Abstract
The aim of this work was to study the biodiversity of yeasts isolated from the autochthonous grape variety called “Uva di Troia”, monitoring the natural diversity from the grape berries to wine during a vintage. Grapes were collected in vineyards from two different geographical areas and spontaneous alcoholic fermentations (AFs) were performed. Different restriction profiles of ITS–5.8S rDNA region, corresponding to Saccharomyces cerevisiae, Issatchenkia orientalis, Metschnikowia pulcherrima, Hanseniaspora uvarum, Candida zemplinina, Issatchenkia terricola, Kluyveromyces thermotolerans, Torulaspora delbrueckii, Metschnikowia chrysoperlae, Pichia fermentans, Hanseniaspora opuntiae and Hanseniaspora guilliermondii, were observed. The yeast occurrences varied significantly from both grape berries and grape juices, depending on the sampling location. Furthermore, samples collected at the end of AF revealed the great predominance of Saccharomyces cerevisiae, with a high intraspecific biodiversity. This is the first report on the population dynamics of ‘cultivable’ microbiota diversity of “Uva di Troia” cultivar from the grape to the corresponding wine (“Nero di Troia”), and more general for Southern Italian oenological productions, allowing us to provide the basis for an improved management of wine yeasts (with both non-Saccharomyces and Saccharomyces) for the production of typical wines with desired unique traits. A certain geographical-dependent variability has been reported, suggesting the need of local based formulation for autochthonous starter cultures, especially in the proportion of the different species/strains in the design of mixed microbial preparations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The indigenous microbiota is very important in winemaking process, in reason of the possible positive or negative effects on wine quality. In particular, yeasts are essential for the carrying out of the alcoholic fermentation (AF), promoting the transformation of grape sugars into ethanol, carbon dioxide and hundreds of other metabolites (Romano et al. 2003a). Starter cultures based on selected strains of Saccharomyces cerevisiae are usually added by oenologists to control the fermentative process, in order to dominate yeasts belonging to the vineyard environment, winery facilities and cellar equipment. The International Organization of Vine and Wine (OIV) affirmed that terroir refers to ‘an area in which collective knowledge of the interactions between the identifiable physical and biological environments and applied viticulture and oenological practices develops, giving distinctive characteristics for the products originating from this area’ (International Organization of Vine and Wine 2010). The definition of "terroir" represents the foundation of the Appellation of Origin, with impact on the wine market and consumer choices. It has been demonstrated that the non-Saccharomyces yeasts contribute to wine qualities (Ciani et al. 2010; Jolly et al. 2014). Different studies have highlighted the important role of the microbiota associated with the “terroir” from which the grapes are grown, able to impart a unique quality to the wine (e.g. Csoma et al. 2010; Di Maio et al. 2012). In the grape/wine environment, Bokulich et al. (2014) have studied the “microbial terroir” and they showed the existence of a close relationship between microbial patterns, region of production and climate. On the above basis, an increasing number of scientific investigations have focused the attention on the cultivable micro-biodiversity connected with spontaneous fermentation, in order to select indigenous strains, displaying positive technological properties and quality traits, for their application in industrial fermentations (e.g. in Apulian region Cappello et al. 2008; Capozzi et al. 2010, 2012; Grieco et al. 2010; Tristezza et al. 2012, 2013, 2014; Garofalo et al. 2015).
During the spontaneous fermentation process, a dynamics of different yeast species occur: the non-Saccharomyces yeasts (mainly belonging Hanseniaspora/Kloeckera, Candida, Pichia, Zygosaccharomyces, Schizosaccharomyces, Torulaspora, Kluyveromyces and Metschnikowia genera) dominate the beginning of AF and then they are replaced by Saccharomyces cerevisiae that complete sugars conversion in ethylic alcohol (Fleet 2008; Ciani et al. 2010; Jolly et al. 2014).
The selection of non-Saccharomyces is very important for the preparation of new starter cultures, since they are able to produce several secondary compounds that can have a positive influence on the quality of the wine (Fleet 2008; Ciani et al. 2010; Bely et al. 2008; De Benedictis et al. 2011). In fact, non-Saccharomyces species may also have an application to improve the wine technological proprieties and to enhance the unique sensorial qualities of typical productions (Fleet 2008; Ciani et al. 2010; Bely et al. 2008; De Benedictis et al. 2011). Non-Saccharomyces can also be used as agents for the biological control of moulds or spoilage microorganism, such as lactic acid bacteria or Brettanomyces bruxellensis (Capozzi et al. 2015; Oro et al. 2014). However, non-Saccharomyces utilization is also associate with such as production of biogenic amines, off-flavors (acetic acid, esters, acetaldehydes, H2S) and with competition for the nutrients availability with S. cerevisiae strains able to complete AF (Capozzi et al. 2015).
Even though, several studies had been already performed in order to characterized autochthonous microbes from Apulian wines (Capozzi et al. 2010, 2012; Grieco et al. 2010; Tristezza et al. 2012, 2013, 2014; Garofalo et al. 2015), the aim of this work was to study, for the first time in a Southern Italian wine, the biodiversity of ‘cultivable’ yeasts isolated from the grapes (“Uva di Troia”, an autochthonous regional variety common denominator of several wines produced in North-Apulian region) up to corresponding wines (so called “Nero di Troia”).
Materials and methods
Yeast isolation from grape berries, musts and wines
Grape berries were directly collected in the vineyard with the aim to avoid contamination yeast the commercial culture strains used in the cellar. For the yeast isolation, 1.00 kg of grape berries were collected aseptically in North Apulia area from two vineyards (Lucera and Ascoli Satriano areas, see Fig. 1) (18° Babo, 0.25 g/L total acidity, 4 g/L malic acid, pH 3.8, free ammonium 165 mg/L and 17° Babo, 0.3 g/L total acidity, 3.6 g/L malic acid, pH 3.8 free ammonium 155 mg/L, respectively for Lucera and Ascoli Satriano area). The grape were pressed for 20 min using a Bag Mixer® (Interscience, France), then spontaneous fermentation of grape juices were carried out in laboratory at 28 °C temperature and monitored over 1 month. Yeast sampling were accomplished at different stages, first from grape berries surface, then during alcoholic fermentation, at the beginning and at the end of fermentation, which were determined on the basis of alcohol content, about 1 %, at the beginning of AF, and 9 %, in the final phases of AF. Yeast from grape surface were isolated according to method of Prakitchaiwattana et al. (2004), Fifty grams of berries were rinsed in 450 ml of 0.1 % peptone water with 0.01 % Tween 80 by orbital shaking in a flask at 150 rpm for 30 min. Aliquots of 0.1 mL from serially diluted samples in physiological solution were plated either on Wallerstein Laboratory (WL) and on nutrient agar (Oxoid, USA) and Lysine medium (Oxoid, USA), both added with 10 mg/L chloramphenicol, that respectively allowed the isolation and identification of non-Saccharomyces and Saccharomyces species. Selection of non-Saccharomyces isolates were chosen on the basis of their different colony morphology, whereas the Saccharomyces strains were isolated randomly.
RFLP analysis and sequencing of 5.8S rRNA gene and the two ribosomal internal transcribed region
The RFLP analysis of 5.8S rRNA gene and the two ribosomal internal transcribed spacer was performed according to method of Esteve-Zarzoso et al. (1999), with some modifications. The Amplification reaction were performed using PCR reaction mix containing 0.5 µM of each primer (ITS1 and ITS4), 200 µM dNTP, buffer 10×, solution Q and 1.25 unit Taq DNA Polymerase (Taq PCR Core; Qiagen, USA). PCR was performed in a thermocycler (I-Cycler, Bio-Rad), using the following program: initial denaturation at 95 °C for 10 min, followed by 35 cycles of denaturing at 94 °C for 1 min, annealing at 55.5 °C for 2 min and extension at 72 °C for 2 min; and a final extension at 72 °C for 10 min, then samples were conserved at 4 °C. Amplification products were previously analysed on 2 % agarose gels, with 1× TBE buffer and stained with ethidium bromide. After electrophoresis, gels were visualized under UV light and photographed (Versa Doc, BIO-RAD). Sizes were estimated by comparison against a DNA length standard (50 bp ladder; Promega, USA) with Quantity One Software (Bio-Rad, USA). Then PCR products were digested without further purification with the fast restriction endonucleases HaeIII, HhaI, HinfI and DdeI (Thermo Scientific, USA), following the manufacture’s instruction. The restriction fragments were separated on 3 % agarose gel with 1X TBE buffer and stained with ethidium bromide. For each sampling point, two PCR products obtained with primers ITS1-ITS4 for each obtained pattern were randomly selected and sequenced (PRIMM, Italy) to confirm the specie assignment.
Genetic characterization of Saccharomyces cerevisiae strains
The genetic variability of S. cerevisiae isolates was evaluated by amplification of δ region, using the primers δ12 (5′TCAACAATGGAATCCCAAC3′) and δ21 (5′-CATCTTAACACCGTATATGA-3′) (Legras and Karst 2003). The protocol described by Capece et al. (2012) was adopted with some modifications. The amplification of δ region was performed directly from the colony, using a reaction mix containing 1 µM primers (δ12 and δ21) and 1.5 unit of Taq DNA Polymerase (Qiagen, USA). The PCR conditions were the following: initial denaturation at 97 °C for 10 min, then reaction mixture was cycled 35 times with 30 s denaturation at 94 °C, 1 min primer annealing at 42 °C and 2 min primer extension at 72 °C, followed by a 10-min final extension step at 72 °C. After electrophoresis gel were visualized under UV light, scanned with (Versadoc System; Bio-Rad, USA) and analysed by using the FP Quest TM software (BioRad, USA). The electrophoresis patterns were grouped, and analysed for the similarity and cophenetic correlations through the Dice coefficient. Cluster analysis was performed using the unweighted pair group method with arithmetic mean (UPGMA). Cophenetic correlation was the measure of how faithfully the tree represents the dissimilarities among observations.
Statistical analyses
Molecular data has been analyzed by One-way ANOVA, Turkey test (p < 0.005). Ecological indices, such as the Shannon-Wiener index of general diversity (H), the richness (S) of the microbial community, Simpson’s diversity indices (D and 1-D) and Evenness (e^H/S) were calculated according to Tristezza et al. (2013). All statistical analyses were performed using Past, version 3.05 (Hammer et al. 2001).
Results
Yeast species identification from grape berries
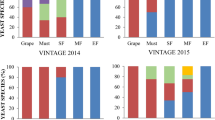
Samples were collected from two different vineyards located in north Apulia region (Fig. 1) during vintage 2012. A total of 136 colonies were isolated from grape berries of Uva di Troia variety and subjected to a PCR–RFLP analysis of the 5.8SITS rDNA region. The yeast species identified and the isolation frequencies obtained are shown in Table 1. The PCR products, showing variations in length ranging from 400 to 880 bp, were digested with HhaI (CfoI), HaeIII, HinfI and DdeI enzymes. The produced fragments were compared with those described previously in literature (Esteve-Zarzoso et al. 1999). In general, we observed 12 different restriction profiles of ITS–5.8S rDNA region, corresponding to Saccharomyces cerevisiae, Issatchenkia orientalis, Metschnikowia pulcherrima, Hanseniaspora uvarum, Candida zemplinina, Issatchenkia terricola, Kluyveromyces thermotolerans, Torulaspora delbrueckii, Metschnikowia chrysoperlae, Pichia fermentans, Hanseniaspora opuntiae and Hanseniaspora guilliermondii (Table 1). Two ITS fragments for each obtained pattern were randomly selected and sequenced and the obtained data were compared with sequences available at the NCBI database (GenBank) using the standard nucleotide_nucleotide homology search Basic Local Alignment Search Tool (BLAST, http://www.ncbi.nlm.nih.gov/BLAST) (corresponding gene accession numbers are reported in Table 1). Several yeast species such as M. pulcherrima, C. zemplinina, H. guilliermondii, H. uvarum and I. terricola represented a common denominator of the two vineyards studied (Table 1). The Figure S1 reportes the frequencies of strains identified from grape berries from the two different vineyards, during vintage 2012.
Among the non-Saccharomyces characterized in this study, the most abundant genera on berries surface were Hanseniaspora (about H. uvarum 22 %, H. guilliermondii 13 %, and H. opuntiae 1 %) and Metschnikowia (35 %, M. pulcherrima 34 % and M. chrysoperlae 1 %) (Figure S1). The analysis of non-Saccharomyces diversity in the two different areas revealed a great variability, showing, in several cases, statistically significant differences among locations (Figure S1). S. cerevisiae, K. thermotolerans, T. delbrueckii, M. chrysoperlae, P. fermentans, I. orientalis and H. opuntiae were isolated only from grape berries collected from Lucera (respectively 5, 7, 1, 1, 3, 1, and 1 %). H. guilliermondii was isolated only from Ascoli Satriano (about 26 %) samples. The frequency of M. pulcherrima showed differences between Lucera (42 %) and Ascoli Satriano (28 %) vineyards. H. uvarum ecotypes have been isolated with higher frequency from Ascoli Satriano (about 36 %), rather than in Lucera vineyards (only 7 %). C. zemplinina and I. terricola frequency did not show significant changes (respectively about 10 and 2 %).
Yeast species identification from fermenting grape juice
A total of 133 colonies were isolated from grape juice at the beginning of alcoholic fermentation (about 1 % EtOH) and subjected to a PCR–RFLP analysis of the 5.8SITS rDNA region as above described. In general, we observed 11 different restriction profiles of ITS–5.8 S rDNA region, corresponding to S. cerevisiae, I. orientalis, M. pulcherrima, H. uvarumi C. zemplinina, I. terricola, K. thermotolerans, T. delbrueckii, P. fermentans, H. opuntiae and H. guilliermondii (Table 2). The differences in yeast frequency and diversity highlighted in the two locations studied might also be addressable to dissimilarities in composition (data reported in material and method section, 2.1), in fact grape juice obtained from grape collected in Lucera area showed higher sugars and free ammonium contents. As described above, two ITS fragments for each obtained pattern were randomly chosen, sequenced and subjected to comparative analysis to confirm species assignation (Table 2). Several yeast species such as S. cerevisiae, M. pulcherrima, C. zemplinina, H. uvarum and I. terricola represented a common denominator between the studied vineyards. Otherwise, some species were isolated only from one vineyard, respectively I. orientalis, K. thermotolerans, T. delbrueckii, P. fermentans, H. guilliermondii and H. opuntiae from Lucera (Table 2). The predominance of non-Saccharomyces yeasts was observed for all the samples analyzed, nevertheless S. cerevisiae strains has been isolated from both vineyards studied, their frequencies were higher in Lucera (about 33 %) than Ascoli Satriano (about 10 %). The Figure S2 describes the frequencies of strains identified from grape juice from the two different vineyards, during vintage 2012.
Among the non-Saccharomyces characterized in this study, the most abundant genera at the beginning of AF were Hanseniaspora (about 38 %, H. uvarum 35 %, H. guilliermondii 1.5 %, and H. opuntiae 1.5 %) and Metschnikowia (M. pulcherrima 25 %) (Figure S2). The analysis of non-Saccharomyces diversity in the two different areas revealed a great variability, showing, in several cases, statistically significant differences among locations (Figure S2). I. orientalis, K. thermotolerans, T. delbrueckii, P. fermentans, H. opuntiae and H. guilliermondii were isolated only from grape juice collected from Lucera (respectively 1.5, 5, 3, 1.5, 3 and 3 %). The presence of M. pulcherrima is different in Lucera and Ascoli Satriano vineyards, respectively 14 and 4 %, furthermore significant differences were reported also with total frequency (about 9 %). H. uvarum ecotypes have been isolated with higher frequency from Ascoli Satriano (about 42 %), contrariwise it frequency was about 28 % in Lucera vineyards. Its frequency was not comparable with those reported for the totality of yeast isolated. C. zemplinina frequency showed significant differences from Lucera and Ascoli Satriano vineyard, respectively 6 and 42 %. As reported in Table 3, the species richness was highest in the yeast population from Lucera (S = 12) than in the population from Ascoli Satriano (S = 6). However, biodiversity not rely merely on the numbers of species but likewise on its relative abundance and dominance. The Shannon diversity index (H), that takes into account the number of individuals as well as number of taxa, was higher for the yeast population from Lucera (H = 1.793) being representative of a more diverse community than that from Ascoli Satriano (H = 1.512) (Table 3). Moreover, the Evenness index measures the uniformity with which individuals are divided among the taxa present in the population. This index was higher in Ascoli Satriano yeast community than in Lucera population, with values of 0.7559 and 0.5005 respectively (Table 3).
Yeast species identification from wines and genetic characterization of Saccharomyces cerevisiae strains
In the final phases of AF (9 % ethanol content), we selected only S. cerevisiae strains (Table S1). A total number of 146 yeast isolates identified as S. cerevisiae were subjected to genotypic characterization by analysis of δ sequences. These ecotypes had been isolated from the first stage of alcoholic fermentation and in the last phases (Tab. S1). PCR analysis of inter-delta region produced 119 different profiles (Table S1). The relationship among strains according to patterns obtained with amplification of inter-delta region was evaluated using cluster analysis. According to the resulting dendrogram (Fig. 2), the strains were distributed in 10 main similarity groups. Only the groups C, D and E include strains of the same isolation area, respectively Lucera and Ascoli Satriano all collected from wine. Contrariwise, other groups contain strains collected from grape berries, grape juice and wine of the two vineyards studied. Cluster B contain only one strain, isolated from wine collected from Ascoli Satriano vineyard. In addition, 5 groups including strains with identical profiles were found. Identical profiles generally has been isolated in the same area, with the exception of profile 16, obtained from two strains collected from both the vineyards studied.
The two S. cerevisiae populations from Lucera and Ascoli Satriano showed low indices of dominance (D = 0.2596) and relative high diversity (H = 1.528 and 1.640, respectively; Table 4).
Discussion
Non-Saccharomyces yeasts isolated from grapes, musts and wines show potential effects on the organoleptic qualities of the final products (Romano et al. 2003a; Ciani et al. 2010). A major understanding of non-Saccharomyces biodiversity in fermenting wines is an essential criterion for quality improvement programs in the oenological productions, and more specifically in the sector of typical wine and oenological geographical indications (Fleet 2008). In the present study, for the first time in a Southern Italian wine, we study the biodiversity of ‘cultivable’ yeasts isolated from the the grapes (“Uva di Troia”, an autochthonous regional grape variety common denominator of several wines produced in North-Apulian region) up to corresponding wines (so called “Nero di Troia”).
The majority of the strains isolated belong to M. pulcherrima, a species common on wine grapes at the time of harvest and in grape must during the early stages of wine fermentation. This species occurs more frequently on damaged berries, on berries used to produce ice wine, and in botrytized (noble-rotted) wines (Oro et al. 2014). Several authors have investigated the potentiality of M. pulcherrima for wine fermentation. In particular, the absence of relevant changes in fermentation rate and chemical composition has been often observed (Jolly et al. 2014; Comitini et al. 2011). Furthermore, Comitini et al. (2011) noted in the final wines a significant decrease in volatile acidity and in total acidity. Other yeast of oenological interest isolated from grape surfaces of “Uva di Troia” belonged to Hanseniaspora spp., mainly H. guilliermondii and H. uvarum. Our results confirmed findings previously reported on literature, showing that the apiculate H. uvarum/K. apiculata may be the predominant species on either the berries and at the beginning of spontaneous must fermentations (e.g. Fleet 2008; Tristezza et al. 2013). All samples collected at the beginning of AF show the predominance of non-Saccharomyces yeast, nevertheless S. cerevisiae have a high frequency, in both Lucera and Ascoli Satriano vineyard. These evidence might be addressable to the presence of damaged grape berries that may be very rich depositories of S. cerevisiae (e.g. Nisiotou et al. 2007; Barata et al. 2012).
The majority of the strains isolated at the beginning of AF belong to Hanseniaspora spp., in particular H. uvarum. Other yeast well represented on grape juice at the beginning of AF are Candida spp. Among Candida spp. the species most important identified is C. zemplinina. Several yeast ecology studies demonstrated the frequent presence of this species in wine fermentations (e.g. Nisiotou et al. 2007; Urso et al. 2008; Zott et al. 2008; Tofalo et al. 2009), is a typical contaminant of botrytized juice fermentations but its presence is also common onto healthy grapes (Barata et al. 2012). In terms of yeast natural biodiversity, strains collected from grape juice are similar to those found in other wine-producing areas. Several authors reported the predominance of Candida and Hanseniaspora genera at the beginning of spontaneous AF (Bezerra-Bussoli et al. 2013; Garofalo et al. 2015), nevertheless Cordero-Bueso et al. (2011) suggested that other non-Saccharomyces yeast such as Lachancea, Wickerhamomyces and Torulaspora can be present as dominant species.
Among the species belonging to the Hanseniaspora genera, our results suggest the dominance of H. uvarum, confirming those reported by Ocón et al. (2010). Contrariwise, Garofalo and coworkers (2015) reported major frequency of H. guilliermondii analyzing Apulian regional wines.
Yeast isolated from wine, at the end of AF, show the predominance of Saccharomyces spp. (i.e. S. cerevisiae). Our findings confirming those reported by other authors (e.g. Tristezza et al. 2009), that suggested the rapidity, reproducibility and sensibility of this method.
Several studies suggested the important role of indigenous non-Saccharomyces and Saccharomyces yeast on wine quality. For this reason, multi-starter cultures designed using autochthonous microbial resources has been suggested as a tool to take advantage of natural biodiversity, enhancing the complexity and specific characteristics of wine (Romano et al. 2003b; Ciani et al. 2006; Ciani et al. 2010; Jolly et al. 2014; Garofalo et al. 2015).
This is the first report on the population dynamics of ‘cultivable’ microbiota diversity of “Uva di Troia” cultivar from the grape to the corresponding wine (“Nero di Troia”), and more general for Southern Italian oenological productions. We also select possible candidates for the design of mixed/multi-strains autochthonous starter cultures for typical Apulian wines, in order to obtain a final product characterized by unique peculiarities as result of the autochthonous virtuous microbial biodiversity. A certain geographical-dependent variability has been reported, suggesting the need of local based formulation for tailored starter cultures for typical wines, especially in the proportion of the different species/strains in the conceiving of mixed microbial preparations.
References
Barata A, Malfeito-Ferreira M, Loureiro V (2012) The microbial ecology of wine grape berries. Int J Food Microbiol 153:243–259
Bely M, Stoeckle P, Masneuf-Pomarède I, Dubourdieu D (2008) Impact of mixed Torulaspora delbrueckii-Saccharomyces cerevisiae culture on high-sugar fermentation. Int J Food Microbiol 122:312–320
Bezerra-Bussoli C, Baffi MA, Gomes E, Da-Silva R (2013) Yeast diversity isolated from grape musts during spontaneous fermentation from a Brazilian winery. Curr Microbiol 67:356–361
Bokulich NA, Thorngate JH, Richardson PM, Mills DA (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA 111:E139–E148
Capece A, Romaniello R, Siesto G, Romano P (2012) Diversity of Saccharomyces cerevisiae yeasts associated to spontaneously fermenting grapes from an Italian “heroic vine-growing area”. Food Microbiol 31:159–166
Capozzi V, Russo P, Beneduce L et al (2010) Technological properties of Oenococcus oeni strains isolated from typical southern Italian wines. Lett Appl Microbiol 50:327–334
Capozzi V, Russo P, Ladero V et al (2012) Biogenic amines degradation by Lactobacillus plantarum: toward a potential application in wine. Front Microbiol 3:122
Capozzi V, Garofalo C, Chiriatti MA et al (2015) Microbial terroir and food innovation: the case of yeast biodiversity in wine. Microbiol Res 181:75–83
Cappello MS, Stefani D, Grieco F et al (2008) Genotyping by amplified fragment length polymorphism and malate metabolism performances of indigenous Oenococcus oeni strains isolated from Primitivo wine. Int J Food Microbiolo 127:241–245
Ciani M, Beco L, Comitini F (2006) Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int J Food Microbiol 108:239–245
Ciani M, Comitini F, Mannazzu I, Domizio P (2010) Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res 10:123–133
Comitini F, Gobbi M, Domizio P et al (2011) Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol 28:873–882
Cordero-Bueso G, Arroyo T, Serrano A et al (2011) Influence of the farming system and vine variety on yeast communities associated with grape berries. Int J Food Microbiol 145:132–139
Csoma H, Zakany N, Capece A et al (2010) Biological diversity of Saccharomyces yeasts of spontaneously fermenting wines in four wine regions: comparative genotypic and phenotypic analysis. Int J Food Microbiol 140:239–248
De Benedictis M, Bleve G, Grieco F et al (2011) An optimized procedure for the enological selection of non-Saccharomyces starter cultures. Antonie Van Leeuwenhoek 99:189–200
Di Maio S, Polizzotto G, Di Gangi E et al (2012) Biodiversity of indigenous Saccharomyces populations from old wineries of south-eastern Sicily (Italy): preservation and economic potential. PLoS One 7:e30428
Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A (1999) Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacteriol 49(1):329–337
Fleet GH (2008) Wine yeasts for the future. FEMS Yeast Res 8:979–995
Garofalo C, El Khoury M, Lucas P et al (2015) Autochthonous starter cultures and indigenous grape variety for regional wine production. J Appl Microbiol 118:1395–1408
Grieco F, Tristezza M, Vetrano C et al (2010) Exploitation of autochthonous micro-organism potential to enhance the quality of Apulian wines. Ann Microbiol 61:67–73
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
International Organization of Vine and Wine (2010) Definition of vitivinicultural ‘‘terroir’’. OIV/VITI 333/2010 Resolution, Tbilisi, 25th June 2010
Jolly NP, Varela C, Pretorius IS (2014) Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 14:215–237
Legras J-L, Karst F (2003) Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol Lett 221:249–255
Nisiotou AA, Spiropoulos AE, Nychas G-JE (2007) Yeast community structures and dynamics in healthy and Botrytis-affected grape must fermentations. Appl Environ Microbiol 73:6705–6713
Ocón E, Gutiérrez AR, Garijo P et al (2010) Presence of non-Saccharomyces yeasts in cellar equipment and grape juice during harvest time. Food Microbiol 27:1023–1027
Oro L, Ciani M, Comitini F (2014) Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J Appl Microbiol 116:1209–1217
Prakitchaiwattana CJ, Fleet GH, Heard GM (2004) Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res 4:865–877
Romano P, Fiore C, Paraggio M et al (2003a) Function of yeast species and strains in wine flavour. Int J Food Microbiol 86:169–180
Romano P, Granchi L, Caruso M et al (2003b) The species-specific ratios of 2,3-butanediol and acetoin isomers as a tool to evaluate wine yeast performance. Int J Food Microbiol 86:163–168
Tofalo R, Chaves-López C, Di Fabio F et al (2009) Molecular identification and osmotolerant profile of wine yeasts that ferment a high sugar grape must. Int J Food Microbiol 130:179–187
Tristezza M, Gerardi C, Logrieco A, Grieco F (2009) An optimized protocol for the production of interdelta markers in Saccharomyces cerevisiae by using capillary electrophoresis. J Microbiol Methods 78:286–291
Tristezza M, Vetrano C, Bleve G et al (2012) Autochthonous fermentation starters for the industrial production of Negroamaro wines. J Ind Microbiol Biotechnol 39:81–92
Tristezza M, Vetrano C, Bleve G et al (2013) Biodiversity and safety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia Region, Italy. Food Microbiol 36:335–342
Tristezza M, Fantastico L, Vetrano C et al (2014) Molecular and technological characterization of Saccharomyces cerevisiae strains isolated from natural fermentation of Susumaniello grape must in Apulia, Southern Italy. Int J Microbiol 2014:897428
Urso R, Rantsiou K, Dolci P et al (2008) Yeast biodiversity and dynamics during sweet wine production as determined by molecular methods. FEMS Yeast Res 8:1053–1062
Zott K, Miot-Sertier C, Claisse O et al (2008) Dynamics and diversity of non-Saccharomyces yeasts during the early stages in winemaking. Int J Food Microbiol 125:197–203
Acknowledgments
This research was supported by the Apulian Region in the framework of Project 6N7AD82 “Autochthonous2Autochthonous: risorse microbiologiche per vini in purezza da vitigni autoctoni (e per produzioni biologiche” and “Biotecnologie degli alimenti per l’innovazione e la competitività delle principali filiere regionali: estensione della conservabilità e aspetti funzionali (BiotecA)”. Vittorio Capozzi is supported by a grant by the Apulian Region in the framework of ‘FutureInResearch’ program (practice code 9OJ4W81).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Frequency (%) of different yeasts species isolated from grape berries of “Uva di Troia”, during vintages 2012. Open bars, Ascoli Satriano vineyards; black bars, Lucera vineyards; light grey bars, total yeast identified. Different letters in superscript bars indicate statistical significance (One-way ANOVA, Turkey test P < 0.005) (JPEG 34 kb)
Figure S2
Frequency (%) of different yeasts species isolated from spontaneous fermentation of grape juice of “Uva di Troia”, during vintages 2012. Open bars, Ascoli Satriano vineyards; black bars, Lucera vineyards; light grey bars, total yeast identified. Different letters in superscript bars indicate statistical significance (One-way ANOVA, Turkey test P < 0.005) (JPEG 54 kb)
Rights and permissions
About this article
Cite this article
Garofalo, C., Tristezza, M., Grieco, F. et al. From grape berries to wine: population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J Microbiol Biotechnol 32, 59 (2016). https://doi.org/10.1007/s11274-016-2017-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2017-4