Abstract
To investigate the effects of temporal variations of process parameters on microbial community structures in the two types of full-scale anaerobic digester treating swine wastewater, three full-scale anaerobic digesters were monitored. An anaerobic filter (AF)-type digester located in Gong-Ju (GJ) showed the highest COD removal among three digesters and maintained stable efficiency. A digester in Hong-Seong (HS) was of the same type as it GJ and showed improved efficiency over the sampling period. A continuously stirred tank reactor (CSTR)-type digester in Soon-Cheon (SC) showed decreasing efficiency due to a high residual concentration of VFAs and NH4+. These process efficiencies were closely correlated to the Simpson indices of the methanogenic communities. Genera Bacillus, Methanosaeta, and Methanospirillum that have filamentous morphology were dominant in both AF-type digesters, but genera Acholeplasma, Methanosarcina, and Methanoculleus that have spherical or coccoid morphology were dominantly abundant in the CSTR-type digester. Correlation between populations suggests a possible syntrophic relationship between genera Desulfobulbus and Methanosaeta in digesters GJ and HS.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Swine wastewater (SWW) is a high-strength liquid organic waste that has a chemical oxygen demand (COD) of 50–150 g/L. In South Korea, more than 30 million tons of SWW were produced in 2012 (MoE 2013). Release of SWW onto the ground or into groundwater causes serious environmental damage. Anaerobic digestion (AD) reduces SWW to a small quantity of innocuous digested sludge solids and generates combustible gas simultaneously, and is therefore a promising treatment technology to treat SWW. Furthermore, owing to its easy biodegradability and high energy-production potential, SWW is more attractive than low-strength organics as a feedstock for the AD process (Kim et al. 2013). Therefore, a plan to construct full-scale digesters for SWW treatment has been instituted in South Korea; 13 full-scale anaerobic livestock wastewater treatment plants are currently operating.
AD is a series of biochemical processes in which a microbial consortium of bacterial and archaeal species decomposes complex organic matter to CH4 and CO2 in the absence of O2 in four main steps: hydrolysis, acidogenesis, acetogenesis, and methanogenesis. Despite the presence of distinct steps, the whole AD process involves complex symbiotic relationships among fast-growing acidogenic bacteria and slow-growing methanogenic archaea. Thus, a balance of populations of these two groups of microorganisms should be sustained to prevent an accumulation of intermediates including volatile fatty acids (VFAs) and H2 and to avoid process instability (Kovács et al. 2013; Pandey et al. 2011). However, full-scale AD plants are susceptible to environmental factors such as heat loss, incomplete mixing, and spatiotemporal difference in substrate characteristics; these fluctuations can impose stress on bacterial and methanogenic communities, eventually leading to process instability and, occasionally, severe disturbance in process efficiency (Madsen et al. 2011). Thus, an understanding of the effects of uncontrollable parameters on process efficiency and microbial community structures in a full-scale AD plant may provide valuable information to predict and improve digester efficiency.
Recent development of high-throughput sequencing methods has enabled investigation of the effects of various environmental factors on the complex microbial community structures in full-scale AD process treating various organic wastes (Cho et al. 2013; Li et al. 2015; Shin et al. 2016; Wan et al. 2013). However, little is known about how process parameters are linked to microbial communities in a full-scale AD plant treating SWW. Moreover, because most full-scale studies have provided only temporal variations of a single plant, only limited information is available concerning regional variations and underlying factors that affect the efficiencies of different types of AD process.
The aim of this study was to quantify how temporal variation in process parameters affects the microbial community structures in two types of full-scale AD plants treating SWW. 454 pyrosequencing was used to analyze the microbial communities of three different full-scale anaerobic digesters treating SWW. Cluster analysis, Spearman’s correlation test, and redundancy analysis (RDA) were performed on samples collected over a year during different seasons and from different locations.
Materials and methods
Digester information, sampling, and physicochemical characterization
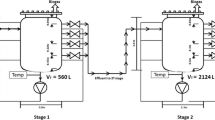
Three full-scale anaerobic digesters treating SWW were located in Gong-Ju, (GJ) Hong-Seong (HS), and Soon-Cheon (SC) in South Korea (Table 1). Digesters GJ and HS are fixed bed reactors equipped with an anaerobic filter (AF). The digester in SC is a continuously stirred tank reactor (CSTR) (Supplementary Fig. 1). All digesters were operated at mesophilic conditions with hydraulic retention times of 9 to 25 days according to the reactor type.
Samples were collected from the influent and the digesters every 3 months for a year at GJ and HS (October 2011 to July 2012), but only two samples could be collected at site SC during this time, because of operation and maintenance problems. Samples are coded by digester and sequence; i.e., HS4 is the fourth sample from HS. Influent samples were taken from pipes conveying mixed substrate into the digester, and digester samples were collected from digestate-circulating pipelines. Samples of 100–200 mL were collected in duplicate sterile plastic containers, stored in a mobile refrigerator at 4 °C, transported to the laboratory within 24 h, and mixed with equal volumes (100 mL) of samples in plastic containers (i.e., total 200 mL). The biogas production and methane contents during the sampling period could not be measured, because all facilities did not measure those biogas production, and all biogas produced was removed by combustion in a flare stack.
COD was measured according to procedures outlined in the Standard Methods (APHA-AWWA-WEF. 2005). Volatile fatty acid (VFA; C2–C6) concentrations were quantified using a gas chromatograph (6890 plus, Agilent, Palo Alto, CA) equipped with an Innowax capillary column and a flame ionization detector. Carbohydrate concentration was quantified using the phenol-sulfuric method (Dubois et al. 1956). Total Kjeldahl nitrogen (TKN) and NH3-N concentrations were measured using the Kjeldahl method (APHA-AWWA-WEF. 2005). Organic nitrogen was estimated as the difference between TKN and NH3-N; protein concentration was determined assuming 6.25 g of protein per gram of organic nitrogen. Lipid concentration was analyzed using gravimetric method after extraction of lipids by solvent (chloroform:methanol, 1:2 v/v) (Bligh and Dyer 1959). All physicochemical analyses were conducted in duplicate.
DNA extraction and 16S rRNA gene amplicon sequencing
Upon arrival from each sampling campaign, 0.2 mL of each digester sample was centrifuged twice at 12,000g for 5 min; the supernatant was removed after each centrifugation to minimize levels of potential PCR inhibitors and DNA from cell debris (Shin et al. 2010). An automated nucleic acid extractor (Magtration System 6GC, Precision System Science, Chiba, Japan) was used to extract DNA from the pelleted samples. The purified DNA was eluted with nuclease-free water and stored at − 20 °C until use.
The V5–V9 hypervariable regions of the 16S rRNA gene were amplified using primers 787f and 1492r (Quince et al. 2009) modified with adapters and barcodes as required by 454 pyrosequencing. The polymerase chain reaction was performed and the purified amplicons were gel-purified as described previously (Kim et al. 2015), with modified thermal cycling conditions: 30 cycles of (1) denaturation at 94 °C for 30 s, (2) annealing at 57 °C for 30 s, and (3) extension at 72 °C for 1 min.
The 454 pyrosequencing was performed by a commercial sequencing service (Macrogen, Seoul, South Korea) according to the manufacturer’s instructions (454 Life Sciences, Branford, CT). To remove the adapter sequences from all amplified reads, an “in-house” program (Macrogen, Seoul, Korea) was used. Low-quality reads (< Q20), short sequences (< 270 bp), and potential chimeras were removed. The resulting sequences were clustered into operational taxonomic units (OTUs) that had ≥ 97% sequence identity by using CD-HIT-OUT (Li et al. 2012). Taxonomic classification was conducted using the SILVA database (http://www.arb-silva.de). All sequences obtained from 454 pyrosequencing were deposited in the National Center for Biotechnology Information (NCBI)’s sequence read archive (PRJNA315957). Accession numbers of samples are GJ1 (SRX1654845), GJ2 (SRX1654846), GJ3 (SRX1654847), GJ4 (SRX1654848), HS1 (SRX1654837), HS2 (SRX1654839), HS3 (SRX1654840), HS4 (SRX1654841), SC1 (SRX1654854), and SC2 (SRX1654855).
Statistical analysis (correlation test and redundancy analysis)
Duncan’s multiple range tests at p = 0.05 were used to compare the means and to group the process parameters. Correlation test and multivariate analysis were performed based on the process parameters and relative abundance of microbial communities obtained in this study. Spearman’s correlation coefficients were calculated using R software, and multivariate analysis was conducted using Canoco 5.0 software (Plant Research International, The Netherlands). RDA was conducted using bacterial and archaeal community data at the genus level and with selected process parameters, which were screened by a forward selection procedure. Bacterial and archaeal diversity indices were calculated using OUT data after rarefying to the smallest sequencing depth (Supplementary Fig. 2).
Results and discussion
Physicochemical properties of three digesters treating SWW
Physicochemical properties of the influent and effluent samples from three full-scale anaerobic digesters treating SWW were analyzed to obtain information on the parameters for use in AD process assessment (Table 2). Clear regional and temporal variations in characteristics of SWW and process efficiencies of digesters were observed.
The average COD concentration of SWW fed to the digesters was 40.0 ± 29.0 g/L, with coefficient of variation (CV) = 72.4%. The influent of digester SC had the highest organic strength (90.0 ± 6.1 g COD/L), and digester GJ had the lowest (16.8 ± 4.9 g COD/L). This large variation in organic strength may be attributable to differences of the scale of swine barns, cleaning method, frequency and water usage, and feces-urine separating method (Choi et al. 2011).
The COD removal in the three digesters varied from 44.0 to 89.2% (Fig. 1) and was significantly negatively correlated (p < 0.01) with residual concentration of NH4+ ion (Spearman’s coefficient ρ = − 0.85), acetate (ρ = − 0.81), and propionate (ρ = − 0.78). Among digesters, GJ showed stable efficiency with the highest COD removal (86.0 ± 4.0%; CV = 4.7%) during the whole investigation period. In digester HS, the COD removal was low (44.0–45.1%) during the first and second sampling (i.e., HS1 and HS2), but thereafter increased to 73.4–77.2%. In contrast, in digester SC, COD removal was high (75.1%) in the first sampling period, but decreased to 55.7% in the second sampling period.
The total concentrations of carbohydrate, protein, and lipid of SWW in the influent of the digester was 6.3 ± 2.3 g/L in GJ, 14.6 ± 3.6 g/L in HS, and 47.8 ± 5.3 g/L in SC. Protein (58.7 ± 19.6%) and lipid (26.3 ± 11.9%) were the major subgroups of total organic components in SWW used in this study. Carbohydrate contribution was low (15.1 ± 9.6%) except in the influent of digester SC. This low concentration may occur because carbohydrate is easily fermented to VFAs during transport and storage (Miron et al. 2000). Thus, the organic strength was clearly stratified along with the location of digester, and regional variations in organic properties of SWW were larger than temporal variations within sites. Average removals over all samples were 54.4 ± 13.0% for carbohydrate and 56.6 ± 18.9% and lipid, but only 43.6 ± 19.8% for protein. The low removal of protein may be partly attributable to the presence of microbes in the effluent, because they consist of 40–50% (dry weight basis) microbial protein (Atkinson and Mavituna 1991). Thus, protein removal efficiency might be underestimated due to the presence or growth of microbes in the digester.
In all effluent samples collected from digester GJ, the concentrations of TVFA (0.10 ± 0.07 g/L) and NH4+ (0.77 ± 0.20 g/L) were lower than in other digesters (Table 2). This stable operation without VFA accumulation was presumably due to the low organic strength of influent and metabolic balance between acidogenic and methanogenic microorganisms in the anaerobic digester (Mulat et al. 2016).
The average residual concentration of TVFA in the effluents was 1.64 ± 0.62 g/L in digester HS and 8.41 ± 5.80 g/L in digester SC. Acetate (68.6 ± 16.2% of TVFA) and propionate (19.3 ± 10.3% of TVFA) were the most abundant intermediates. The concentration of NH4+ in the effluent of was 1.81 ± 0.28 g/L in digester HS and 4.56 ± 0.95 g/L in digester SC. High concentration (1.7–14 g/L) of NH4+ can inhibit the activity of bacteria and archaea (Chen et al. 2008). In digester HS, the concentration of NH4+ ranged from 1.91 to 2.14 g/L during the first and second sampling periods, during which coincided COD removal efficiency was poor (44.0–45.1%) (Table 2, Fig. 1). Thereafter, the concentration of NH4+ decreased to 1.51 g/L and COD removal increased to 77.2% in the fourth sampling period (HS4).
In contrast, in digester SC, the concentration of NH4+ was 3.73 g/L at first sampling period showed good COD removal (75.1%), but the COD removal rapidly decreased in as NH4+ concentration increased to 5.60 g/L. Because susceptibility to NH4+ concentration can vary widely depending on substrate and inoculum, degree of acclimation, and other environmental conditions (Chen et al. 2008), the inhibition by NH4+ might differ between the two digesters.
Microbial community structures in the three digesters
Bacterial community structures
A total 111,127 raw bacterial and archaeal sequence reads were obtained from ten effluents of three anaerobic digesters by using 454 pyrosequencing analysis. Twenty-four phyla accounted for 96.2 ± 1.8% the total bacterial reads. Ten phyla that had relative abundance > 1.0% in all samples were considered significant groups; the remaining 14 phyla collectively accounted for 1.2 ± 0.7% of bacterial reads and were considered as minor groups (Supplementary Fig. 3). Phyla Proteobacteria (29.6 ± 16.9%), Firmicutes (37.7 ± 11.0%), and Bacteroidetes (11.2 ± 2.7%) were dominant in all samples; these groups are commonly detected with high relative abundance in anaerobic digesters that treat various substrates (Li et al. 2010; Lim et al. 2013; Liu et al. 2015; Shin et al. 2016; Wan et al. 2013).
At the genus level, 59.2 ± 7.9% of the total bacteria were classified into 500 genera; 38 genera that showed relative abundance > 1.0% in at least one of the ten effluent samples were designated as significant groups (Table 3). Among these 38 genera, Acholeplasma (4.8 ± 5.3%), Alkaliphilus (3.1 ± 2.5%), Arcobacter (2.9 ± 4.2%), Bacillus (4.2 ± 3.3), Clostridium (1.1 ± 0.9%), Fastidiosipila (6.0 ± 2.3%), Geobacter (1.4 ± 3.6%), Pseudomonas (7.3 ± 6.5%), Thauera (5.2 ± 10.2%), Tissierella (1.2 ± 1.2%), and Truepera (2.2 ± 3.3%) were dominant, with ≥ 1.0% average abundance in all samples.
Genus Arcobacter (6.8 ± 4.3%) and Clostridium (2.0 ± 0.6%) were more abundant in digester HS than in GJ and SC. Arcobacter cannot ferment carbohydrate nor produce acidic intermediates, but can use amino acids as energy sources (George Garrity et al. 2005). Many species of genus Clostridium can use amino acids, and may help to degrade protein in SWW (Vos et al. 2009).
Genus Fastidiosipila can grow in anaerobic and microaerobic conditions; it was detected evenly in all digesters with high relative abundance from 2.1 to 9.9% (Falsen et al. 2005). This genus cannot ferment carbohydrates and only produces small amounts of acids in Fastidious Anaerobe Broth with meat granules (Falsen et al. 2005). In a study of an anaerobic membrane reactor treating landfill leachate genus Fastidiosipila was the most abundant regardless of NH4+ concentration in the reactor (Xie et al. 2014). Thus, in this study, genus Fastidiosipila may tolerate the high level of NH4+ up to 5.60 g/L and partly use proteins for growth.
Thauera was the most dominant bacterial genus in GJ1; this bacterium is an aerobe and can be detected in wastewater treatment plants (George Garrity et al. 2005). The highest abundance (30.3% in GJ1) of genus Thauera was likely due to re-inoculation with anaerobic sludge taken from a full-scale anaerobic digester treating primary and secondary sludge 3 months before the GJ1. As AD progressed, the relative abundance of genus Thauera decreased to < 0.5% by GJ4.
The relative abundance of genera Geobacter and Tissierella suddenly increased in GJ4. These genera are obligate anaerobes that can obtain energy for growth from oxidation of multi-carbon compounds such as aromatic hydrocarbons with reduction of Fe(III) (Lovley and Anderson 2000). Genus Geobacter can be abundant in anaerobic microbial fuel cells due to its exoelectrogenic characteristics (Lu et al. 2012; Vargas et al. 2013). Genus Tissierella typically produces acetate, ammonia, and carbon dioxide; Tissierella praeacuta can hydrolyze gelatin. Tissierella has been identified in other digesters treating agricultural wastes and microalgal biomass (Jaenicke et al. 2011; Zhao et al. 2016). Thus, during AD of SWW, Geobacter may contribute to decomposition of hydrocarbons, and Tissierella may contribute to decomposition of proteins.
Genera Acholeplasma, Alkaliphilus, Caldicoprobacter, and Genus Candidatus Cloacamonas were more abundant in digester SC (14.3 ± 4.2, 6.5 ± 1.9, 2.2 ± 1.3, and 3.2 ± 1.6%, respectively) than in GJ and HS. Genus Acholeplasma is facultative anaerobe and most species in it can utilize various sugars as major energy sources, so they partly contribute to degradation of carbohydrate (Parte et al. 2011). Also, many stains in genus Acholeplasma can synthesize fatty acids from acetate (Bittman 1993); by this process, these strains may compete for acetate with aceticlastic methanogens (AMs), and this interaction may explain lower abundance of AM in digester SC (5.1 ± 1.7%) than in GJ and HS (Fig. 2b). Genus Alkaliphilus is a strict anaerobe that can ferment various organic substrates in the presence or absence of electron acceptors such as thiosulfate, fumarate, crotonate, iron, cobalt, or chromium for growth (Vos et al. 2009). Thus, the higher abundance of genus Alkaliphilus in digester SC than in GJ and HS may be due to the higher concentration of inorganic elements in SWW fed to SC than to GJ and HS. Genus Caldicoprobacter is composed of thermophilic and neutrophilic bacteria that can ferment a wide range of sugars, but not proteins (Bouanane-Darenfed et al. 2014). Genus Candidatus Cloacamonas is considered to be a hydrogen-producing syntroph (Gao et al. 2015), so the higher abundance of Candidatus Cloacamonas in digester SC may influence the dominance of genus Methanoculleus, which scavenges hydrogen gas. In contrast to those genera, genus Bacillus was more abundant in digester GJ (3.9 ± 2.4%) and HS (6.5 ± 3.0%) than in digester SC. Several species of genus Bacillus can utilize NH4+ as their nitrogen source, and thereby reduce ammonia inhibition of anaerobes in digesters GJ and HS (Vos et al. 2009).
Archaeal community structures and cluster analysis
454 pyrosequencing analysis obtained 20,043 archaeal sequence reads; 99.1 ± 1.3% of them belonged to four orders: Methanomicrobiales (75.6 ± 10.4%), Methanosarcinales (16.0 ± 12.9%), Thermoplasmatales (4.5 ± 4.0%), and Methanobacteriales (3.0 ± 2.7%) (Fig. 2a). Methanomicrobiales are hydrogenotrophic methanogens (HMs); this order was greatly dominant with an average relative abundance of 75.6 ± 10.4% over all samples. This result concurs with previous studies that reported the dominance of HMs during AD of SWW (Cho et al. 2013; Da Silva et al. 2015; Song et al. 2010).
Sixteen genera of methanogens accounted for 93.8 ± 4.6% of archaeal reads. Methanimicrococcus (1.3 ± 2.0%), Methanobrevibacter (2.2 ± 1.7%), Methanoculleus (22.0 ± 24.8%), Methanogenium (10.5 ± 13.2%), Methanosaeta (11.8 ± 13.8%), Methanosarcina (1.9 ± 1.8%), and Methanospirillum (40.9 ± 21.9%) showed ≥ 1.0% average abundance in all samples and were considered major groups (Table 4).
In GJ, genera Methanospirillum and Methanosaeta accounted for 87.2% of the archaeal community in GJ1, but the relative abundance of Methanosaeta dramatically decreased to 5.6% in GJ2, and stayed < 6% until GJ4. In contrast, Methanoculleus and Methanogenium had abundance < 2.1% in GJ1, but their numbers suddenly increased to 12.8–17.0% (Methanoculleus) and 25.1–36.7% (Methanogenium) in GJ2, but decreased to 8.1% (Methanoculleus) and 6.0% (Methanogenium) in GJ4.
In HS, the dominant archaeal genus in the digester HS was Methanospirillum (61.5 ± 6.8%), followed by Methanosaeta (19.5 ± 12.9%); Methanoculleus decreased from 17.1% in HS1 to 5.5% in HS4. Genus Methanosarcina (1.4 ± 0.9%) was less abundant in GJ and HS than in SC because filamentous microorganisms (i.e., Methanosaeta) are better adapted to attached growth processes such as anaerobic filters than spherical microbes are (i.e., Methaosarcina) (Cheng et al. 1991).
In SC, genus Methanoculleus (69.3 ± 4.8%) was the most dominant, and Methanosarcina (5.0 ± 1.8%) was more abundant than Methanosaeta (0.2 ± 0.1%). Genus Methanosaeta generally has a lower maximum specific growth rate and half-saturation constant than Methanosarcina (De Vrieze et al. 2012; Yu et al. 2006). Thus, the predominance of Methanosarcina over Methanosaeta in SC, in contrast to digester GJ can be explained by the much higher concentration of acetate in SC (1694.7–4050.5 mg COD/L) than in GJ (27.6–208.1 mg COD/L). Nevertheless, genus Methanosaeta was more dominant than Methanosarcina even at high concentration of acetate (843.2–1988.2 mg COD/L) in digester HS. This dominance may occur because the filamentous morphology of Methanosaeta contributes to immobilization and granulation of cells and the good efficiency of the reactors (Kita et al. 2016; Zhou et al. 2015).
To visualize the similarity and dissimilarity of microbial community structures among the three digesters, cluster analysis was conducted separately for bacterial and archaeal communities based on the relative abundance of each out. Both bacterial and archaeal communities were clearly divided into two clusters at the 20% level of the information remaining criterion (Supplementary Fig. 4). Interestingly, eight samples in Cluster 1 were taken from AF-type digesters (i.e., GJ, HS), but the two samples in Cluster 2 were collected from the CSTR-type digester (SC). Genera Bacillus, Methanosaeta, and Methanospirillum that have filamentous morphology could proliferate with the fixed bed in the digester and are more dominant in AF-type digesters than in in the CSTR-type (Oren 2014a, b; Vos et al. 2009). In contrast, genera Acholeplasma, Methanosarcina, and Methanoculleus that have spherical or coccoid morphology are more abundant in CSTR-type digesters than in AF-type digesters (Oren 2014a, c; Parte et al. 2011). Thus, the type of anaerobic digester likely had a significant effect on the formation of different microbial communities.
Diversity indices and correlation analysis
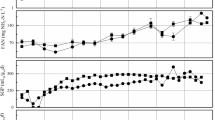
The average bacterial diversities (Fig. 3a) were calculated for the three digesters. Shannon indices were 3.42 ± 0.43 in GJ, 3.52 ± 0.29 in HS, and 3.83 ± 0.16 in SC. Simpson indices were 0.91 ± 0.04 in GJ, 0.93 ± 0.02 in HS, and 0.96 ± 0.01 in SC. The difference in bacterial diversity indices among the three digesters were not significant for either index (p > 0.05; Duncan’s multiple range test).
The average archaeal diversities (Fig. 3b) were also calculated. Shannon diversity indices were 1.54 ± 0.41 in GJ, 1.03 ± 0.16 in HS, and 1.12 ± 0.08 in SC. Simpson indices were 0.66 ± 0.12 in GJ, 0.49 ± 0.08 in HS, and 0.48 ± 0.05 in SC. The differences in both indices were not significant (p > 0.05) between the archaeal communities in digesters HS and SC, but both indices for digester GJ were statistically different from other digesters.
The Shannon and Simpson indices had opposite relations to the process parameters of the two microbial domains. For bacterial communities, the Shannon indices correlated more closely than Simpson indices with the process parameters; bacterial Shannon indices were positively correlated with the residual concentrations of VS, acetate, and TVFA (ρ = 0.66; all p < 0.05). In contrast, for archaeal communities, Simpson indices correlated more significantly than Shannon indices with the process parameters; archaeal Simpson index was positively related with COD removal efficiency (ρ = 0.73; p < 0.05) but negatively correlated with residual concentrations of VS (ρ = − 0.66; p < 0.05) and acetate and TVFA (ρ = − 0.68; p < 0.05).
To investigate the possible interactions between bacterial and archaeal communities, Spearman’s correlation was conducted using the relative abundances of bacterial and archaeal genera. To remove noisy data, major methanogenic genera that showed a clear shift were first selected, then major bacterial genera that showed statistically significant correlation with more than three major methanogenic genera were selected. Genus Desulfobulbus (0.4 ± 0.4%) showed a high positive correlation with Methanosaeta, Methanospirillum, and Methanolinea (ρ > 0.7; p < 0.05), but a high negative correlation with Methanoculleus (ρ = − 0.62; p < 0.1). Desulfobulbus is a sulfate-reducing bacterium that can oxidize propionate to acetate in the presence of sulfate (Harmsen et al. 1996). Accordingly, a syntrophic consortium between AMs and propionate-oxidizing bacteria can be established, and Methanosaeta has been mainly detected as a partner (Harmsen et al. 1996; Oude Elferink et al. 1998; Ziganshin et al. 2011). Thus, a syntrophic relationship between Desulfobulbus and Methanosaeta might be constructed, despite the low relative abundance of Desulfobulbus (≤ 1.0%) in GJ1 and HS1–2. However, a decrease of Desulfobulbus to 0.07% in GJ2–3 and 0.15% in HS3 may cause changes in this syntrophic relationship and result in decrease in numbers of Methanosaeta. In addition, because filamentous Methanosaeta is more vulnerable to ammonia inhibition than HMs are, varying concentrations of NH4+ may affect the growth and activity of Methanosaeta. Methanosarcina underwent smaller changes in abundance than Methanosaeta. The difference may be attributable to differences in phenotype such as size and shape. Methanosarcina cells are large and spherical, so they have a high volume-to-surface ratio, and they form clusters; this behavior increases their resistance to ammonia inhibition (De Vrieze et al. 2012).
Ordination of process parameters and microbial community structures
RDA was used to produce an ordination diagram (Fig. 3) of environmental parameters with regard to bacterial and archaeal communities in the samples. The result of ordination had a significant eigenvalue = 0.618 with p < 0.05, which means that the environment variables explained 61.8% of the variance in the species variables at the 95% confidence level. The RDA plot indicated that the microbial community structure tended to vary among localities and seasons. COD removal efficiency was positively correlated with protein removal, but negatively correlated with residual concentration of TVFA and NH4+. No correlation was observed between protein removal and residual concentration of TVFA or NH4+. Even though some differences in statistical significance existed, the correlation trend among process parameters corresponded to the results of Spearman’s correlation.
All samples from digesters GJ and HS were located along the vertical axis of the ordination diagram; this pattern means that COD and protein removal had a greater influence than the concentration of TVFA and NH4+ on the arrangement of species variables of both digesters. The ten samples from three digesters can be divided into two groups based on the transverse axis, which means that the effects of TVFA and NH4+ on the placement of species data was less significant among samples from digester GJ and HS due to a much lower concentration of TVFA and NH4+ than in samples from SC.
These findings regarding the influence of physicochemical efficiency, the statistical view of ecological parameters, and the minute examination of bacterial and methanogenic communities may guide development of an optimal strategy to stabilize field operation and to develop AD processes by improving efficiencies and improving the understanding of processes that occur within AD reactors.
Conclusion
AD efficiency was monitored at three sites. The AF-type digester GJ showed the highest COD removal efficiencies among three digesters and maintained stable digestion efficiency. HS also an AF-type digester; it showed improved digestion efficiency during the sampling period. SC was a CSTR-type digester; it had the highest residual concentration of VFAs and NH4+ so its digestion efficiency was relatively low. These process efficiencies corresponded well to the Simpson indices of the archaeal communities. In both AF-type digesters, genera Bacillus, Methanosaeta, and Methanospirillum that have filamentous morphology were dominant, but in the CSTR-type digester, genera Acholeplasma, Methanosarcina, and Methanoculleus that have spherical or coccoid morphology were most abundant.
References
APHA-AWWA-WEF (2005) Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C., 1167 pp
Atkinson B, Mavituna F (1991) Biochemical engineering and biotechnology handbook. The Nature Press, New York
Bittman R (1993): Mycoplasma membrane lipids chemical composition and transbilayer distribution. In: Rottem S, Kahane I (Editors), Mycoplasma cell membranes. Springer US, Boston, MA, pp. 29–52, DOI: https://doi.org/10.1007/978-1-4615-2924-8_2
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(1):911–917. https://doi.org/10.1139/y59-099
Bouanane-Darenfed A, Fardeau M-L, Ollivier B (2014) The family Caldicoprobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: Firmicutes and Tenericutes. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 13–17
Chen Y, Cheng JJ, Creamer KS (2008) Inhibition of anaerobic digestion process: a review. Bioresour Technol 99(10):4044–4064. https://doi.org/10.1016/j.biortech.2007.01.057
Cheng P, Mueller RE, Jaeger S, Bajpai R, Iannotti EL (1991) Lactic acid production from enzyme-thinned corn starch using Lactobacillus amylovorus. J Ind Microbiol Biotechnology 7:27–34
Cho K, Lee J, Kim W, Hwang S (2013) Behavior of methanogens during start-up of farm-scale anaerobic digester treating swine wastewater. Process Biochem 48(9):1441–1445. https://doi.org/10.1016/j.procbio.2013.04.016
Choi EH, Yoon YM, Kim CH (2011) Generation and characteristics analysis of swine manure for introducing biogas system. 한국관개배수위원회 18:25–32
Da Silva MLB, Cantão ME, Mezzari MP, Ma J, Nossa CW (2015) Assessment of bacterial and archaeal community structure in swine wastewater treatment processes. Microb Ecol 70(1):77–87. https://doi.org/10.1007/s00248-014-0537-8
De Vrieze J, Hennebel T, Boon N, Verstraete W (2012) Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol 112:1–9. https://doi.org/10.1016/j.biortech.2012.02.079
Dubois M, Gilles M, Hamilton J, Godess P, Smith F (1956) Colorimetric method for determination of sugars related substances. Anal Chem 28(3):350–356. https://doi.org/10.1021/ac60111a017
Falsen E, Collins MD, Welinder-Olsson C, Song Y, Finegold SM, Lawson PA (2005) Fastidiosipila sanguinis gen. nov., sp. nov., a new Gram-positive, coccus-shaped organism from human blood. Int J Syst Evol Microbiol 55(2):853–858. https://doi.org/10.1099/ijs.0.63327-0
Gao S, Zhao M, Chen Y, Yu M, Ruan W (2015) Tolerance response to in situ ammonia stress in a pilot-scale anaerobic digestion reactor for alleviating ammonia inhibition. Bioresour Technol 198:372–379. https://doi.org/10.1016/j.biortech.2015.09.044
George Garrity, Don J. Brenner, Noel R. Krieg, Staley JR (2005) Bergey’s Manual® of Systematic Bacteriology. Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria. Springer US, United States of America
Harmsen HJ, Akkermans AD, Stams AJ, de Vos WM (1996) Population dynamics of propionate-oxidizing bacteria under methanogenic and sulfidogenic conditions in anaerobic granular sludge. Appl Environ Microbiol 62(6):2163–2168
Jaenicke S, Ander C, Bekel T, Bisdorf R, Dröge M, Gartemann K-H (2011) Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS One 6(1):e14519. https://doi.org/10.1371/journal.pone.0014519
Kim W, Cho K, Lee S, Hwang S (2013) Comparison of methanogenic community structure and anaerobic process performance treating swine wastewater between pilot and optimized lab scale bioreactors. Bioresour Technol 145:48–56. https://doi.org/10.1016/j.biortech.2013.02.044
Kim W, Shin SG, Han G, Cho K, Hwang S (2015) Structures of microbial communities found in anaerobic batch runs that produce methane from propionic acid—seeded from full-scale anaerobic digesters above a certain threshold. J Biotechnol 214:192–198. https://doi.org/10.1016/j.jbiotec.2015.09.040
Kita A, Suehira K, Miura T, Okamura Y, Aki T, Matsumura Y, Tajima T, Nishio N, Nakashimada Y (2016) Characterization of a halotolerant acetoclastic methanogen highly enriched from marine sediment and its application in removal of acetate. J Biosci Bioeng 121(2):196–202. https://doi.org/10.1016/j.jbiosc.2015.05.018
Kovács E, Wirth R, Maróti G, Bagi Z, Rákhely G, Kovács KL (2013) Biogas production from protein-rich biomass: fed-batch anaerobic fermentation of casein and of pig blood and associated changes in microbial community composition. PLoS One 8(10):e77265. https://doi.org/10.1371/journal.pone.0077265
Li L, He Q, Ma Y, Wang X, Peng X (2015) Dynamics of microbial community in a mesophilic anaerobic digester treating food waste: relationship between community structure and process stability. Bioresour Technol 189:113–120. https://doi.org/10.1016/j.biortech.2015.04.015
Li P, Wang Y, Wang Y, Liu K, Tong L (2010) Bacterial community structure and diversity during establishment of an anaerobic bioreactor to treat swine wastewater. Water Sci Technol 61(1):243–252. https://doi.org/10.2166/wst.2010.807
Li W, Fu L, Niu B, Wu S, Wooley J (2012) Ultrafast clustering algorithms for metagenomic sequence analysis. Briefings in Bioinformatics
Lim JW, Chen CL, Ho IJR, Wang JY (2013) Study of microbial community and biodegradation efficiency for single- and two-phase anaerobic co-digestion of brown water and food waste. Bioresour Technol 147:193–201. https://doi.org/10.1016/j.biortech.2013.08.038
Liu A-C, Chou C-Y, Chen L-L, Kuo C-H (2015) Bacterial community dynamics in a swine wastewater anaerobic reactor revealed by 16S rDNA sequence analysis. J Biotechnol 194:124–131. https://doi.org/10.1016/j.jbiotec.2014.11.026
Lovley DR, Anderson RT (2000) Influence of dissimilatory metal reduction on fate of organic and metal contaminants in the subsurface. Hydrogeol J 8(1):77–88. https://doi.org/10.1007/PL00010974
Lu L, Xing D, Ren N, Logan BE (2012) Syntrophic interactions drive the hydrogen production from glucose at low temperature in microbial electrolysis cells. Bioresour Technol 124:68–76. https://doi.org/10.1016/j.biortech.2012.08.040
Madsen M, Holm-Nielsen JB, Esbensen KH (2011) Monitoring of anaerobic digestion processes: a review perspective. Renew Sust Energ Rev 15(6):3141–3155. https://doi.org/10.1016/j.rser.2011.04.026
Miron Y, Zeeman G, van Lier JB, Lettinga G (2000) The role of sludge retention time in the hydrolysis and acidification of lipids, carbohydrates and proteins during digestion of primary sludge in CSTR systems. Water Res 34(5):1705–1713. https://doi.org/10.1016/S0043-1354(99)00280-8
MoE (2013) Statistics of livestock wastewater treatment, Seoul, South Korea
Mulat DG, Jacobi HF, Feilberg A, Adamsen APS, Richnow H-H, Nikolausz M (2016) Changing feeding regimes to demonstrate flexible biogas production: effects on process performance, microbial community structure, and methanogenesis pathways. Appl Environ Microbiol 82(2):438–449. https://doi.org/10.1128/AEM.02320-15
Oren A (2014a) The family Methanosarcinaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: other major lineages of bacteria and the archaea. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 259–281
Oren A (2014b) The family Methanospirillaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: other major lineages of bacteria and the archaea. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 283–290
Oren A (2014c) The family Methanomicrobiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: other major lineages of bacteria and the archaea. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 231–246. https://doi.org/10.1007/978-3-642-30197-1_257
Oude Elferink SJWH, Vorstman WJC, Sopjes A, Stams AJM (1998) Characterization of the sulfate-reducing and syntrophic population in granular sludge from a full-scale anaerobic reactor treating papermill wastewater. FEMS Microbiol Ecol 27(2):185–194. https://doi.org/10.1111/j.1574-6941.1998.tb00536.x
Pandey PK, Ndegwa PM, Soupir ML, Alldredge JR, Pitts MJ (2011) Efficacies of inocula on the startup of anaerobic reactors treating dairy manure under stirred and unstirred conditions. Biomass Bioenergy 35(7):2705–2720. https://doi.org/10.1016/j.biombioe.2011.03.017
Parte A, Krieg NR, Ludwig W, Whitman W, Hedlund BP, Paster BJ, Staley JT, Ward N, Brown D (2011) Bergey’s Manual of Systematic Bacteriology: Volume 4: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. Springer New York
Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Meth 6(9):639–641. https://doi.org/10.1038/nmeth.1361
Shin SG, Han G, Lim J, Lee C, Hwang S (2010) A comprehensive microbial insight into two-stage anaerobic digestion of food waste-recycling wastewater. Water Res 44(17):4838–4849. https://doi.org/10.1016/j.watres.2010.07.019
Shin SG, Koo T, Lee J, Han G, Cho K, Kim W, Hwang S (2016) Correlations between bacterial populations and process parameters in four full-scale anaerobic digesters treating sewage sludge. Bioresour Technol 214:711–721. https://doi.org/10.1016/j.biortech.2016.05.021
Song M, Shin SG, Hwang S (2010) Methanogenic population dynamics assessed by real-time quantitative PCR in sludge granule in upflow anaerobic sludge blanket treating swine wastewater. Bioresour Technol 101(1):S23–S28. https://doi.org/10.1016/j.biortech.2009.03.054
Vargas IT, Albert IU, Regan JM (2013) Spatial distribution of bacterial communities on volumetric and planar anodes in single-chamber air-cathode microbial fuel cells. Biotechnol Bioeng 110(11):3059–3062. https://doi.org/10.1002/bit.24949
Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman W (2009) Bergey’s Manual of Systematic Bacteriology: Volume 3: The Firmicutes. Springer New York
Wan S, Sun L, Sun J, Luo W (2013) Biogas production and microbial community change during the co-digestion of food waste with Chinese silver grass in a single-stage anaerobic reactor. Biotechnol Bioprocess Eng 18(5):1022–1030. https://doi.org/10.1007/s12257-013-0128-4
Xie Z, Wang Z, Wang Q, Zhu C, Wu Z (2014) An anaerobic dynamic membrane bioreactor (AnDMBR) for landfill leachate treatment: performance and microbial community identification. Bioresour Technol 161:29–39. https://doi.org/10.1016/j.biortech.2014.03.014
Yu Y, Kim J, Hwang S (2006) Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol Bioeng 93(3):424–433. https://doi.org/10.1002/bit.20724
Zhao B, Liu J, Frear C, Holtzapple M, Chen S (2016) Consolidated bioprocessing of microalgal biomass to carboxylates by a mixed culture of cow rumen bacteria using anaerobic sequencing batch reactor (ASBR). Bioresour Technol 222:517–522. https://doi.org/10.1016/j.biortech.2016.09.120
Zhou L, Yu H, Ai G, Zhang B, Hu S, Dong X (2015) Transcriptomic and physiological insights into the robustness of long filamentous cells of Methanosaeta harundinacea, prevalent in upflow anaerobic sludge blanket granules. Appl Environ Microbiol 81(3):831–839. https://doi.org/10.1128/AEM.03092-14
Ziganshin A, Schmidt T, Scholwin F, Il’inskaya O, Harms H, Kleinsteuber S (2011) Bacteria and archaea involved in anaerobic digestion of distillers grains with solubles. Appl Microbiol Biotechnol 89(6):2039–2052. https://doi.org/10.1007/s00253-010-2981-9
Funding
This work was supported by the International Joint R&D project for Energy Technology of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) and the Ministry of Trade, Industry & Energy (MOTIE) of the Republic of Korea (No. 2012T100100621) and by the “Human Resources Program in Energy Technology” of the KETEP grant funded by the MOTIE, Republic of Korea (No. 20144030200460).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Gerald Thouand
Electronic supplementary material
ESM 1
(PDF 2.77 mb)
Rights and permissions
About this article
Cite this article
Han, G., Shin, S.G., Cho, K. et al. Temporal variation in bacterial and methanogenic communities of three full-scale anaerobic digesters treating swine wastewater. Environ Sci Pollut Res 26, 1217–1226 (2019). https://doi.org/10.1007/s11356-017-1103-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-1103-y