Abstract
Bacteria that harbor the mer operon in their genome are able to enzymatically reduce mercury (II) to the volatile form of mercury Hg (0). Detoxification of contaminated waste by using these bacteria may be an alternative to conventional methods for mercury removal. Residual glycerol from the biodiesel industry can be used as a carbon source to accelerate the process. This work shows for the first time the feasibility of using residual glycerol as a carbon source for Hg removal by bacteria prospected from contaminated environments. Eight bacterial isolates were able to remove mercury and degrade glycerol in mineral medium and residual glycerol. Mercury removal was monitored by atomic absorption spectroscopy and glycerol degradation by high performance liquid chromatography. The best results of mercury removal and glycerol degradation were obtained using isolates of Serratia marcescens M25C (85 and 100 %), Klebsiella pneumoniae PLB (90 and 100 %), Klebsiella oxytoca (90 and 100 %), and Arthrobacter sp. U3 (80 and 75 %), with addition of 0.5 g L−1 yeast extract. The Arthrobacter sp. U3 isolate is common in soils and has proven to be a promising candidate for environment applications due to its low pathogenicity and higher Hg removal and glycerol degradation rates.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Mercury (Hg) occurs naturally in the environment in chemical species with different solubility, reactivity, and toxicity, causing several impacts on ecosystems and human health (UNEP). Mercury is emitted into the environment by numerous natural and anthropogenic sources, through complex combinations involving chemical, physical, and biological reactions (Maxson 2005; Driscoll et al. 2013). The most common anthropogenic sources of mercury are the industrial establishments with different production profile such as petrochemical, electronic, measuring equipments, inks, and chlorine soda, besides the extraction of gold and dental amalgam (Li et al. 2009; Pirrone et al. 2010) .
Although mercury is toxic to both eukaryotic cells and prokaryotic cells, some microorganisms possess metal resistance mechanisms, including efflux pumps (Silver and Phung 1996; Waldron and Robinson 2009), chelation by enzymatic polymers, precipitation, biomethylation (Boening 2000), and enzymatic reduction related to mer operon (Silver and Walderhaug 1992; Ma et al. 2009). Bacteria that harbor mer operon are able to enzymatically reduce mercury (II) to the volatile form of mercury Hg (0) (Nies 1999; Barkay et al. 2003; Barkay and Wagner-Döbler 2005; Zhang et al. 2006; Dash and Das, 2012). The merA genes are part of an operon comprising regulatory genes and other encoding genes of transport proteins (Summers 1992; Silver and Phung 1996; Narita et al. 2003; Dash and Das 2012). In general, many mercury-resistant isolates possess the merR, merP, and merA genes encoding proteins for regulatory function, transport and extracellular binding, and mercuric (II) reductase, respectively (Silver and Hobman 2007; Hobman 2007).
Mercuric (II) reductase (MerA) is a flavin oxidoreductase (~120 kDa), which acts as a dimer and is composed of three domains. The three-dimensional structure of the enzyme indicates that the active site is formed by the interaction of the central domain of a subunit with another C-terminal domain (Barkay and Wagner-Döbler 2005). The central domain, described as a pyridine nucleotide oxidoreductase disulfide group, is where catalysis and the transfer of two electrons from NADPH to Hg (II) via FAD occur. In this case, there is a pair of active redox cysteines that carry out the charge-transfer reaction and the Hg (II) is reduced by electron transference to the volatile form of mercury Hg (0). The core of the MerA enzyme is a C-terminal domain found only in the mercuric reductase enzyme, together with the central domain (Silver and Hobman 2007). An N-terminal domain has the function of directing the Hg (II) to the active site of MerA. This domain is essential in cells with low concentrations of sulfide agents, where the supply of thiolated Hg (II) may be limited. Thus, the mer system has evolved to perform metal regulation, transport, and enzymatic catalysis, converting the highly reactive form of the mercury cation to volatile Hg (0) (Barkay and Wagner-Döbler 2005). This adaptive microbial response to Hg is an alternative for remediation of Hg-contaminated sites; in addition, the genus Pseudomonas has been explored for potential use in contaminated environments (Wagner-Döbler 2003; Barkay and Wagner-Döbler 2005; Mortazavi et al. 2005; Pepi et al. 2010, Giovanella et al. 2011; Cabral et al. 2013).
Conventionally, remediation of contaminated environments by toxic metal ions has employed physicochemical techniques including precipitation, filtration, and electrochemical recovery as well as membrane separation, excavation, waste disposal in landfills, or covering of the contaminated site. These techniques are ineffective, costly, and often produce additional waste with greater potential for contamination (Camargo et al. 2007; Naja and Volesky 2010).
Biodiesel is considered as potential substitute for diesel. Accordingly, its main by-product is glycerol; once for every 9 kg of biodiesel produced, about 1 kg of crude glycerol by-product is formed (Dasari et al. 2005; Johnson and Taconi, 2007; Zeng and Sabra, 2011). Glycerin derived from biodiesel production has low value because of the impurities, as it contains residues of soda, free fatty acids, methanol, salts of fatty acids, esters, sulfur compounds, proteins, and minerals (Thompson and He 2006); thus, it demands costs for disposal (Yazdani and Gonzales 2007; Escapa et al. 2009). However, residual glycerol is considered a highly compact and assimilable carbon source for bacteria to obtain energy for metabolic processes under aerobic and anaerobic conditions (Silva et al. 2009). Concerning the treatment of effluents, the use of residual glycerol as a carbon source for biological denitrification and phosphorus removal has been studied (Bodík et al. 2009; Yuan et al. 2010; Torà et al. 2011; Guerrero et al. 2012), aiming to reduce the cost of treatment of these effluents.
Although bioremediation of mercury-contaminated effluents has been reported in literature, technologies aimed at distinct metabolic pathways as well as the use of alternative carbon sources (residual glycerol) for application of microorganisms in metal biotransformation have not been studied. Thus, the study on methodologies that join removal efficiency and low cost of application may effectively expand bioremediation technology for large-scale models.
This study aimed to investigate growth and the ability to remove Hg2+ of eight Gram-negative bacteria cultivated in medium and the feasibility of Hg removal using residual glycerol as a carbon source. Additionally, the degradation of residual glycerol by microorganisms, the identification of merA genes in Gram-negative isolates by PCR, and the inhibitory and biocide concentration of Hg (II) on bacteria will be assessed.
2 Material and Methods
2.1 Microorganisms
Bacteria were previously isolated from metal-contaminated industrial effluents, according to the methodology described by Giovanella et al. 2011.
Thirty-five isolates resistant to Hg were evaluated for growth on glycerol as the only carbon source. Indirect turbidimetric method was used, and the optical density was measured at 600 nm in the culture medium MM1 Richard & Vogel7 (KCl 0.7 g L−1, KH2PO4 2 g L−1, Na2HPO4 3 g L−1, NH4NO3 1 gL−1, MgSO4 4 mg L−1, FeSO4 0.2 mg L−1, MnCl2 0.2 mg L−1, CaCl2 0.2 mg L−1, containing HgCl2 (10 μM), and 10 g L−1 commercial glycerol (99 %). After 48 h of incubation at 30 °C, the optical density at 600 nm was measured, and the isolates presenting cell growth (B50A, CP1.0, M50C, M25C, PLA, PLB, U3, and U14) were selected for the other experiments.
2.2 Molecular Identification of Mercury-Resistant Bacteria
The bacterial genomic DNA was extracted using Wizard Genomic DNA Purification Kit (Promega, WI). The 16S ribosomal DNA (rDNA) was amplified in a PCR using the following universal primers for bacteria: 27 F (5′-AGATTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′). The amplification reaction was based on Sambrook and Russel (2001): buffer (50 mM of Tris–HCl, pH 9.0, 50 mM of KCl, 2.5 % of Triton × 100), dNTPs (200 μM from each), 0.2 μM of MgCl2, 0.25 pmol of each primer, 0.8 ng/μL of DNA sample, and 0.02 U of Taq DNA polymerase. The amplification was performed in a thermal cycler (MJ Research Inc., Watertown, MA, USA), and the program consisted of 35 cycles (initial denaturation at 95 °C for 5 min, subsequent denaturation at 95 °C for 30 s, annealing at 50 °C for 1 min, extension at 72 °C for 90 s, and final extension for 5 min at 72 °C). The PCR products were purified by standard method of precipitation with PEG 8000 (polyethylene glycol). For the sequencing reactions of PCR fragments, kits of labeled terminators (GE Healthcare), with primer 519r (5′-GWATTACCGCGGCKGCTG-3′), was used. All generated sequences were submitted to the Genbank/NCBI database.
2.3 PCR Detection of merA Genes
For detection of merA gene, DNA of all bacterial isolates were obtained as described above for amplification of 16S rDNA genes. The PCR reaction for merA was performed using primers A1 F (ACC ATC GGC GGC ACC TGC GT) and A5 R (ACC ATC GTC AGG TAG GGG AAC) (Liebert et al. 1997). Each reaction contained 5 μL of 10× PCR buffer, 1.5 μL of 25 mM MgCl2, 1 μL of 10 mM dNTPs, 0.02 U of Taq DNA polymerase (Denville, NJ), 0.5 μL of 20 pmol μL−1 of each primer (forward e reverse), and 16.75 μL of ultrapure water. The following PCR conditions were used for merA: hot start at 95 °C for 5 min, followed by 29 cycles of 95 °C for 1 min, 64 °C for 2 min, and 90 °C for 3 min and a final extension for 5 min at 72 °C. Each PCR reaction included two bacterial Gram-positive as a negative control (B. cereus CP1.0 and Arthrobacter sp. U3) that were prepared in identical conditions as described above. The reactions were run in GeneAmp PCR System 9700 (Applied Biosystems, CA). The products of PCR reactions were separated on 1 % agarose gel (Fisher Scientific, MA) and observed using a Geldoc 2000 system (Bio-Rad, CA).
2.4 Preparation of Cells
Different culture media were used to standardize the trials, as described below.
Trials in LB medium: each mercury-resistant isolate was inoculated onto LB medium (bacteriological tryptone 10 g L−1, yeast extract 5 g L−1, NaCl 10 g L−1) pH 6.7, containing HgCl2 (10 μM), and incubated for 24 h at 30 °C under agitation at 100 rpm. Initial bacterial counts of 107 CFU/mL were standardized by turbidimetry at 600 nm (0.05 to 0.45) and the corresponding CFU/mL in Petri plates. Uninoculated culture medium was used as negative control.
Trials in mineral medium MM1 and residual glycerol: each mercury-resistant isolate was inoculated onto MM1 (KCl 0.7 gL−1, KH2PO4 2 g L−1, Na2HPO4 3 g L−1, NH4NO3 1 g L−1, MgSO4 4 mg L−1, FeSO4 0.2 mg L−1, MnCl2 0.2 mg L−1, CaCl2 0.2 mg L−1) pH 6.7, containing HgCl2 (10 μM) and residual glycerol (10 g L−1) and incubated for 48 h at 30 °C under agitation at 100 rpm. Initial bacterial counts of 107 CFU mL−1 were standardized by turbidimetry at 600 nm (0.05 to 0.45) and the corresponding CFU mL−1 in Petri plates. Uninoculated culture medium was used as negative control.
2.5 Minimal Inhibitory Concentrations
The bacterial isolates were evaluated for their ability to grow in LB medium containing different concentrations of HgCl2. All tests were performed in 96-multiwell plates with each well being filled with 200 μL of the growth medium. Each well was inoculated with 5 μL of overnight cultures following incubation at 30 °C for 48 h. The lowest concentration of mercury at which no visible growth was observed was considered as the minimal inhibitory concentration (MIC) (Ball et al. 2007). Uninoculated LB medium served as negative controls, and inoculated LB medium without mercury served as positive controls.
2.6 Minimal Biocide Concentration
The isolates were tested for minimal biocide concentration of mercury in 96-well polystyrene plates on LB medium containing HgCl2 at concentrations varying from 125 to 1500 μM, incubated for 48 h at 30 °C under agitation at 100 rpm. After the incubation period, 25 μL culture was inoculated in Petri dishes containing Agar nutrient. The biocide concentration was determined after 72 h of incubation at 30 °C by the presence or absence of microbial growth.
2.7 Mercury Removal by the Isolates in LB Medium
The essay was performed according to Kannan and Krishnamoorthy (2006). Briefly, about 2 × 107 CFU mL−1 of the bacterial isolates were inoculated in LB medium containing 10 μM of HgCl2 at 30 °C for 24 h. At these time, samples were taken was growth was determined as OD600 as well the mercury removal capacity (%) as the mercury remaining and pH. Uninoculated LB medium served as negative controls, and inoculated LB medium without mercury served as positive controls.
2.8 Mercury Removal by Isolates Using Alternative Carbon Source (Residual Glycerol)
The mercury removal capacity was determined in mineral medium MM1 containing 10 μM of HgCl2, 10 g L−1 of residual glycerol plus 0.5 g L−1 yeast extract as a nitrogen source. The initial cell concentration was 107 CFU mL−1 and the culture medium was incubated for 24, 48, and 72 h at 30 °C and 100 rpm. After the incubation period, aliquots were sampled for analysis of the remaining mercury and glycerol, pH, and optical density at 600 nm. The culture medium MM1 containing 10 μM of HgCl2, 10 g L−1 of residual glycerol, and 0.5 g L−1 yeast extract not inoculated were used as a negative control.
In addition, the mercury removal capacity was evaluated in MM1 medium containing 10 μM HgCl2 and 10 g L−1 residual glycerol without addition of yeast extract as a nitrogen source. The initial cell concentration was 107 CFU mL−1, and the culture medium was incubated for 24, 48, 72, 96, 120, and 144 h at 30 °C and 100 rpm. After the incubation period, aliquots were sampled for analysis of the remaining mercury and glycerol, pH, and optical density at 600 nm. The culture medium MM1 containing 10 μM HgCl2 and 10 g L−1 of residual glycerol not inoculated were used as a negative control.
2.9 Mercury Analysis by Cold Vapor Atomic Absorbance Spectroscopy
The concentration of mercury remaining in the medium inoculated with isolates and uninoculated was determined following digestion as proposed by EPA method 7471B. All glassware used during Hg (II) analyses was cleaned in 30 % HNO3 and rinsed several times in ultrapure water prior to use. An aliquot of supernatant (0.1 mL) was collected and added to 5 and 2 mL of concentrated sulfuric and concentrated nitric acids, respectively. Then, 10 mL of 5 % potassium permanganate was added to the tubes, following autoclavation for 15 min at 121 °C. The remaining potassium permanganate was reduced with 6 mL of 12 % hydroxylamine hydrochloride. Mercury analysis was performed in an atomic absorption spectrometry/cold vapor (Analyst 100, Perkin Elmer, Waltham, MA, USA).
2.10 Chromatography Analysis
The culture samples were prepared by centrifugation at 10,000 g for 5 min to pellet the cell, followed by filtration using 0.22 μm pore size cellulose acetate membrane filters. Glycerol concentration in media samples was analyzed using a Shimadzu HPLC (Shimadzu Corp., Japan) equipped with a RID-10A refractive index detector. The stationary and mobile phases were an Aminex HPX-87H column (300 × 7.8 mm, Bio-Rad, USA) and 0.005 M H2SO4 solution at 0.8 mL min−1, respectively. The column temperature was controlled at 65 °C. For the HPLC analyses, standards of HPLC grade of glycerol was used (>99.5 purity).
3 Results and Discussion
Thirty-five Hg-resistant strains previously isolated were tested for growth on medium containing purified glycerol as a carbon source. Of these, only eight isolates grew in culture medium MM1 with glycerol after 48 h of incubation and thus were selected for further experiments (data not shown).
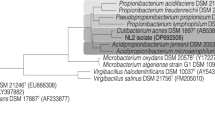
Analysis of 16S rRNA genes showed that the eight isolates capable to utilize glycerol as a carbon source belong to distinct species, six being Gram-negative and two Gram-positive bacteria (Table 1). Pseudomonas, Serratia, Citrobacter, Klebsiella, and Bacillus are reported as glycerol-degrading bacteria forming bioproducts (Silva et al. 2009), while bacteria of the genera Proteus and Arthrobacter were not reported in other studies as glycerol-degrading bacteria (Fig. 1).
Once glycerol has been used for production of different products, various microorganisms have been evaluated for their glycerol bioconversion ability (Sarma et al. 2012). Tao et al. (2005) investigated Serratia marcescens strains for the production of the natural red pigment prodigiosin and observed higher yield when using glycerol as a carbon source. An isolate from the genus Bacillus promoted glycerol degradation and produced higher quantities of ethanol, as reported by Nakas et al. 1983. Several studies on the ability of the bacteria Citrobacter and Klebsiella to metabolize glycerol have been found in literature (Homann et al. 1990; Daniel et al. 1995; Seifert et al. 2001; Forage and Foster, 1982; Tong et al. 1991; Menzel et al. 1997; Németh et al. 2003). These processes can be oxidative or reductive (Zhu et al. 2002) and involve enzymes that catalyze the conversion of glycerol into assimilable products by bacterial cells (Daniel et al. 1995; Luers et al. 1997; Macis et al. 1998).
Mercury resistance is frequent among Gram-negative microorganisms. Nakamura et al. (2001) have isolated 55 mercury-resistant bacteria and found only four Gram-positive. Several studies have shown that Pseudomonas have high resistance to different forms of mercury (Fortunato et al. 2005; Ní Chadhain et al. 2006; Zhang et al. 2012; Sone et al. 2013; Cabral et al. 2014). Members of the genera Arthrobacter, Bacillus, Citrobacter, Klebsiella, Proteus, and Serratia are less reported in literature but have been characterized as Hg-tolerant bacteria (Ní Chadhain et al. 2006; Zeroual et al. 2001; Keramati et al. 2011; Chien et al. 2012; Dash et al. 2013; Bafana et al. 2010).
After 48 h of incubation in LB medium supplemented with Hg at concentrations ranging from 0 to 1500 μM, higher resistance level was observed in all isolates, but with different tolerance capabilities (Table 2). Isolates showed MIC between 250 and 920 μM and minimal biocide concentration (MBC) between 550 and 1000 μM Hg. Klebsiella oxytoca U14, Pseudomonas sp. B50A, and Serratia marcescens M25C were the most resistant bacteria, since they were extinct in 1000 μM Hg. A similar result was obtained by Chatziefthimou et al. (2007), who evaluated chemolithotrophic thermophilic microorganisms from a hot spring with high Hg concentrations. These results indicate that the efficiency of mercury detoxification system may vary according to the microorganisms involved.
All isolates removed Hg from LB medium (Fig. 2). The isolates Citrobacter freundii PLA and Klebsiella pneumoniae PLB were the most efficient and statistically similar in their ability to remove Hg, with percent remotion higher than 90 %. The isolate Arthrobacter sp. U3 exhibited slower growth rates and reduced ability to remove Hg (35 %) in LB medium. Bafana et al. 2010 investigated Arthrobacter strains in a nutrient medium and found similar results of Hg removal after 24 h of incubation. The lower growth rate may be due to the culture medium used, since high population growth rate was observed when the isolate was inoculated onto MM1 (Fig. 3g) within 24 h of cultivation. Hozzein et al. 2008 found greater number of actinomycetes in mineral medium, when compared to conventional culture media, and (Jones and Keddie, 2006) have reported more suitable culture media for isolation and selection of bacteria of the genus Arthrobacter. An intermediate group of four microorganisms (Pseudomonas sp. B50A, Bacillus cereus CP1.0, Proteus mirabilis M50C, and Klebsiella oxytoca U14) was able to remove about 80 % of the mercury present in the culture medium. Sadhukhan et al. (1997) studied Hg-resistant microorganisms and obtained similar results on Hg removal after 24 h of incubation. The authors found that the Hg removal ability of the microorganisms was related to their MIC values, corroborating the findings of Ghosh et al. (1996). However, in the present study, this relationship was not observed because the isolates with higher MIC were not the most efficient in removing Hg. Despite the isolate Serratia marcescens M25C presented the largest MIC among the isolates, its percent removal was only 52 %.
Removal of Hg2+ (%) (bars), glycerol consumed (crosshatched bars), and optical density (triangle down) by resistant strains after incubation at 30 °C in MM1 culture medium containing 10 μM of HgCl2, 10 g L−1 of residual glycerol without yeast extract. Isolate Pseudomonas sp. B50A (a), P. mirabilis M50C (b), K. pneumoniae PLB (c). Each point represents mean values of triplicate experiments, and error bars are the standard deviation of mean
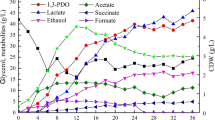
Removal of Hg2+ (%) (bars), glycerol consumed (crosshatched bars), and optical density (triangle down) by resistant strains after incubation at 30 °C in MM1 culture medium containing 10 μM of HgCl2, 10 g L−1 of residual glycerol plus 0.5 g L−1 yeast extract. Isolate Pseudomonas sp. B50A (a), B. cereus CP1.0 (b), S. marcescens M25C (c), P. mirabilis M50C (d), C. freundii PLA (e), K. pneumoniae PLB (f), Arthrobacter sp. U3 (g), K. oxytoca U14 (h). Each point represents mean values of triplicate experiments, and error bars are the standard deviation of mean
The differences found in the present study may be due to the presence of metallothionein in some isolates. These proteins have cysteine and methionine residues rich in sulfhydryls, which complex metals, protecting the cells from the toxicity of these elements (Erbe et al. 1995). However, the presence of metallothionein has not been evaluated in this study to confirm this hypothesis. Furthermore, it is possible that the mer operon genes were not induced sufficiently by the low Hg concentration in the culture medium, once the mercuric reductase activity is inducible rather than constitutive. Another hypothesis is that the microorganisms have different resistance behavior that was not evaluated in this study.
PCR amplification of merA genes from the eight bacteria under study are presented in Fig. 1. The merA genes amplification from Pseudomonas sp. B50A and S. marcesens M25C isolates had the fragment amplified of an expected size. However, for the isolates P. mirabilis M50C, C. freundii PLA, and K. pneumoniae PLB, the amplicon sizes were smaller than 1238 bp, possibly due to variations in the size of the merA genes sequence, which may be related to the high diversity of merA genes (Ní et al. 2006; Oregaard and Sørensen 2007). The amplification of K. pneumoniae PLB isolate showed two extra bands, evidencing that the primer may have low specificity (Felske et al. 2003). Although merA genes were not detected in K. oxytoca U14 isolate, they may be present, once mer system is the most common mode of bacterial mercury resistance (Osborn et al. 1997; Barkay et al. 2003); thus, additional studies using other primers may be carried out. Once the primers were specific for Gram-negative bacteria, the B. cereus CP1.0 and Arthrobacter sp. U3 isolates used as negative control were not amplified because they are Gram-positive bacteria.
Only the isolates Pseudomonas sp. B50A, P. mirabilis M50C, and K. pneumoniae PLB grew in culture medium containing residual glycerol as a carbon source without the addition of yeast extract (Fig. 2a–c). Despite these three isolates have degraded glycerol, and removed Hg from the medium, the bacterial growth was negatively affected by the culture medium when compared to LB medium. Although the lag phase was longer under these conditions, bacterial growth and glycerol degradation were observed during the incubation period, suggesting that the three isolates were tolerant to the impurities from residual glycerol present in the culture medium. Residual glycerol contains substances such as sodium hydroxide and methanol, which may inhibit bacterial growth (Thompson and He 2006).
Mercury removal by the Pseudomonas sp. B50A isolate was 80 % within only 24 h of incubation, reaching 95 % after 96 h, not depending on high population growth rates (Fig. 1a). Other authors (Vetriani et al. 2005; Chatziefthimou et al. 2007) observed that mercury II removal preceded high cell concentrations in the culture medium, since the glycerol degradation was dependent on high bacterial population growth. The isolates entered the stationary phase after 72 h of incubation probably due the low pH of the culture medium that reached 4.7 (data not shown), due to the synthesis of by-products during glycerol degradation (Biebl et al. 1999).
For the isolates P. mirabilis M50C and K. pneumoniae PLB, the Hg removal was observed when the population reached an optical density (OD) of 0.9 and 1.3 (Fig. 2b, c), with percent removal of 95 and 80 % after 72 and 96 h of incubation, respectively.
The addition of yeast extract to the culture medium MM1 had a positive effect on the microbial growth, once only three isolates grew on MM1 without yeast extract, while eight isolates grew on the medium containing 0.5 g L−1 yeast extract (Fig. 3a, b, c, d, e, f, g, and h). Besides providing extra nitrogen source and vitamins, yeast extract also provides a carbon source that can be used for biomass production (Nikel et al. 2010). For Escherichia coli, the need for supplementation of culture medium with nutrients such as yeast extract and tryptone for cell growth is well documented (Dharmadi et al. 2006; Murarka et al. 2008; Durnin et al. 2009; Nikel et al. 2010). Choi et al. 2011 assessed the biomass and production of metabolites in Kluyvera cryocrescens and found a positive effect when the yeast extract was added to the culture medium.
The eight isolates entered into the logarithmic phase in the first 24 h, degrading glycerol and removing Hg (Fig. 3a, b, c, d, e, f, g, and h). With respect to Hg, the most part was removed during the first 24 h of incubation, which is a positive response from the practical point of view, once the glycerol degradation was dependent on incubation time and cell growth.
K. oxytoca U14, K. pneumoniae PLB, and S. marcescens M25C were more effective for glycerol degradation, showing 100 % degradation at the end of the experiment. Numerous studies have reported bacteria of the genus Klebsiella using residual glycerol as a carbon source and their ability to degrade glycerol (Mu et al. 2006; Zeng and Sabra 2011; Rodriguez et al. 2012; Rossi et al. 2012).
Interestingly, Pseudomonas sp. B50A, S. marcescens M25C, P. mirabilis M50C, Arthrobacter sp. U3, and K. oxytoca U14 removed greater amount of Hg from MM1 containing yeast extract than from LB medium (Fig. 2a, c, d, g, and h). This positive response may be related to the constitution of the medium, since the LB medium has a greater amount of complex organic compounds capable to complex mercury. Several common constituents in culture media bind to metals, influencing the interaction of these ions with microorganisms (Ramamoorthy and Kushner 1975). Farrell et al. (1993) also showed that the culture medium may have a significant effect on Hg bioavailability, such that the interpretation of data may be obscured by the effects of the composition of the culture medium.
Despite the Arthrobacter sp. U3 isolate did not show the best response for Hg removal and glycerol degradation, an effective result was observed, with 80 % Hg removal after 24 h incubation, and 75 % glycerol degradation from the culture medium (Fig. 3g). It is worth mentioning that this isolate exhibited higher cell density in MM1 medium than in LB culture medium, possibly due to improper composition of LB medium for this microorganism. Bacteria of the genus Arthrobacter rarely cause disease in humans due to its low pathogenicity (Bernasconi et al. 2004). These bacteria can be found in soil and grow better than other bacterial species in stressed environments, such as alkaline and metal-contaminated sites (Camargo et al. 2004). This is due to their metabolic versatility and the ability to detoxify/degrade organic xenobiotics and metals (Sajjaphan et al. 2004). However, there is little information on the use of these bacteria in bioremediation of mercury-contaminated effluents. Thus, this isolate is a promising candidate for further studies aimed at application in mercury-contaminated wastewater with low rates of metabolizable carbon.
4 Conclusion
All isolates in this study have high mercury resistance ability and potential to remove Hg from effluents. The presence of the merA genes in five of the six Gram-negative isolates has shown that mercury removal possibly occur in these isolates by volatilization. The bacterial growth is negatively affected by the culture medium containing residual glycerol as a carbon source when compared to LB medium. The best results were obtained with the addition of 0.5 g L−1 yeast extract to the culture medium, evidencing the need for supplementation of culture medium with nutrients. Although supplementation is required, its concentration is very low, thus maintaining the economic viability of the proposal. The best results of mercury removal and glycerol degradation were obtained using isolates of Serratia marcescens M25C, Klebsiella pneumoniae PLB, Klebsiella oxytoca, and Arthrobacter sp. U3. The Arthrobacter sp. U3 isolate is common in soils and has proven to be a promising candidate for environment applications due to its low pathogenicity and higher Hg removal and glycerol degradation rates. This study evidenced the viability of using residual glycerol as a carbon source for microbial mercury removal from mercury-contaminated environments with low levels of metabolizable carbon.
References
Bafana, A., Krishnamurthi, K., Patil, M., & Chakrabarti, T. (2010). Heavy metal resistance in Arthrobacter ramosus strain G2 isolated from mercuric salt-contaminated soil. Journal of Hazardous Materials, 177, 481–486.
Ball, M. M., Carrero, P., Castro, D., & Yarzabal, L. A. (2007). Mercury resistance in bacterial strains isolated from tailing ponds in a gold mining area near El Callao (Bolivar State, Venezuela). Current Microbiology, 54, 149–154.
Barkay, T., & Wagner-Döbler, I. (2005). Microbial transformations of mercury: potentials, challenges, and achievements in controlling mercury toxicity in the environment. Advances in Applied Microbiology, 57, 1–52.
Barkay, T., Miller, S. M., & Summers, A. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiology Reviews, 27, 355–384.
Bernasconi, et al. (2004). Arthrobacter woluwensis subacute infective endocarditis: case report and review of the literature. Clinical Infectious Diseases, 38, 27–31.
Biebl, H., Menzel, K., Zeng, A. P., & Deckwer, W. D. (1999). Microbial production of 1,3-propanediol. Applied Microbiology and Biotechnology, 52, 289–297.
Bodík, I., Blastáková, A., Sedlácek, S., & Hutnan, M. (2009). Biodiesel waste as source of organic carbon for municipal WWTP denitrification. Bioresource Technology, 100, 2452–2456.
Boening, D. W. (2000). Ecological effects, transport, and fate of mercury: a general review. Chemosphere, 40, 1335–1351.
Cabral, L., Giovanella, P., Gianello, C., Bento, F. M., Andreazza, R., & Camargo, F. A. O. (2013). Isolation and characterization of bacteria from mercury contaminated sites in Rio Grande do Sul, Brazil, and assessment of methylmercury removal capability of a Pseudomonas putida V1 strain. Biodegradation, 24, 319–331.
Cabral, L., et al. (2014). Impact of selected anions and metals on the growth and in vitro removal of methylmercury by Pseudomonas putida V1. International Biodeterioration & Biodegradation, 91, 29–36.
Camargo, F. A. O., Bento, F. M., Okeke, B. C., & Frankenberger, W. T. (2004). Hexavalent chromium reduction by an actinomycete, Arthrobacter acrytallopoietes ES 32. Biological Trace Element Research, 97, 183–194.
Camargo, F. A. O., Bento, F. M., & Jacques, R. J. S. (2007). Uso de microrganismos para remediação de metais. In C. A. Ceretta, L. S. Silva, & J. M. Reichert (Eds.), Tópicos em ciência do solo (pp. 468–496). Minas Gerais: Sociedade Brasileira de Ciência do Solo.
Chatziefthimou, A., Crespo-Medina, M., Wang, Y., et al. (2007). The isolation and initial characterization of mercury resistant chemolithotrophic thermophilic bacteria from mercury rich geothermal springs. Extremophiles, 11, 469–479.
Chien, M., Nakahata, R., Ono, T., Miysuke, K., & Endo, G. (2012). Mercury removal and recovery by immobilized Bacillus megaterium MB1. Frontiers of Chemical Science and Engineering, 6, 192–197.
Choi, W. J., Hartono, M. R., Chan, W. H., & Yeo, S. S. (2011). Ethanol production from biodiesel-derived crude glycerol by newly isolated Kluyvera cryocrescen. Applied Microbiology and Biotechnology, 89, 1255–1264.
Daniel, R., Stuertz, K., & Gottschalk, G. (1995). Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. Journal of Bacteriology, 177, 4392–4401.
Dasari, M. A., Kiatsimkul, P. P., Sutterlin, W. R., & Suppes, G. J. (2005). Low-pressure hydrogenolysis of glycerol to propylene glycol. Applied Catalysis A: General, 281, 225–231.
Dash, H. R., & Das, S. (2012). Bioremediation of mercury and importance of bacterial mer genes. International Biodeterioration & Biodegradation, 75, 207–213.
Dash, H. R., Mangwani, N., & Das, S. (2013). Characterization and potential application in mercury bioremediation of highly mercury-resistant marine bacterium Bacillus thuringiensis PW-05. Environmental Science and Pollution Research International, 21, 2642–2653.
Dharmadi, Y., Murarka, A., & Gonzalez, R. (2006). Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnology and Bioengineering, 94, 821–829.
Driscoll, C. T., Mason, R. P., Chan, H. M., Jacob, D. J., & Pirrone, N. (2013). Mercury as a global pollutant: sources, pathways, and effects. Environmental Science & Technology, 47, 4967–4983.
Durnin, G., Clomburg, J., Yeates, Z., Alvarez, P. J. J., Zygourakis, K., Campbell, P., & Gonzalez, R. (2009). Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnology and Bioengineering, 103, 148–161.
Erbe, J. L., Taylor, K. B., & Hall, L. M. (1995). Metalloregulation of the cyanobacterial smt locus: identification of SmtB binding sites and direct interaction with metals. Nucleic Acids Research, 23, 2472–2478.
Escapa, A., Manuel, M. F., Morant, A., Gomez, X., Guiot, S. R., & Tartakovsky, B. (2009). Hydrogen production from glycerol in a membraneless microbial electrolysis cell. Energy & Fuels, 23, 4612–4618.
Farrell, R. E., Germida, J. J., & Huang, P. M. (1993). Effects of chemical speciation in growth media on the toxicity of mercury (II). Applied and Environmental Microbiology, 59, 1507–1514.
Felske, A. D. M., Fehr, W., Pauling, B. V., Von Canstein, H., & Wagner-Döbler, I. (2003). Functional profiling of mercuric reductase (merA) genes in biofilm communities of a technical scale biocatalyzer. BMC Microbiology, 3, 22.
Forage, R. G., & Foster, A. M. (1982). Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. Journal of Bacteriology, 149, 413–419.
Fortunato, R., Crespo, J. G., & Reis, M. A. M. (2005). Biodegradation of thiomersal containing effluents by a mercury resistant Pseudomonas putida strain. Water Research, 39, 3511–3522.
Ghosh, S., Sadhukhan, P. C., Ghosh, D. K., Chaudhuri, J., & Mandal, A. (1996). Studies on the effect of mercury and organomercurial on the growth and nitrogen fixation by mercury-resistant Azotobacter strains. Journal of Applied Microbiology, 80, 319–326.
Giovanella, P., Bento, F., Cabral, L., Gianello, C., & Camargo, F. A. O. (2011). Isolamento e seleção de micro-organismos resistentes e capazes de volatilizar mercúrio. Quim Nova, 34, 231–236.
Guerrero, J., Tayà, C., Guisasola, A., & Baeza, J. A. (2012). Glycerol as a sole carbon source for enhanced biological phosphorus removal. Water Research, 46, 2983–2991.
Hobman, J. L. (2007). MerR family transcription activators: similar designs different specifities. Molecular Microbiology, 65, 1275–1278.
Homann, T., Tag, C., Biebl, H., Deckwer, W. D., & Schink, B. (1990). Fermentation of glycerol to 1,3-propanediol by Klebsiella and Citrobacter strains. Applied Microbiology and Biotechnology, 33, 121–126.
Hozzein, W. N., Ali, M. I. A., & Rabie, W. (2008). A new preferential medium for enumeration and isolation of desert actinomycetes. World Journal of Microbiology and Biotechnology, 24, 1547–1552.
Johnson, D. T., & Taconi, K. A. (2007). The glycerin glut: options for the value-added conversion of crude glycerol resulting from biodiesel production. Environmental Progress, 26, 338–348.
Jones, D., & Keddie, R. M. (2006). The genus Arthrobacter. Prokaryotes, 3, 945–960.
Kannan, S. K., & Krishnamoorthy, R. (2006). Isolation of mercury resistant bacteria and influence of abiotic factors on bioavailability of mercury: a case study in Pulicat lake north of Chennai, South East Índia. Science of the Total Environment, 367, 341–353.
Keramati, P., Hoodaji, M., & Tahmourespour, A. (2011). Multi-metal resistance study of bacteria highly resistant to mercury isolated from dental clinic effluent. African Journal of Microbiology Research, 5, 831–837.
Li, J., Zhang, L., Wu, Y., Liu, Y., Zhou, P., Wen, S., et al. (2009). A national survey of polychlorinated dioxins, furans (PCDD/Fs) and dioxin-like polychlorinated biphenyls (DL-PCBs) in human milk in China. Chemosphere, 75, 1236–1242.
Liebert, C. A., Wireman, J., Smith, T., & Summers, A. O. (1997). Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Applied and Environmental Microbiology, 63, 1066–1076.
Luers, F., Seyfried, M., Daniel, R., & Gottschalk, G. (1997). Glycerol conversion to 1,3-propanediol by Clostridium pasteurianum: cloning and expression of the gene encoding 1,3-propanediol dehydrogenase. FEMS Microbiology Letters, 154, 337–345.
Ma, Z., Jacobsen, F. E., & Giedroc, D. P. (2009). Coordination chemistry of bacterial metal transport and sensing. Chemistry Review, 109, 4644–4681.
Macis, L., Daniel, R., & Gottschalk, G. (1998). Properties and sequence of the coenzyme B12-dependent glycerol dehydratase of Clostridium pasteurianum. FEMS Microbiology Letters, 164, 21–28.
Maxson, P. (2005). Dynamics of mercury pollution on regional and global scales. In N. Pirrone & K. R. Mahaffey (Eds.), Atmospheric processes and human exposures around the world (pp. 25–50). New York: Springer.
Menzel, K., Zeng, A. P., & Deckwer, W. D. (1997). Enzymatic evidence for an involvement of pyruvate dehydrogenase in the anaerobic glycerol metabolism of Klebsiella pneumoniae. Journal of Biotechnology, 56, 135–142.
Mortazavi, S., Rezaee, A., Khavanin, A., Varmazyar, S., & Jafarzadeh, M. (2005). Removal of mercuric chloride by a mercury resistant Pseudomonas putida strain. Journal of Biological Sciences, 5, 269–273.
Mu, Y., Teng, H., Zhang, D. J., Wang, W., & Xiu, Z. L. (2006). Microbial production of 1, 3-propanediol by Klebsiella pneumoniae using crude glycerol from biodiesel preparations. Biotechnological Letters, 28, 1755–1759.
Murarka, A., Dharmadi, Y., Yazdani, S. S., & Gonzalex, R. (2008). Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Applied and Environmental Microbiology, 74, 1124–1135.
Naja, G. M., & Volesky, B. (2010). Treatment of metal-bearing effluents: removal and recovery. In L. K. Wang, J. P. Chen, Y. T. Hung, & N. K. Shammas (Eds.), Handbook on heavy metals in the environment (pp. 247–291). Boca Raton: CRC Press.
Nakamura, K., Iwahara, M., & Furukama, K. (2001). Screening of organomercurial-volatilizing bacteria in the mercury-polluted sediments and seawater of Minamata Bay in Japan. Clean Product and Process, 3, 104–107.
Nakas, J. P., Schaedle, M., Parkinson, C. M., Coonley, C. E., & Tanenbaum, S. W. (1983). System development of linked-fermentation production of solvents from algal biomass. Applied and Environmental Microbiology, 46, 1017–1023.
Narita, M., Chiba, K., Nishizawa, H., et al. (2003). Diversity of mercury resistance determinants among Bacillus strains isolated from sediment of Minamata Bay. FEMS Microbiology Letters, 223, 73–82.
Németh, A., Kupcsulik, B., & Sevella, B. (2003). 1,3-Propanediol oxidoreductase production with Klebsiella pneumoniae DSM2026. World Journal of Microbiology and Biotechnology, 19, 659–663.
Ní Chadhain, S. M., Schaefer, J. K., Crane, S., Zylstra, G. J., & Barkay, T. (2006). Analysis of mercuric reductase (merA) gene diversity in an anaerobic mercury-contaminated sediment enrichment. Environmental Microbiology, 8, 1746–1752.
Nies, D. H. (1999). Microbial heavy-metal resistance. Applied Microbiology and Biotechnology, 51, 730–750.
Nikel, P. I., Ramirez, M. C., Pettinari, M. J., Méndez, B. S., & Galvagno, M. A. (2010). Ethanol synthesis from glycerol by Escherichia coli redox mutants expressing adhE from Leuconostoc mesenteroides. Journal of Applied Microbiology, 109, 492–504.
Oregaard, G., & Sørensen, S. J. (2007). High diversity of bacterial mercuric reductase genes from surface and sub-surface floodplain soil (Oak Ridge, USA). The ISME Journal, 1, 453–467.
Osborn, A. M., Bruce, K. D., Strike, P., & Ritchie, D. A. (1997). Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiology Review, 19, 239–262.
Pepi, M., et al. (2010). Mercury-resistant bacterial strains Pseudomonas and Psychrobacter spp. isolated from sediments of Orbetello Lagoon (Italy) and their possible use in bioremediation processes. International Biodeterioration & Biodegradation, 65, 85–91.
Pirrone, N., Cinnirella, S., Feng, X., et al. (2010). Global mercury emissions to the atmosphere from natural and anthropogenic sources. Atmos. Chemical Physics, 10, 5951–5964.
Ramamoorthy, S., & Kushner, D. J. (1975). Binding of mercury and other heavy metal ions by microbial growth media. Microbiology Ecology, 2, 162–176.
Rodriguez, M. A. A., Gozzi, A., Abreu Filho, B. A., Zanin, G. M., & Moraes, F. F. (2012). Potential of Klebsiella oxytoca for 1,3-propanediol production from glycerol under excess substrate conditions. African Journal of Biotechnology, 11, 12675–12681.
Rossi, D. M., Costa, J. B., Souza, E. A., Peralba, M. C. R., & Ayub, M. A. Z. (2012). Bioconversion of residual glycerol from biodiesel synthesis into 1,3-propanediol and ethanol by isolated bacteria from environmental consortia. Renewable Energy, 39, 223–227.
Sadhukhan, S., Ghosh, J., Chaudhuri, D. K., & Ghosh, A. (1997). Mandal mercury and organomercurial resistance in bacteria isolated from freshwater fish of wetland fisheries around Calcutta. Environmental Pollution, 97, 71–78.
Sajjaphan, K., Shapir, N., Wackett, L. P., Palmer, M., Blackmon, B., Tomkins, J., & Sadowsky, J. (2004). Arthrobacter aurescens TC1 atrazine catabolism genes trzN, atzB, and atzC are linked on a 160-kilobase region and are functional in Escherichia coli. Applied and Environmental Microbiology, 70, 4402–4407.
Sambrook, J., & Russel, D. W. (2001). Molecular cloning: a laboratory manual (3rd ed.). New York: Cold Spring Arbor.
Sarma, S. J., Brar, S. K., Sydney, E. B., Bihan, Y. L., Buelna, G., & Soccol, C. R. (2012). Microbial hydrogen production by bioconversion of crude glycerol: a review. International Journal of Hydrogen Energy, 37, 6473–6490.
Seifert, C., Bowien, S., Gottschalk, G., & Daniel, R. (2001). Identification and expression of the genes and purification and characterization of the genes products involved in reactivation of coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. European Journal of Biochemistry, 268, 2369–2378.
Silva, G. P., Mack, M., & Contiero, J. (2009). Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnology Advances, 27, 30–39.
Silver, S., & Hobman, J. L. (2007). Mercury microbiology: resistance systems, environmental aspects, methylation, and human health. Microbiology Monographs, 6, 357–370.
Silver, S., & Phung, L. T. (1996). Bacterial heavy metal resistance: new surprises. Annual Review of Microbiology, 50, 753–789.
Silver, S., & Walderhaug, M. (1992). Gene regulation of plasmid- and chromosomal determined inorganic ion transport in bacteria. Microbiological Reviews, 56, 195–228.
Sone, Y., Mochizuki, Y., Koizawa, K., Nakamura, R., Pan-Hou, H., Itoh, T., & Kiyono, M. (2013). Mercurial-resistance determinants in Pseudomonas strain K-62 plasmid pMR68. AMB Express, 3, 41.
Summers, A. O. (1992). Untwist and shout: a heavy metal responsive transcriptional regulator. Journal of Bacteriology, 174, 3097–3101.
Tao, J., Wang, X., Shen, Y., & Wei, D. (2005). Strategy for the improvement of prodigiosin production by a Serratia marcescens mutant through fed-batch fermentation. World Journal of Microbiology and Biotechnology, 21, 969–972.
Thompson, J. C., & He, B. B. (2006). Characterization of crude glycerol from biodiesel production from multiple feedstocks. Applied Engineering in Agriculture, 22, 261–265.
Tong, I. T., Liao, H. H., & Cameron, D. C. (1991). 1,3-propanediol production by Escherichia coli expressing genes from the Klebsiella pneumoniae dha regulon. Applied and Environmental Microbiology, 57, 3541–3546.
Torà, J. A., Baeza, J. A., Carrera, J., & Oleszkiewicz, J. A. (2011). Denitritation of a high-strength nitrite wastewater in a sequencing batch reactor using different organic carbon sources. Chemical Engineering Journal, 172, 994–998.
UNEP. http://www.chem.unep.ch/mercury/report/GMA-report-TOC.htm. Accessed 23 Sep 2013.
Vetriani, C., et al. (2005). Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Applied and Environmental Microbiology, 71, 220–226.
Wagner-Döbler, I. (2003). Pilot plant for bioremediation of mercury-containing industrial wastewater. Applied Microbiology and Biotechnology, 62, 124–133.
Waldron, K. J., & Robinson, N. J. (2009). How do bacterial cells ensure that metalloproteins get the correct metal? Nature Reviews. Microbiology, 7, 25–35.
Yazdani, S. S., & Gonzales, R. (2007). Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Current Opinion in Biotechnology, 18, 213–219.
Yuan, Q., Sparling, R., Lagasse, P., Lee, Y. M., Taniguchi, D., & Oleszkiewicz, J. A. (2010). Enhancing biological phosphorus removal with glycerol. Water Science and Technology, 61, 1837–1843.
Zeng, A. P., & Sabra, W. (2011). Microbial production of diols as platform chemicals: recent progress. Current Opinion in Biotechnology, 22, 749–757.
Zeroual, Y., Moutaouakkil, A., & Blaghen, M. (2001). Volatilization of mercury by immobilized bacteria (Klebsiella pneumoniae) in different support by using fluidized bed bioreactor. Current Microbiology, 43, 322–327.
Zhang, R., Wang, Y., & Gu, J. D. (2006). Identification of environmental plasmid-bearing Vibrio species isolated from polluted and pristine marine reserves of Hong Kong and resistance to antibiotics and mercury. Antonie van Leeuwenhoek, 89, 307–315.
Zhang, W., Chen, L., & Liu, D. (2012). Characterization of a marine-isolated mercury resistant Pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Applied Microbiology and Biotechnology, 93, 1305–1314.
Zhu, M. M., Lawman, P. D., & Cameron, D. C. (2002). Improving 1.3-propanediol production from glycerol in a metabolically engineered Escherichia coli by reducing accumulation of sn-glycerol-3-phosphate. Biotechnology Progress, 18, 694–699.
Acknowledgments
The authors wish to thank the Brazilian Bureau for Science and Technology (CNPq) and CAPES for the financial support for this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Giovanella, P., Costa, A.P., Schäffer, N. et al. Detoxification of Mercury by Bacteria Using Crude Glycerol from Biodiesel as a Carbon Source. Water Air Soil Pollut 226, 224 (2015). https://doi.org/10.1007/s11270-015-2480-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-015-2480-9