Abstract
Decachlorobiphenyl (DCB) is one of the 209 polychlorinated biphenyls congeners characterized by its high toxicity and chemical stability. It is produced by industrial activities. A possible strategy to eliminate DCB is by bacterial degradation. The main objective of this study was to define the optimal conditions for biodegradation and bioaccumulation of DCB by Pseudomonas extremaustralis ADA-5 isolated from a worm intestine. Bacterial growth kinetics were determined in minimal medium with added biphenyl and DCB. By GC coupled to mass spectrometry, we found that the strain had the ability to degrade 9.75% of available DCB, using it as a carbon source and was able to accumulate 19.98% of this pollutant in biomass. Membrane lipids may be altered by DCB. Phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL) were identified by thin-layer chromatography as the membrane lipids of the cell. At 250 mg L−1 of DCB in the culture medium, membranes showed a 30% decrease in the PE concentration, an 18% increase in the PG, and a 12% increase in CL. ADA-5 was able to catabolize DCB and may be used for bioremediation of highly chlorinated toxic compounds in soil.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Persistent organic pollutants (POPs) have increased significantly in recent times, causing irreversible damage in the composition, structure, and functionality of ecosystems as well as in terrestrial biomes. POPs are a group of toxic synthetic compounds, highly dangerous to human health and to the environment. Among the main pollutants released, as products of human and industrial activity, polychlorinated biphenyls (PCBs) stand out for their high toxicity (She et al., 2007). PCBs are POPs that comprise various congeners that vary in the amount and position of the chlorine atoms that are attached to biphenyl rings (Polak et al., 2016). Many PCB congeners are found in soil, water, and sediments, even though their use in recent times has been restricted. Decachlorobiphenyl (DCB) is a highly chlorinated PCB, characterized by its lipophilicity, stability, and biomagnification (Han et al., 2009). Due to its high chemical stability, the biodegradation process of DCB is slow (Costabeber et al., 2006). For this reason, different physical and chemical methods have been proposed, for its rapid elimination (Huang et al., 2014; Zhao et al., 2012). Its incineration has been approved and it is used as the standard method for the removal of DCB in soil and sediment (Hatamian-Zarmi et al., 2009). Nevertheless, the contaminants released by this procedure generate other environmental problems (Tharakan et al., 2006). Bioremediation technologies have gained relevance in recent years as an efficient alternative for the degradation of recalcitrant toxic compounds. Bioremediation is a biological process that uses microorganisms to remove contaminants or hazardous organic compounds from the environment (Vidali, 2001). Bioremediation of biphenyl in soil, sediments, and water has been reported (Anyasi & Atagana, 2011; Bedard et al., 1987). Pseudomonas bacteria have the capacity to tolerate and remove PCBs in arochlor mixtures (Mathews & Sithebe, 2018). Pseudomonas fluorescens strain F113 has been studied for degradation of PCBs. It grows with biphenyl as its only carbon source and has the capacity to degrade PCB congeners including the commercial mixture Delor 103 (Villacieros et al., 2005). It has been reported that biphenyl increases the aerobic degradation capacity of PCBs in P. pseudoalcaligenes KF707 (Sandri et al., 2017). Recently, we reported that the Pseudomonas extremaustralis ADA-5 isolated from the digestive tract of the earthworm Eisenia fetida showed a high potential for the removal of DCB (219.7 mg L−1) at an initial concentration of 1500 mg L−1 (Zenteno-Rojas et al., 2019). ADA-5 is genetically similar to several Pseudomonas species that have biochemical and genetic potential for the biodegradation of organochlorine compounds, such as polychlorinated biphenyls (Chakraborty & Das, 2016). Due to its lipophilicity, DCB may cause damage to bacterial membranes and compromise cell survival (Murínová & Dercová, 2014). In fact, solvents affect cell membranes and biochemical processes of adaptation directly related to the maintenance of the membrane fluidity and the stability. Flexibility is a characteristic of membrane structure and could be regulated by change of the fatty acid profile that form membrane lipids (Denich et al., 2003). A Pseudomonas sp. strain resistant to tributyltin (TBT) was isolated from an environment contaminated by car filter (Bernat et al., 2014) and evaluated for its adaptation to TBT and for its degradation capacity. The composition of membrane lipids and cellular proteins was also analyzed in the presence of TBT and variations in the composition and concentration of phospholipids were found in the presence of toxic contaminants (Bernat et al., 2013). The main objective of this research was to evaluate the biodegradation and bioaccumulation of decachlorobiphenyl by native strain P. extremaustralis ADA-5 and to determine the effects of DCB on its membrane lipid composition.
2 Materials and Methods
2.1 Chemical Reagents
Decachlorobiphenyl (DCB) (98% purity) and biphenyl (99% purity) were purchased from Sigma-Aldrich® (USA). Pentane was chromatographic grade (Sigma-Aldrich®) and organic reagents were of analytical grade (Thermo Fischer Scientific®, USA). For the analysis of ADA-5 membrane lipid composition, sodium acetate-1-14C (99% purity) obtained of Amersham Biosciences® (USA). Chloroform (98% purity) and high-performance TLC plates were provided by Merck® (USA) were used.
2.2 Bacterium Strain
Strain ADA-5 was previously isolated from the digestive tract of E. fetida cultivated in a vermicomposting system contaminated with a high concentration of DCB (Zenteno-Rojas et al., 2019). ADA-5 was identified as Pseudomonas extremaustralis according to the sequence analysis of the 16S rRNA gene. The sequence was deposited in GenBank with accession number KY110419.1. This strain was initially reactivated by aseptic cultures in Brain Heart Infusion (BHI) medium (Bioxon®, USA). Then it was adapted to a minimal medium AOB [2.0 g L−1 (NH4)2SO4, 1.0 g L−1 K2HPO4, 0.3 g L−1 NaCl, 0.3 g L−1 MgSO4.7H2O, and 0.03 g L−1 FeSO4.7H2O] with glucose (3.0 g L−1) and yeast extract (1.0 g L−1).
2.3 Master Cell Bank (MCB)
A pure strain of P. extremaustralis ADA-5 grown in a minimum medium with DCB was used to elaborate a master cell bank, according to the methodology recommended by Del Puerto et al. (2009). When the bacterial culture was in the middle of the logarithmic phase, aliquots of 10 mL were collected and then centrifuged at 10,000g for 5 min at 25 °C. The biomass pellet was placed in microtubes that contained a mixture of minimal medium-glycerol 7:3 (v/v) and then cryopreserved at −20 °C.
2.4 Optimization of a Culture Medium to Increase Pseudomonas extremaustralis ADA-5 Growth and Exopolysaccharides
Box-Behnken experimental design was performed to evaluate cell growth of P. extremaustralis ADA-5. Three factors at three levels (+1, 0, −1) were evaluated: X1 = yeast extract (0, 1, and 2 g L−1); X2 = ammonium sulfate (0, 1, and 2 g L−1); and X3 = biphenyl (0, 250, and 500 mg L−1). The combination of the levels of each factor allowed an evaluation of a total of 15 treatments (Table 1). Bottles with 100 mL of AOB medium were inoculated with ADA-5 (10% v/v), maintained at 37 °C, and shaking at 120 rpm for 72 h. Biomass (CFU mL−1) and exopolysaccharides (mg L−1) were determined. To measure biomass, 5 mL aliquots of bacterial suspension were taken for each treatment and inoculated in Petri dishes with AOB medium for the quantification of CFU (cell mL−1) using a colony counter. To quantify exopolysaccharides produced by the bacteria, the methodology of Geel-Schutten et al. (1998) was implemented. One milliliter of the culture medium of each treatment was centrifuged at 11,000g for 5 min. The supernatant was collected in a 50-mL tube and was mixed with ice cold ethanol at a 2:1 (v/v) ratio overnight at 4 °C. Then the precipitates were separated by centrifugation (2000g for 15 min) and these were resuspended in 1 mL of demineralized water. Polysaccharides obtained were brought to constant weight in test tubes, incubated at 55 °C for 4 days. The results were reported in g L−1 of exopolysaccharides. With Box-Behnken experimental design, we obtained the optimization values of principal effects, double factor, and quadratic factor of the independent variables. A second order polynomial model was adjusted to the response data obtained from the design. The polynomial equation proposed for the two responses (Y1 and Y2) was:
where Yi (i = 1 to 2) are the predicted response for increasing biomass (BIOM) and exopolyssacharides (EPS), respectively. b0 = constant, b1 = yeast extract, b2 = ammonium sulfate, and b3 = biphenyl; b1, b2, and b3 = linear coefficients; b11, b22, and b33 = quadratic coefficients; b12, b13, b23, and b123 = cross product coefficients.
2.5 Evaluation of the Ability of Pseudomonas extremaustralis ADA-5 to Grow on DCB
From the optimization test results, the treatment that allowed the best growth and a greater production of cell biomass was selected to further evaluate the ability of the strain to grow in the presence of DCB contaminant. For this purpose, the bacterial strain was taken from a preculture of the minimal AOB medium [2.0 g L−1 (NH4)2SO4, 1.0 g L−1 K2HPO4, 0.3 g L−1 NaCl, 0.3 g L−1 MgSO4.7H2O, 0.03 g L−1 FeSO4.7H2O] enriched with glucose (3.0 g L−1) and yeast extract (1.0 g L−1) as carbon sources amended with 250 mg L−1 of biphenyl. Subsequently, the strain was cultivated in the AOB medium but using biphenyl (250 mg L−1) as sole carbon source (formulation 1). Then, the strain was adapted to the AOB medium containing both biphenyl (250 mg L−1) and DCB (250 mg L−1) (formulation 2). The two formulations were compared with a control medium AOB without any carbon source. The strain was kept in a 250-mL flask containing 25 mL of culture medium during 336 h of growth kinetics. Growth was determined by colony-forming unit (CFU) quantification every 12 h. The experiments were carried out in triplicate.
2.6 Evaluation of the Capacity for Biodegradation and Bioaccumulation of DCB by P. extremaustralis ADA-5
The capacity of P. extremaustralis ADA-5 to biodegrade and bioaccumulate DCB was evaluated in flasks containing 25 mL of the AOB medium with 250 mg L−1 of DCB as the unique carbon source [2.0 g L−1 (NH4)2SO4, 1.0 g L−1 K2HPO4, 0.3 g L−1 NaCl, 0.3 g L−1 MgSO4.7H2O, 0.03 g L−1 FeSO4.7H2O, and 250 mg L−1 DCB] (formulation 3) compared to a control without carbon source. The strain was inoculated in each of the formulations at a proportion of 10% (v/v) and incubated at 30 °C with constant shaking at 150 rpm during 336 h. Growth was determined by CFUs every 12 h and all experiments were performed in triplicate. The extraction and quantification of DCB was carried out at the initial time (To = 0 h) and at the final time (Tf = 336 h), both in the culture medium (CM) and in the cellular biomass (CB). A total of 5 ml of liquid medium was obtained and pentane was added in a 1:1 proportion (v/v). The mixture was vigorously stirred for 20 min and then treated for 30 min in an ultrasonic device. The supernatant obtained in the organic part was divided and concentrated in a 1 mL solution. A total of 20 mL of the culture at the Tf was centrifuged at 4000 rpm for 5 min to form a CB pellet (Valenzuela-Encinas et al., 2008). The concentration of DCB in both CM and CB samples and the products of the removal process were determined according to the method reported by Villalobos-Maldonado et al. (2015).
2.7 Analysis of the Membrane Lipid Profiles of P. extremaustralis ADA-5 Grown in DCB
ADA-5 lipid composition was determined after labeling with [1-14C] acetate. One milliliter of culture of the strain adapted in AOB minimal medium [2.0 g L−1 (NH4)2SO4, 1.0 g L−1 K2HPO4, 0.3 g L−1 NaCl, 0.3 g L−1 MgSO4.7H2O, 0.03 g L−1 FeSO4.7H2O with 3.0 g L−1 glucose and 1.0 g L−1 yeast extract] (treatment A) or AOB minimal medium with DCB as carbon source [2.0 g L−1 (NH4)2SO4, 1.0 g L−1 K2HPO4, 0.3 g L−1 NaCl, 0.3 g L−1 MgSO4.7H2O, 0.03 g L−1 FeSO4.7H2O, and 250 mg L−1 of DCB] (treatment B) was used. Later, 0.5 μCi [14C] acetate (60 mCi mmol−1) was added to each culture. Cultures were incubated for 24 h and 72 h. Cells were obtained by centrifugation, washed with water (Green & Joseph, 2012), and resuspended in 100 μl of water. Lipids were extracted according to the methodology reported by Bligh and Dyer (1959). The chloroform phase was used for lipid analysis on thin-layer chromatography (TLC) plates (high-performance TLC aluminum sheets, silica gel 60; Merck®). After two-dimensional separation using the solvent systems described by De Rudder et al. (1997), individual lipids were quantified using a Phosphorimager (Storm 820; Molecular Dynamics). This assay was performed in triplicate.
2.8 Statistical Analysis
In this study, Box-Behnken fractional factorial design was applied using the response surface methodology for the optimization of growth and production of bacterial biomass. Data obtained were analyzed by ANOVA with a significance level of α = 0.05 and the results obtained were analyzed for each variable and a regression equation was constructed to study the relationship between the variables evaluated. t-Student’s test (P < 0.05) was used to analyze the results of biodegradation and bioaccumulation tests with Statgraphics Centurion XV Plus for Windows.
3 Results
3.1 Optimization of the Culture Medium
The results of the Box-Behnken design for evaluating the adaptation to the culture medium of P. extremaustralis ADA-5 are presented in Table 1. ANOVA (Table 2) showed that yeast extract (P < 0.0035) and yeast extract plus biphenyl (P < 0.0146) had significant effects on biomass production. The components that showed significant effects on the production of exopolysaccharides (Table 3) were yeast extract (P < 0.0008), the interaction of yeast extract and ammonium sulfate (P < 0.0046), and the quadratic components of ammonium sulfate, under the hypothesis that the variables examined induced cell growth and adaptability of P. extremaustralis to PCBs. The following equation with R = 0.90 and P < 0.05 for biomass production was obtained by regression analysis.
The effects of X1, X2, and X3 variables and their combined effects on the YBIOM response were modeled with a polynomial equation (Eq. 2). A positive value represents an effect that favors optimization, while a negative value indicates an antagonistic effect. The values of X1, X2, and X3 were substituted in the equation to obtain the theoretical values and biomass, finding that the predicted values and the observed values were in agreement. Treatments 6 and 11 showed the highest production of biomass (Table 1). The growth of the strain in treatment 6 (without biphenyl) was optimal with a high cell growth of 2.22 × 108 CFU mL−1 of medium during the 72 h. Cell growth obtained in treatment 11 was 6.0 × 108 CFU mL−1 where 250 mg L−1 of biphenyl was added to the minimal medium as sole carbon source. The methodology of response surface (Fig. 1a) revealed highest biomass production with the highest concentration of yeast extract (2 g L−1); however, yeast extract and biphenyl interaction also showed high biomass production. In this experiment, treatment 11 had the highest production of exopolysaccharides (EPS). The response surface plot (Fig. 1b) revealed that the highest exopolysaccharides production corresponded to high levels of yeast extract and biphenyls. The equation was obtained by a regression analysis with R = 0.95 and P< 0.05 for the production of exopolysaccharides.
In this case, X1, X1X2, and X22 are significant model terms. The theoretical and observed values (3) for YEPS were shown to be in agreement. In both experiments (YBIOM and YEPS), yeast extract and biphenyl significantly influenced the growth of P. extremaustralis. According to the results obtained, Box-Behnken design is an efficient tool to design parameters that promote the production ADA-5 strain biomass and exopolysaccharides.
3.2 Bacterium Strain Adaptation
The adaptive capacity of P. extremaustralis ADA-5 was evaluated using the optimal culture medium (formulation 1), amended with biphenyl as the sole carbon source (Fig. 2). In this condition, bacterial growth exceeded 9.0 × 107 CFU mL−1 of culture during 336 h, showing a stationary phase between 48 and 72 h and a μmax = 0.0079 h−1, as well as a doubling time (td) = 87.74 h. The adaptation of this strain to formulation 2, where biphenyl and DCB were the carbon source, showed a diauxic behavior, due to the sequential assimilation of the carbon source used by the strain. In the first stage, bacterial growth exceeded 8.0 × 107 CFU mL−1 between 24 and 48 h, while in the second stage, growth increased to 1.1 × 108 CFU mL−1 at 168 h with μmax = 0.016 h−1 and a td = 43.32 h (2.2).
3.3 Quantification of Biodegradation and Bioaccumulation of DCB
Biodegradation and bioaccumulation of DCB was analyzed by means of culture formulation 3 (Fig. 3), using the conditions of the optimized AOB medium modified with 250 mg L−1 DCB as sole carbon source. Bacterial growth was greater than 2.0 × 108 CFU mL−1 of medium during the 72 h of experimentation, where the μmax and the td in the stationary phase were 0.0371 h−1 and 18.68 h, respectively. Both the adaptability and the bacterial growth obtained with formulation 3 were greater than those obtained with formulations 1 and 2. With formulation 3, strain ADA-5 was shown to be fully adapted to DCB.
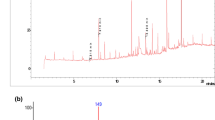
In this study, residual DCB was quantified. In Fig. 4, GC-MS chromatograms show DCB with a retention time of 14.23 min. The identification was carried out in the culture medium (CM) (Fig. 4a) and in the cellular biomass (CB) (Fig. 4b) where the highest abundance of the contaminant is shown. DCB quantification was performed using a standard reagent with concentrations determined in standard curves. The results showed that the concentration of the remaining contaminant in broth was 23.33 mg L−1, which represents 89.77% removal by strain ADA-5, while 182.66 mg L−1 bioaccumulated in the bacterial biomass represents 80.02 % of the total DCB used in formulation 3. As a result of DCB balance, 22.26 mg L−1 of the contaminant biodegraded by the strain was quantified, representing 9.75% biodegradation of DCB in the system evaluated (Table 4). Also, the presence of 1,3 bis-(1,1 dimethyl ethyl) benzene and benzoates, which can be products of DCB biodegradation, was detected by gas chromatography (Table ESM 1_ supplementary data).
3.4 Membrane Lipid Profiles of P. extremaustralis ADA-5 During the Biodegradation and Bioaccumulation of DCB
The identification and quantification of the membrane lipids of P. extremaustralis ADA-5 was carried out at 24 and 72 h during the biodegradation and bioaccumulation of DCB under the conditions given in formulation 3, comparing it with the lipid structure when the strain was grown in AOB medium without the contaminants (Table 5). When the strain was grown in the AOB medium without DCB (treatment A), a greater abundance of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL) was found. In this condition, strain ADA-5 showed a 5% and 10% increase in the abundance of PE and CL, respectively, and a 15% decrease in PG at the evaluation times (24 h and 72 h). In AOB medium in the presence of DCB (250 mg L−1 of DCB), there was a 20% decrease in PE and 20% increase in CL and no change in the abundance of PG (38%). ADA-5 membrane lipids at 72 h under treatment B culture conditions had a lipid composition of 30% PE, 38% PG, and 32% CL (Table 5).
4 Discussion
Research on the use of bacteria for the removal of toxic organic compounds in the environment has provided new sustainable alternatives for bioremediation. Martínez-Toledo and Rodríguez-Vázquez (2011) evaluated different formulations of a culture medium to improve the phenanthrene degradation process, using Pseudomonas putida strain CB-100. Tandlich et al. (2011) reported that Pseudomonas stutzeri had a high growth rate when cultivated in crystalline biphenyl at concentrations of 1.5 g L−1. They showed that the mechanism of absorption and assimilation of biphenyl by the bacterial strain depends on the concentration of the contaminant in the medium or in the soil.
In a different study, Pseudomonas pseudoalcaligenes strain KF707 was able to degrade pentafluorobiphenyl. The strain used this toxic chemical as a carbon source and showed growth exceeded of 1 × 107 CFU mL−1 after 72 h of incubation (Hughes & Clark, 2011). In other experiments, it has been observed that the EPS may chelate polychlorinated biphenyls due to its physicochemical characteristics (Mishra & Jha, 2013) or be used for synthesis of nanoparticles in the degradation of some PCB congeners (Escárcega-González et al., 2018). In some Pseudomonas species, such as P. aeruginosa, biphenyl is easily assimilated and induces a stimulant effect for the assimilation of PCB by the production of dioxygenase enzymes that catabolize the carbon structure of PCB (biphenyl) and lead to a diauxic growth (Chakraborty & Das, 2016). This behavior can be attributed to the fact that this bacterium requires biphenyl as an inducer for the assimilation of the contaminant. Our results show that P. extremaustralis ADA-5 was able to use DCB as a carbon source for its growth. Similar studies have shown the importance of adapting bacterial strains to culture media enriched with DCB as a way of increasing their the contaminant assimilation and elimination capacity (Sánchez-Pérez et al., 2019). It has also been reported that bacteria isolated from contaminated sites, when grown in culture media amended with PCBs, are efficient in the elimination of these compounds (Weiland et al., 2017). De Limae Silva et al. (2018), evaluated the bacterial community present in methanogenic bioreactors, quantifying 5.26 × 1012 g/TVS cells which were not affected by the addition of arochlor 1260 (mixture of highly chlorinated PCBs) and suggested that this group of bacteria developed mechanisms for the biodegradation of organic compounds.
There are few studies focused on the elimination of highly chlorinated contaminants such as decachlorobiphenyl and BDE-209 (decabromobiphenyl ether) using strains isolated from contaminated sites (Qiu et al., 2016); Yu et al., 2020). Qiu et al. (2016) evaluated the ability of a cold-resistant bacterium Comamonas testosteroni (phylum: Proteobacteria) to degrade decachlorobiphenyl. This strain could tolerate and degrade up to 500 μg L−1 of DCB, and a bacterial growth of 7.0 × 108 cell mL−1 was reported. Furthermore, the compounds produced by the possible co-metabolism of the pollutant DCB are structurally similar to those reported by Hughes and Clark (2011), where 3-pentafluorophenyl-cyclohexa-3,5-diene-1,2-diol and 3-pentafluorophenyl-benzene-1,2-diol were detected in the supernatants of resting cells cultured with biphenyl and 2,3,4,5,6-pentafluorobiphenyl.
During the biodegradation of DCB by ADA-5 in the culture media, there is a phase of assimilation of DCB in which the contaminant increases in the cellular biomass produced by the strain. Strain ADA-5 was able to bioaccumulate high concentrations of DCB. This biological mechanism has been previously shown in different Pseudomonas species isolated from sites contaminated with DCB. Sanchez-Pérez et al. (2019) reported the capacity of different bacterial strains isolated from rhizosphere soil to remove and bioaccumulate up to 200 mg L−1 of DCB in modified culture media. In a previous work, we evaluated the ability of P. extremaustralis ADA-5 to bioaccumulate DCB. We found that this bacterial species was able to bioaccumulate up to 219.7 mg L−1 of DCB (Zenteno-Rojas et al., 2019). Lipid composition can change during the assimilation process, both in the bioaccumulation process and in the biodegradation of DCB (Murínová & Dercová, 2014). Here, our results suggest that the P. extremaustralis ADA-5 has biological mechanisms for the biodegradation and bioaccumulation of DCB that are important for the elimination of the contaminant.
The phospholipids of the cytoplasmic membrane of bacteria help to maintain cell viability and, in the presence of alterations, cells modify their phospholipid structure (Dercová et al., 2018). Thus, the effects of phospholipid composition on membrane fluidity have been studied. It has been shown that PE is responsible for providing lateral pressure to the membrane bilayer and maintaining the position of amino acids (Berg et al., 2006). Alterations in phospholipids can stabilize the membrane by reducing its fluidity, as in the response of P. putida S-12 to toluene, which reduced the concentration of PE and increased the amount of PG and CL (Weber & de Bont, 1996). The role of PG in the translocation of membrane proteins that participate in the synthesis of CL in bacterial cells contaminated with COP has been demonstrated (Donato et al., 1997). In the present work, membrane changes in P. extremaustralis ADA-5 were observed during the process of adaptation to biphenyl and DCB (formulations 1 and 2) that induced the assimilation of DCB. Similar results were obtained by Murínová and Dercová (2014), who showed an increase in PE and a decrease in PG in the membrane of bacterial strain exposed to PCBs. The substantial increase in the content of CL is a known adaptive mechanism in stressful environments (Prossnigg et al., 2010), with the enzyme cardiolipin synthase in the cytoplasmic membrane participating in the synthesis of CL. The synthase uses two PG molecules (donor and acceptor) for the transfer of phosphatidyl groups (Dercová et al., 2018). Recent studies have reported that bacteria with low levels of cardiolipin synthase in their cytoplasmic membrane are more affected by organic solvents (Bernal et al., 2007) and the physical properties of their cytoplasmic membrane are modified. Rühl et al. (2012) showed that P. putida uses specific physical properties of glycerophospholipids, to readjust the membrane barrier to environmental stresses, suggesting different strategies of the genus Pseudomonas to maintain the barrier function of cell membranes. Zorádová et al. (2011) obtained results similar to those found here with P. extremaustralis ADA-5, showing a significant decrease in cell biomass and changes in the fatty acid profile of the membrane lipids of P. stutzeri caused by the addition of PCBs to cultures. Our results showed that DCB caused variations in the abundance of membrane lipids (PE, PG, and CL). The combined presence of biphenyl and DCB in the bacterial adaptation phase leads to the gradual increase in cellular biomass. This could explain the capacity for biodegradation and bioaccumulation of DCB by P. extremaustralis ADA-5.
5 Conclusions
In this study, a Box-Behnken experimental design was used to evaluate culture media formulations and conditions to improve growth and the production of biomass and exopolysaccharides by P. extremaustralis ADA-5 in the presence of decachlorobiphenyl (DCB). Growth kinetics showed the ability of the strain to grow and biodegrade 9.75% of DCB, by using it as a carbon source and to bioaccumulate 19.98% of DCB in the cellular biomass of 250 mg L−1. Based on this, we infer that this bacterium has the biochemical machinery to use these highly chlorinated compounds as a carbon source. Likewise, it was confirmed that the presence of the contaminant in the culture medium significantly influences the composition of the lipids of cell membrane. Therefore, due to these biological characteristics and its metabolic capacity, this native strain was found to be a promising degrader of DCB.
References
Anyasi, R. O., & Atagana, H. I. (2011). Biological remediation of polychlorinated biphenyls (PCB) in the environment by microorganisms and plants. African Journal of Biotechnology, 10, 18916–18938. https://doi.org/10.5897/AJB10.557.
Bedard, D. L., Haberl, M. L., May, R. J., et al. (1987). Evidence for novel mechanisms of polychlorinated biphenyl metabolism in Alcaligenes eutrophus H850. Applied and Environmental Microbiology, 53, 1103–1112. https://doi.org/10.1128/AEM.53.5.1103-1112.1987.
Berg, J. M., Tymoczko, J. L., & Stryer, L. (2006). Biochemistry (5th ed.p. 1514). WH Freeman and Company.
Bernal, P., Muñoz-Rojas, J., & Hurtado, A. (2007). Pseudomonas putida cardiolipine synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environmental Microbiology Reports, 9, 135–1145. https://doi.org/10.1111/j.1462-2920.2006.01236.x.
Bernat, P., Gajewska, E., Szewczyk, R., et al. (2013). Tributyltin (TBT) induces oxidative stress and modifies lipid profile in the filamentous fungus Cunninghamella elegans. Environmental Science and Pollution Research International, 21, 4228–4235. https://doi.org/10.1007/s11356-013-2375-5.
Bernat, P., Siewiera, P., Soboń, A., et al. (2014). Phospholipids and protein adaptation of Pseudomonas sp. to the xenoestrogen tributyltin chloride (TBT). World Journal of Microbiology and Biotechnology, 9, 2343–2350. https://doi.org/10.1007/s11274-014-1659-3.
Bligh, E. G., & Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology, 37, 911–917. https://doi.org/10.1139/o59-099.
Chakraborty, J., & Das, S. (2016). Characterization of the metabolic pathway and catabolic gene expression in biphenyl degrading marine bacterium Pseudomonas aeruginosa JP-11. Chemosphere, 144, 1706–1714. https://doi.org/10.1016/j.chemosphere.2015.10.059.
Costabeber, I., Sifuentes, D. S. J., Odorissi, X. A., et al. (2006). Levels of polychlorinated biphenyls (PCBs) in meat and meat products from the state of Rio Grande do Sul, Brazil. Food and Chemical Toxicology, 44, 1–7. https://doi.org/10.1016/j.fct.2005.01.005.
De Limae Silva, M. R., Correa, R. C., Sakamoto, I. K., et al. (2018). Microbial characterization of methanogenic and iron-reducing consortium in reactors with polychlorinated biphenyls. Current Microbiology, 75, 666–676. https://doi.org/10.1007/s00284-018-1431-2.
De Rudder, K. E., Thomas-Oates, J. E., & Geiger, O. (1997). Rhizobium meliloti mutants deficient in phospholipid N-methyltransferase still contain phosphatidylcholine. Journal of Bacteriology, 179, 6921–6928. https://doi.org/10.1128/jb.179.22.6921-6928.1997.
Del Puerto, C., Iglesias, E., Morales, T., et al. (2009). Organización y manejo de la colección de cepas de referencia del Instituto Finlay. VacciMonitor, 18, 20–24 Available on-line at: https://www.redalyc.org/articulo.oa?id=203414612004.
Denich, T. J., Beaudette, L. A., Lee, H., et al. (2003). Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. Journal of Microbiological Methods, 52, 149–182. https://doi.org/10.1016/S0167-7012(02)00155-0.
Dercová, K., Murínová, S., Dudášová, H., et al. (2018). The adaptation mechanisms of bacteria applied in bioremediation of hydrophobic toxic environmental pollutants: How indigenous and introduced bacteria can respond to persistent organic pollutants-induced stress? Persistent organic pollutants, Stephen Kudom Donyinah. IntechOpen. https://doi.org/10.5772/intechopen.79646.
Donato, M. M., Jurado, A. S., & Antunes-Madeira, M. C. (1997). Effects of a lipophilic environmental pollutant (DDT) on the phospholipid and fatty acid contents of Bacillus stearothermophilus. Archives of Environmental Contamination and Toxicology, 33, 341–349. https://doi.org/10.1007/s002449900263.
Escárcega-González, C. E., Garza-Cervantes, J. A., Vázquez-Rodríguez, A., et al. (2018). Bacterial exopolysaccharides as reducing and/or stabilizing agents during synthesis of metal nanoparticles with biomedical applications. International Journal of Polymer Science, 7045852, 15. https://doi.org/10.1155/2018/7045852.
Geel-Schutten, G., Flesch, F., Ten, B. B., et al. (1998). Screening and characterization of Lactobacillus strains producing large amounts of exopolysaccharides. Applied Microbiology and Biotechnology, 50, 697–703. https://doi.org/10.1007/s002530051353.
Green, M. R., & Joseph, S. (2012). Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press.
Han, X., O’Connor, J. C., Donner, E. M., et al. (2009). Noncoplanar 2,2′, 3,3′, 4,4′, 5,5′, 6,6′-decachlorobiphenyl (PCB 209) did not induce cytochrome P450 enzyme activities in primary cultured rat hepatocytes, was not genotoxic, and did not exhibit endocrine-modulating activities. Toxicology, 255, 177–186. https://doi.org/10.1016/j.tox.2008.10.013.
Hatamian-Zarmi, A., Shojaosadati, S. A., Vasheghani-Farahani, E., et al. (2009). Extensive biodegradation of highly chlorinated biphenyl and Aroclor 1242 by Pseudomonas aeruginosa TMU56 isolated from contaminated soils. International Biodeterioration and Biodegradation, 63, 788–794. https://doi.org/10.1016/j.ibiod.2009.06.009.
Huang, L., Su, G., & Liu, Y. (2014). Effect of NiFe2O4 on PCDF by products formation during termal degradation of decachlorobiphenyl. RSC Advances, 4, 25453–25460. https://doi.org/10.1039/c4ra02580f.
Hughes, D., & Clark, B. R. (2011). Biodegradation of polyfluorinated biphenyl in bacteria. Biodegradation, 22, 741–749. https://doi.org/10.1007/s10532-010-9411-7.
Martínez-Toledo, A., & Rodríguez-Vázquez, R. (2011). Response surface methodology (Box-Behnken) to improve a liquid media formulation to produce biosurfactant and phenanthrene removal by Pseudomonas putida. Annals of Microbiology, 61, 605–613. https://doi.org/10.1007/s13213-010-0179-0.
Mathews, S., & Sithebe, P. (2018). The role of bacteria on the breakdown of recalcitrant polychlorinated biphenyls (PCBs) compounds in wastewater. Waste Water and Water Quality. https://doi.org/10.5772/intechopen.75400.
Mishra, A., & Jha, B. (2013). Microbial Exopolysaccharides. In E. Rosenberg, E. F. De Long, S. Lory, E. Stackebrandt, & F. Thompson (Eds.), The Prokaryotes. Springer. https://doi.org/10.1007/978-3-642-31331-8-25.
Murínová, S., & Dercová, K. (2014). Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. International Journal of Microbiology, 873081, 16. https://doi.org/10.1155/2014/873081.
Polak, M. L., Zlatic, E., Demšar, L., et al. (2016). Degradation of PCBs in dry fermented sausages during drying/ripening. Food Chemistry, 213, 246–250. https://doi.org/10.1016/j.foodchem.2016.06.051.
Prossnigg, F., Hickel, A., Pabst, G., et al. (2010). Packing behaviour of two predominant anionic phospholipids of bacterial cytoplasmic membranes. Biophysical Chemistry, 50, 129–135. https://doi.org/10.1016/j.bpc.2010.04.004.
Qiu, L., Wang, H., & Wang, X. (2016). Isolation and characterization of a cold-resistant PCB209-degrading bacterial strain from river sediment and its application in bioremediation of contaminated soil. Journal of Environmental Science and Health Part C, 51, 204–212. https://doi.org/10.1080/10934529.2015.1094324.
Rühl, J., Hein, E. M., & Hayen, H. (2012). The glycerophospholipid inventory of Pseudomonas putida is conserved between strains and enables growth condition-related alterations. Microbial Biotechnology, 5, 45–58. https://doi.org/10.1111/j.1751-7915.2011.00286.x.
Sánchez-Pérez, B. N., Zenteno-Rojas, A., Rincón-Molina, C. I., et al. (2019). Rhizosphere and endophytic bacteria associated to Ocimum basilicum L. with decaclorobiphenyl removal potential. Water, Air, & Soil Pollution, 231, 134. https://doi.org/10.1007/s11270-020-04481-6.
Sandri, F., Fedi, S., & Zannoni, D. (2017). Biphenyl modulates the expression and function of respiratory oxidases in the polychlorinated-biphenyls degrader Pseudomonas pseudoalcaligenes KF707. Frontiers in Microbiology, 8, 1223. https://doi.org/10.3389/fmicb.2017.01223.
She, J., Holden, A., Sharp, M., et al. (2007). Polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) in breast milk from the Pacific Northwest. Chemosphere, 67, S307–S317. https://doi.org/10.1016/j.chemosphere.2006.05.154.
Tandlich, R., Vrana, B., & Balaz, S. (2011). Biodegradation mechanism of biphenyl by a strain of Pseudomonas stutzeri. Journal of Environmental Science and Health, Part C, 46, 337–344. https://doi.org/10.1080/10934529.2011.542383.
Tharakan, J., Tomlinson, D., Addagada, A., et al. (2006). Biotransformation of PCBs in contaminated sludge: potential for novel biological technologies. Engineering in Life Sciences, 6, 43–50. https://doi.org/10.1002/elsc.200620117.
Valenzuela-Encinas, C., Neria-Gonzalez, I., Alcantara-Hernández, R. J., et al. (2008). Phylogenetic analysis of the archaeal community in an alkaline saline soil of the former lake Texcoco (México). Extremophiles, 12, 247–254. https://doi.org/10.1007/s00792-007-0121-y.
Vidali, M. (2001). Bioremediation an overview. Pure and Applied Chemistry, 73, 1163–1172. https://doi.org/10.1351/pac200173071163.
Villacieros, M., Whelan, C., Mackova, M., et al. (2005). Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Applied and Environmental Microbiology, 71, 2687–2694. https://doi.org/10.1128/AEM.71.5.2687-2694.2005.
Villalobos-Maldonado, J. J., Meza-Gordillo, R., Mancilla-Margalli, N. A., et al. (2015). Removal of decachlorobiphenyl in vermicomposting process amended with rabbit manure and peat moss. Water, Air, & Soil Pollution, 226, 159. https://doi.org/10.1007/s11270-015-2400-z.
Weber, F. J., & de Bont, J. A. M. (1996). Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochimica et Biophysica Acta, 1286, 225–245. https://doi.org/10.1016/S0304-4157(96)00010-X.
Weiland, B. N., Fischer, M. A., Schramm, K. W., et al. (2017). Polychlorinated biphenyl (PCB)-degrading potential of microbes present in a cryoconite of jamtalferner glacier. Frontier of Microbiology, 8, 110.5. https://doi.org/10.3389/fmicb.2017.01105.
Yu, Y., Yin, H., Peng, H., et al. (2020). Proteomic mechanism of decabromodiphenyl ether (BDE-209) biodegradation by Microbacterium Y2 and its potential in remediation of BDE-209 contaminated water-sediment system. Journal of Hazardous Materials, 387, 0304–3894. https://doi.org/10.1016/j.jhazmat.2019.121708.
Zenteno-Rojas, A., Martínez-Romero, E., Rincón-Molina, C. I., et al. (2019). Removal of high concentrations decachlorobiphenyl of earthworm Eisenia fetida and its symbiotic bacteria in a vermicomposting system. Water, Air, & Soil Pollution, 230, 116. https://doi.org/10.1007/s11270-019-4170-5.
Zhao, L., Hou, H., & Shimoda, K. (2012). Formation pathways of polychlorinated dibenzofurans (PCDFs) in sediments contaminated with PCBs during the thermal desorption process. Chemosphere, 88, 1368–1374. https://doi.org/10.1016/j.chemosphere.2012.05.042.
Zorádová, S., Dudášová, H., Lukáčová, L., et al. (2011). The effect of polychlorinated biphenyls (PCBs) on the membrane lipids of Pseudomonas stutzeri. International Biodeterioration and Biodegradation, 65, 1019–1023. https://doi.org/10.1016/j.ibiod.2011.03.012.
Acknowledgements
We thank the Biochemical Engineering Postgraduate Department-ITTG and CONACYT for granting a scholarship to Miguel Ángel Gómez López. We thank MA. Celina Lujan Hidalgo from the ITTG laboratory for technical assistance. We thank Michael Dunn for reading the manuscript.
Funding
Miguel Ángel Gómez López had a Conacyt scholarship during his Master of Science in Biochemical Engineering. Financial support was from Tecnologico Nacional de México 7676.20-P.
Author information
Authors and Affiliations
Contributions
All authors contributed to this study and have approved the final manuscript. RRR designed the study. MAGL and AZR performed laboratory experiments and data analysis. CIRM, LAMG, and MAVG contributed new reagents and analytical tools. VMRV and FARM data analysis. EMR, RRR, and AZR wrote the manuscript.
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 12 kb)
Rights and permissions
About this article
Cite this article
López, M.A.G., Zenteno-Rojas, A., Martinez-Romero, E. et al. Biodegradation and Bioaccumulation of Decachlorobiphenyl (DCB) by Native Strain Pseudomonas extremaustralis ADA-5. Water Air Soil Pollut 232, 192 (2021). https://doi.org/10.1007/s11270-021-05122-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-021-05122-2