTG-3′). PCRs were conducted under the following conditions: an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C (bacteria) or 55 °C (archaea) for 30 s, elongation at 72 °C for 90 s, followed by a final elongation of 10 min at 72 °C. The amplicons were purified using the QIAquick PCR Purification kit (Qiagen, Valencia, CA, USA), and quantified using a spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The purified PCR products (~1 µg of each sample) were used for pyrosequencing. All the pyrosequencing procedures, including the construction of a single-stranded DNA library, emulsion PCRs, and pyrosequencing reactions, were performed by Chunlab Inc. (Seoul, Korea) using a Roche/454 GS Junior system according to the manufacturer’s instructions.
Pyrosequencing data analyses
Pyrosequencing data were analyzed according to previously published methods (Jeon et al. 2013). Briefly, raw data from each sample were separated by unique barcodes in the demultiplexing step, and low-quality reads, based on the average quality score, were excluded from further analysis. The primer sequences were trimmed based on the profile of the 16S rRNA V1–V3 regions by pairwise sequence alignments and the hmm-search program of the HMMER 3.0 package (Eddy 2011). To correct sequencing errors, the representative sequences in each cluster of trimmed sequences were selected for taxonomic identification. The taxonomic positions of individual reads were determined according to the highest pairwise similarity among the top five BLASTN hits against the EzTaxon-e database (http://eztaxon-e.ezbiocloud.net), and chimeric sequences were removed by UCHIME (Edgar et al. 2011). Alpha diversity indices were calculated by the MOTHUR Package (Schloss et al. 2009). The pyrosequencing dataset have been deposited into the MG-RAST server under accession numbers listed in Table S1. The compositions of bacterial species from each sample were calculated with CL community software (Chunlab Inc., Seoul, Korea). Canonical correspondence analysis (CCA; XLSTAT version 2012, Addinsoft, New York, NY, USA) was conducted to describe the correlations between microbial populations and the operational conditions, including temperature, pH, and VFAs, as well as anaerobic digester performance, including methane production.
Design of modified primer set specific for 7th order of methanogens and construction of clone libraries
A primer set specific for 7th order of methanogens was designed from alignment study. For construction of the library, PCR with the modified primer set was conducted, which used the total DNA from the MD samples as template. PCR conditions included an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 90 s, followed by a final elongation for 10 min at 72 °C. The amplified products were ligated into the pGEM-T vector (Promega, Madison, WI, USA) and transformed into Escherichia coli DH5α according to the manufacturer’s instructions. Clones were randomly selected and their plasmids were extracted, purified, and sequenced by Cosmo Genetech Co., Ltd., Seoul, Korea, on ABI 3730 capillary sequencers using BigDye v. 3.1 sequencing chemistry (Applied Biosystems, Foster City, CA, USA).
The sequences were aligned with representative reference sequences with CLUSTAL X (version 1.83) (Thompson et al. 1997). Phylogenetic trees were constructed by MEGA software (version 69.05). An evolutionary distance matrix was generated according to (Jukes and Cantor 1969), and inferred using the neighbor-joining method (Saitou and Nei 1987). The neighbor-joining tree topology was evaluated by a bootstrap analysis based on 1000 replicates (Felsenstein 1985).
Results
Anaerobic digestion performance
During three successive transfers of the preliminary study (150 mL), the mesophilic (37 °C) ADs were more stable and produced higher methane yields in all batch cultures than the thermophilic (55 °C) ADs. Gracilaria inoculated with marine sediment produced the highest methane yield, 293 ± 13.3 mL/g VS and 236.4 ± 71.2, under mesophilic and thermophilic conditions, respectively. The average methane production during three successive transfers is shown in Table S2.
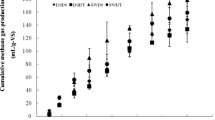
For the larger scale (2 L), the methane, VFAs, and pH profiles in the MD and TD are shown in Figs. 1 and 2. During the digestion process, methane production consistently increased and appeared to reach saturation phase by d 35 in the MD and TD. The average cumulative methane yields during three successive transfers in the MD and TD were 222.6 ± 17.3 and 246.1 ± 11 mL/g VS (Figs. 1, 2), respectively.
Cumulative methane production in the mesophilic digester (MD). Open triangles pH, closed circles cumulative methane production. The black arrows represent the times at which the samplings for the microbial community analysis were conducted (d 5, 15, 25, and 35). Volatile fatty acids include acetate (black vertical bars), propionate (gray vertical bars), and butyrate (dark gray vertical bars)
Cumulative methane production in the thermophilic digester (TD). Open triangles pH, closed circles cumulative methane production. The black arrows represent the times at which the samplings for the microbial community analysis were conducted (d 5, 15, 25, and 35). Volatile fatty acids include acetate (black vertical bars), propionate (gray vertical bars), and butyrate (dark gray vertical bars)
Both digesters produced higher concentrations of VFAs during the early phase, which subsequently decreased after d 15 and continued to decrease until the end of the digestion process. Although both digesters exhibited similar profiles, the VFA concentrations were more than twice higher in the TD (Fig. 2) than in the MD (Fig. 1). VFAs were totally degraded by the end of the digestion process in the MD. Likewise, acetate and butyrate were also almost completely consumed in the TD, except for the propionate whose concentration was maintained at 1.64 mM.
The pH profiles from the MD and TD are shown in Figs. 1 and 2, respectively. The pH initially decreased in the early phase and increased slightly until the end of the AD process. A continuous pH decrease from 7.8 after the inoculation to 6.6 at d 15 was observed in the MD, which was followed by a rapid recovery. Meanwhile, the pH suddenly decreased to 6.4 on d 5 in the TD, which was followed by a prolonged recovery.
Comparison of microbial communities
A total of 154,938 sequences were obtained from the collected samples, and 60.5 and 39.5 % of the sequences were assigned to the bacteria and archaea domains, respectively.
A comparison of the microbial richness and diversity in the MD and TD is shown in Table 1. As indicated by the Ace and Chao 1 values, bacterial richness during each sampling period was higher in the MD than in the TD over the course of the AD process. Similarly, archaeal richness during each sampling period was always higher in the MD than in the TD according to the Chao1 values. In addition, slightly lower sequence coverage in the MD, along with a higher Shannon index, indicated that the microbial diversity of the MD was higher than that of the TD. The higher microbial diversity in the MD was also supported by the number of operational taxonomic units (OTUs), which was clearly higher in the MD than in the TD.
Changes in bacterial diversity
Seven major bacterial phyla with relative abundances greater than 0.5 % were obtained from each sample in the MD and TD (Fig. 3a). The dynamic profiles of bacterial phyla during the digestion period differed significantly between the MD and TD (Fig. 3a). Proteobacteria was the predominant phylum in the original marine sediment, accounting for 51 % of total sequence. However, Firmicutes was the most predominant phylum during the MD, and its maximum proportion was 67 % at d 5. Bacteroides was another dominant phylum in the MD, and its relative abundance reached 22 % at d 5. In comparison, phyla Synergistetes, Proteobacteria, Planctomycetes, and Cloacamonas were present in lesser percentage and were present only in the MD. Interestingly, the phylum Thermotogae, which is commonly found in thermophilic environments, was observed in the MD. In contrast, Firmicutes was the only bacterial phylum found in TD.
A total of 367 bacterial genera were detected in both digesters, and the taxonomic compositions of their bacterial communities at the genus level with abundance greater than 0.01 % are shown in Table 2.
Clostridium was the most abundant genus in the MD, where it reached the maximum proportion of 38.45 % on d 15. Among the genera containing hydrolytic bacteria that can degrade cellulose and/or pectin, Cellulosilyticum was the most prevalent, having the highest proportion during the early phase (13.7 %), while uncultured Ruminococcaceae, uncultured Lachnospiraceae, and Ruminococcus had lower proportions (4.2, 3.3, and 1.2 %, respectively) (Cai and Dong 2010; Desvaux 2005; Rode et al. 1981; Schink and Zeikus 1980). The relative abundance of the genus Acetobacteroides, which includes carbohydrate-fermenting bacteria (Su et al. 2014), was highest on d 5 and then decreased. The genus Bacteroidales uncultured showed a reverse pattern, as its proportion was lowest during the initial phase and then increased. From the sequencing data, the genera Aminobacterium (Baena et al. 2000), Aminivibrio (Honda et al. 2013), and Cloacamonas (Pelletier et al. 2008), which are composed of syntrophic bacteria that interact with methanogens, were found, and the genus Aminobacterium was highly abundant during the late phases. The presence of the genus Desulfovibrio, which contains sulfate reducers, was confirmed, proving that it competes with methanogens in the MD. The genus Mesotoga, which belongs to the phylum Thermotogae, was identified, and it may degrade various polysaccharides in the MD (Nesbø et al. 2012). The proportion of the genus Phycisphaera, which is likely to be involved in the degradation of complex heteropolysaccharides (Wang et al. 2015), was the lowest.
Coprothermobacter and Defluviitalea were the two most predominant genera in the TD. The genus Defluviitalea was dominant initially (62.7 %) and then nearly disappeared, while the genus Coprothermobacter increased to 90.75 % during the late phase. The bacteria in the genus Coprothermobacter are proteolytic hydrogen producers that are associated with hydrogenotrophic methanogens such as Methanothermobacter, which use casein, gelatin, and bovine serum albumin as protein sources (Sasaki et al. 2011).
The genus Caldicoprobacteraceae, which comprises xylanolytic bacteria (Yokoyama et al. 2010), was highly abundant during the early phase and then gradually declined. Hydrogenispora, a genus containing carbohydrate-fermenting bacteria (Liu et al. 2014), was another abundant genus in the TD, and it had the highest proportion on d 15. Unlike the above cases, the genera Thermacetogenium (Hattori et al. 2000), Tepidanaerobacter (Westerholm et al. 2011), Caloramator, and Ruminococcaceae uncultured were present at low proportions in the TD. The pyrosequencing information indicates that bacterial succession, in which distinctive bacteria play unique roles during the digestion phases, occurred in the anaerobic digesters.
Changes in archaeal diversity
Euryarchaeota was the major archaeal phylum, constituting 94.5 and 99.8 % of the total sequences in the MD and TD, respectively (Table S3), while the phylum Crenarchaeota was a minor one, and it was limited to the MD. The distributions of archaeal sequences at the order level from each sample are shown in Fig. 3b. The dynamic profiles of the archaeal community compositions in the MD and TD were significantly different; the diversity was higher in the MD than in the TD. Miscellaneous Crenarchaeotal Group (MCG), accounting for 47 % of total sequence, was the predominant archaeal group in the original marine sediment, but it nearly disappeared during anaerobic digestion process. At the order level, four methanogen orders, Methanobacteriales, Methanomassiliicoccales, Methanococcales, and Methanosarcinales, were found in the anaerobic digesters. The order Methanobacteriales dominated the TD, representing 95–99 % of the total archaeal sequences. Meanwhile, an archaeal group that is related to the seventh methanogen order, named Methanomassiliicoccales, dominated the MD and increased in abundance from 66 % on d 5–90 % on d 35.
A total of 98 archaeal genera were obtained from both digesters, and the taxonomic compositions of the archaeal communities at the genus level with abundance greater than 0.01 % are shown in Table 3.
Methanothermobacter was the most predominant archaeal genus in the TD (95.03–99.47 %), and the genus Methanomassiliicoccus was the most predominant one in the MD (90.07 % in the final phase). The other identified archaeal genera with low abundances in the MD were Methanobacterium and Methanococcus.
Correlations between microbial communities, operational conditions, and digester performance
Canonical correspondence analysis (CCA) results provided further evidence of correlations between microbial communities and environmental factors that include operational conditions such as temperature, pH, butyrate, acetate, and propionate, as well as digester performance factors such as methane production (Green 1989) (Fig. 4a, b). Considering that AD is driven by both bacteria and methanogen orders, CCA was performed using the major genera of bacteria and methanogens detected in this study. The results of the CCA analysis showed that environmental factors accounted for more than 85 % of the variations in the relative abundances of bacteria and archaea, suggesting that environmental factors are substantially responsible for the distribution of the major orders. Both bacterial and archaeal CCA analyses showed that VFAs were positively correlated with temperature, but inversely correlated with pH. Bacterial orders, such as Coprothermobacter and Hydrogenispora, which were only found in the TD, were positively correlated with temperature, and a similar phenomenon was also observed for the methanogen genus Methanothermobacter. Significance analysis on the environmental variables revealed that temperature accounted for the greatest difference in both archaeal and bacterial community composition observed between the MD and TD and had a statistically significant correlation with microbial composition (P < 0.05).
Canonical correspondence analysis (CCA) ordination diagrams. The correlations between bacterial community profiles (a), archaeal community profiles (b) (at the order level, pyrosequencing data), and the operational conditions and anaerobic digester performance are represented as black vectors; bacterial orders fitting greater than 85 % are displayed. Solid circles represent the samples (blue for the mesophilic digester (MD), and red for the thermophilic digester (TD))
Design of modified primer set specific for 7th order of methanogens and construction of clone libraries
The sequences of the modified PCR primer set specific for 7th order of methanogens are: 21b-F (5′-TCCGGTTGATCCTGCCGGC-3′) (DeLong 1992) and 1492c-R (5′-TACAGATACCTTGTTACGACTT-3′) (Lane 1991). Almost full-length 16S rDNA gene sequence (1471 bp) was obtained from sequencing of clones in the library constructed with PCR amplicons generated by the modified primer set. The randomly selected five clones showed the highest sequence similarities (97–98 %) to the environmental clones derived from wastewater sludge (AF424770). The nearest cultivated neighbor of the clones was Methanomassiliicoccus luminyensis B10T with (91–92 %) sequence identity. In the phylogenetic tree of 16S rDNA gene sequence, all clones were placed among Lake Pavin Cluster (Fig. S1).
Discussion
One of most reliable indicators for process imbalance during AD is the accumulation of VFAs, which is followed by a decrease in pH (Franke-Whittle et al. 2014). Higher VFA concentrations and pH decrease in ADs have been attributed to higher hydrolysis reaction rates under thermophilic conditions and the high activity of acidogenic bacteria, which are acid producers, respectively. Despite of the higher VFA and lower pH levels of the TD, inhibition of methanogens by VFAs was not observed considering the methane yields. One of the previous AD study showed that the inhibition occurs under high propionate concentrations (>12.32 mM) (Wang et al. 2009), which is several fold higher than the TD. In addition, digester acidification, which can deteriorate the AD process (Akuzawa et al. 2011), was not detected in the TD despite the prolonged pH recovery. In fact, the pH in the TD was still in the near-neutral range for methanogen activity. The methane yields in this study are comparable to those of previous anaerobic digesters that also used seaweeds as substrates (Saccorhiza polyschides, 255 mL/g VS; Laminaria digitata, 246 mL/g VS; Saccharina latissima, 335 mL/g VS; and Ulva spp., 191 mL/g VS (Vanegas and Bartlett 2013) under mesophilic condition (35 °C), while methane yield of 128.8 mL/g VS was produced by Laminaria digitate at 45 °C (Vanegas and Bartlett 2012). These data indicate that the operational stability and performance of the MD and TD were comparable in terms of methane yields despite their different VFA concentrations and pH profiles.
A comparison between the bacterial and archaeal communities identified in this study and in previous studies is summarized in Table S3. Our and other studies including temporal succession during the solid state (SS) AD of corn stover (Li et al. 2015) and anaerobic digestion of carrot pomace under mesophilic condition (Garcia et al. 2011) found that Firmicutes and Bacteroidetes were predominant throughout the processes. The AD in which the brown macroalgae S. latissima and wastewater were used as the substrate and inoculum (Pope et al. 2013), respectively, the phyla Spirochaetes and Chloroflexi, which were not identified in our study, were major groups during AD. However, the phylum Firmicutes, which was abundant in other AD processes, accounted for a minor proportion of the microbes. The phylum Bacteroides was constantly predominant in the studies under mesophilic condition. The identification of a core bacterial community during the AD of seaweeds is still hampered by limited data, and thus further studies are required.
Euryarchaeota was the predominant archaeal phylum in both MDs and TDs, accounting for more than 90 % of the total archaeal sequences. However, the archaeal distributions, at the order level, differed among the studies. An earlier anaerobic digester, which used the brown macroalgae S. japonica as a substrate and marine sediment as the inoculum, was dominated by the order Methanosarcinales during the primary culture and by the order Methanococcales during the subculture (Miura et al. 2014). Other MDs employing the brown macroalgae S. latissima and marine sediment showed that the most prevalent archaeal order was the Methanosarcinales. The orders Methanomicrobiales and Methanobacteriales were also present at significant proportions in a MD (Pope et al. 2013).
The dominance of the hydrogenotrophic methanogens Methanothermobacter and Methanomassiliicoccales over acetoclastic methanogens (order Methanosarcinales) (<1 % of the total archaeal community) in the MD and TD was demonstrated. The dominance of H2-oxidizing Methanothermobacter in the TD might be due to the fact that thermophilic conditions favor hydrogenotrophs, as they are more capable of adapting to higher temperatures compared with acetoclastic methanogens (Chen 1983). The predominance of the hydrogenotrophic methanogen pathway in the TD was also supported by the high abundance of Coprothermobacter, along with well-defined syntrophic, hydrogen-producing bacteria belonging to the genera Hydrogenispora, Thermacetogenium, and Tepidanaerobacter. The seventh methanogen order Methanomassiliicoccales has recently been validated by the International Committee on Systematics of Prokaryotes (Oren and Garrity 2013), which recognized a hydrogenotrophic methanogen that utilizes an external H2 source to reduce methyl compounds to methane (Borrel et al. 2014). To the best of our knowledge, this is the first report to describe the abundance of the seventh methanogen order in a mesophilic anaerobic digester.
Methanomassiliicoccales can be divided into three large clusters: the Methanomassiliicoccus luminyensis cluster, which is mainly composed of sequences from soils and sediments and, to a lesser extent, from digestive tracts; the “Candidatus Methanomassiliicoccus alvus” cluster, which mostly contains sequences retrieved from animal digestive tracts; and the Lake Pavin cluster, which comprises sequences retrieved from diverse environments, but not digestive tracts (Borrel et al. 2013). Notably, the majority of Methanomassiliicoccales related sequences in our pyrosequencing data were phylogenetically related to multiple 16S rRNA sequences from the water column of the meromictic Lake Pavin in France, and distantly related to the M. luminyensis cluster (see Fig. S1). Of all the Methanomassiliicoccales related sequences, more than 95 % were closely related (higher than 97 % similarity) to AF424770, an uncultured archaea that has previously been retrieved from wastewater sludge and that belongs to the Lake Pavin cluster. Meanwhile, the sequences are distantly related to the cultured representative, M. luminyensis, ranging from 88.59 to 90.28 % 16S rRNA gene sequence similarities. These data suggest that the methanogens in the MD are members of the Lake Pavin cluster.
To confirm the abundance of Methanomassiliicoccales in the MD, a modified PCR set was designed and clone libraries were constructed. Previously, PCR bias has been reported when using different methods to identify the seventh order of methanogens (Snelling et al. 2014). In the study, 16S rRNA clone libraries constructed using the primer set Arch f364 and Arch r1386, as well as metagenome sequencing of the 16S rRNA and mcrA genes, failed to detect the Methanomassiliicoccales, but amplicon sequencing of 16S rRNA using primers Ar915aF and Ar1386R identified the methanogens, which comprised 13 % of the total archaea. Thus, it was determined to verify the abundance of Methanomassiliicoccales in the MD using different primer set other than 21F-519R, which was used for the pyrosequencing. For this purpose, we designed a new modified primer set (21b-F, 1492c-R) specific for the seventh order of methanogens from the alignment study. The sequencing of the clones constructed from the PCR products using the primer set showed that 16S rDNA gene for 7th order of methanogens was dominant. These data strongly suggest the predominance of the Methanomassiliicoccales in the MD sample.
Conclusions
This study showed that the MD and TD produced a comparable cumulative methane yield. The MD had a lower VFAs concentration and faster pH recovery than those of the TD, which did not experience process deterioration, regardless of the higher accumulations of VFAs. According to pyrosequencing data, the MD had greater microbial diversity than the TD in which a few specific microbes dominated. The bacterial community in both digesters was dominated by phylum Firmicutes, while hydrogenotrophic methanogens, Methanomassiliicoccales and Methanobacteriales, were two dominant archaeal orders in the MD and TD, respectively. The CCA analysis showed that temperature is the environmental factor that is most responsible for distinctive and adapted microbial communities under both conditions. Further study is needed to elucidate the seventh methanogen order, Methanomassiliicoccales, and its possible roles during anaerobic digestion process.
References
Adekunle KF, Okolie JA (2015) A review of biochemical process of anaerobic digestion. Adv Biosci Biotechnol 6:205–212
Akuzawa M, Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y (2011) Distinctive responses of metabolically active microbiota to acidification in a thermophilic anaerobic digester. Microbial Ecol 61:595–605
Amani T, Nosrati M, Sreekrishnan TR (2010) Anaerobic digestion from the viewpoint of microbiological, chemical, and operational aspects: a review. Environ Rev 18:255–278
APHA (1995) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, Washington
Baena S, Fardeau ML, Labat M, Ollivier B, Garcia JL, Patel BKC (2000) Aminobacterium mobile sp. nov., a new anaerobic amino-acid-degrading bacterium. Int J Syst Evol Microbiol 50:259–264
Balch WE, Fox CE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296
Borrel G, Harris HMB, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O’Toole PW, Brugère J-F (2013) Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1:e00453-13. doi:10.1128/genomeA.00453-13
Borrel G, Parisot N, Harris HMB, Peyretaillade E, Gaci N, Tottey W, Bardot O, Raymann K, Gribaldo S, Peyret P, O’Toole PW, Brugère JF (2014) Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics BioMed Central 15(1):679
Cai S, Dong X (2010) Cellulosilyticum ruminicola gen. nov., sp. nov., isolated from the rumen of yak, and reclassification of Clostridium lentocellum as Cellulosilyticum lentocellum comb. nov. Int J Syst Evol Microbiol 60:845–849
Cantrell KB, Ducey T, Ro KS, Hunt PG (2008) Livestock waste–to- bioenergy generation opportunities. Bioresour Technol 99:7941–7953
Chen M (1983) Adaptation of mesophilic anaerobic sewage fermentor populations to thermophilic temperatures. Appl Environ Microbiol 45:1271–1276
Chynoweth DP, Owens JM, Legrand R (2001) Renewable methane from anaerobic digestion of biomass. Renew Energy 22:1–8
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
Desvaux M (2005) Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol Rev 29(4):741–764
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7(10):e1002195
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15
Franke-Whittle IH, Walter A, Ebner C, Insam H (2014) Investigation into the effect of high concentrations of volatile fatty acids in anaerobic digestion on methanogenic communities. Waste Manag 34(11):2080–2089
Garcia LS, Jangid K, Whitman WB, Das KC (2011) Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol 102:7249–7256
Green RH (1989) Power analysis and practical strategies for environmental monitoring. Environ Res 50:195–205
Hattori S, Kamagata Y, Hanada S, Shoun H (2000) Thermacetogeniumphaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int J Syst Evol Microbiol 50:1601–1609
Honda T, Fujita T, Tonouchi A (2013) Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int J Syst Evol Microbiol 63:3679–3686
Hughes AD, Kelly MS, Black KD, Stanley MS (2012) Biogas from macroalgae: is it time to revisit the idea? Biotechnol Biofuels 5:86
Jeon YS, Chun J, Kim BS (2013) Identification of household bacterial community and analysis of species shared with human microbiome. Curr Microbiol 67(5):557–563
Jukes TH, Cantor CR (1969) Evolution of protein molecules. Academic Press, New York, pp 21–132
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Li WW, Yu HQ (2011) Biohydrogen production with high-rate bioreactors. In: Pandey A et al (eds) Biofuels alternative feedstocks and conversion processes. Elsevier, San Diego
Li YF, Nelson MC, Chen PH, Graf J, Li Y, Yu Z (2015) Comparison of the microbial communities in solid-state anaerobic digestion (SS-AD) reactors operated at mesophilic and thermophilic temperatures. Appl Microbiol Biotechnol 99(2):969–980
Liu Y, Qiao JT, Yuan XZ, Guo RB, Qiu YL (2014) Hydrogenispora ethanolica gen. nov., sp. nov., an anaerobic carbohydrate-fermenting bacterium from anaerobic sludge. Int J Syst Evol Microbiol 64:1756–1762
Mhuantong W, Charoensawan V, Kanokratana P, Tangphatsornruang S, Champreda V (2015) Comparative analysis of sugarcane bagasse metagenome reveals unique and conserved biomass-degrading enzymes among lignocellulolytic microbial communities. Biotechnol Biofuels 8:16
Migliore G, Alisi C, Sprocati AR, Massi E, Ciccoli R, Lenzi M, Wang A, Cremisini C (2012) Anaerobic digestion of macroalgal biomass and sediments sourced from the Orbetello lagoon, Italy. Biomass Bioenergy 42:69–77
Miura T, Kita A, Okamura Y, Aki T, Matsumura Y, Tajima T, Kato J, Nakashimada Y (2014) Evaluation of marine sediments as microbial sources for methane production from brown algae under high salinity. Bioresour Technol 169:362–366
Nesbø CL, Bradnan DM, Adebusuyi A, Dlutek M, Petrus AK, Foght J, Doolittle WF, Noll KM (2012) Mesotoga prima gen. nov., spnov., the first described mesophilic species of the Thermotogales. Extremophiles 16:387–393
Oren A, Garrity GM (2013) List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 63:3931–3934
Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R, Rivière D, Ganesan A, Daegelen P, Sghir A (2008) ‘‘Candidatus Cloacamonas acidaminovorans’’: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190(7):2572–2579
Pope PB, Vivekanand V, Eijsink VGH, Horn SJ (2013) Microbial community structure in a biogas digester utilizing the marine energy crop Saccharina latissimi. Biotech 3:407–414
Rode LM, Genthner BR, Bryant MP (1981) Syntrophic association by cocultures of the methanol- and CO(2)-H(2)-utilizing species Eubacterium limosum and pectin-fermenting Lachnospiramultiparus during growth in a pectin medium. Appl Environ Microbiol 42:20–22
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sasaki K, Morita M, Sasaki D, Nagaoka J, Matsumoto N, Ohmura N, Shinozaki H (2011) Syntrophic degradation of proteinaceous materials by the thermophilic strains Coprothermobacter proteolyticus and Methanothermobacter thermautotrophicus. J Biosci Bioeng 112:469–472
Schink B, Zeikus J (1980) Microbial methanol formation: a major end product of pectin metabolism. Curr Microbiol 4:387–389
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Schramm W, Lehnberg W (1984) Mass culture of brackish-water-adapted seaweeds in sewage-enriched seawater II. Fermentation for biogas production. Hydrobiologia 116–117(1):276–281
Shi J, Wang Z, Stiverson JA, Yu Z, Li Y (2013) Reactor performance and microbial community dynamics during solid-state anaerobic digestion of corn stover at mesophilic and thermophilic conditions. Bioresour Technol 136:574–581
Snelling TJ, Genc B, McKain N, Watson M, Waters SM, Creevey CJ, Wallace RJ (2014) Diversity and community composition of methanogenic archaea in the rumen of Scottish upland sheep assessed by different methods. PLoS ONE 9(9):e106491
Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, Qiu YL (2014) Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol 64(9):2986–2991
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vanegas CH, Bartlett J (2012) Anaerobic digestion of Laminaria digitata: the effect of temperature on biogas production and composition. Waste Biomass Valor 4:509–515. doi:10.1007/s12649-012-9181-z
Vanegas CH, Bartlett J (2013) Green energy from marine algae: biogas production and composition from the anaerobic digestion of Irish seaweed species. Environ Technol 34(15):2277–2283
Wang Y, Zhang Y, Wang J, Meng L (2009) Effects of volatile fatty acid concentrations on methane yield and methanogenic bacteria. Biomass Bioenergy 33(5):848–885
Wang X, Sharp CE, Jones GM, Grasby SE, Brady AL, Dunfield PF (2015) Stable-isotope-probing identifies uncultured Planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl Environ Microbiol 81:4607–4615
Wei N, Quarterman J, Jin YS (2013) Marine macroalgae: an untapped resource for producing fuels and chemicals. Trends Biotechnol 31:70–77
Westerholm M, Roosand S, Schnürer A (2011) Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Syst Appl Microbiol 34:260–266
Yokoyama H, Wagner ID, Wiegel J (2010) Caldicoprobacter oshimai gen. nov., sp. nov., an anaerobic, xylanolytic, extremely thermophilic bacterium isolated from sheep faeces, and proposal of Caldicoprobacteraceae fam. nov. Int J Syst Evol Microbiol 60:67–71
Acknowledgments
This study was supported by the Marine Biotechnology Research Division, Korea Institute of Ocean Science and Technology (KIOST) in-house program (PE99413) and C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning in the Republic of Korea (2015M3D3A1A01064884).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict interests
The authors have declared that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Azizi, A., Kim, W. & Lee, J.H. Comparison of microbial communities during the anaerobic digestion of Gracilaria under mesophilic and thermophilic conditions. World J Microbiol Biotechnol 32, 158 (2016). https://doi.org/10.1007/s11274-016-2112-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2112-6