Abstract
NADPH-dependent thioredoxin reductase (NTR), which is a flavin protein family of disulfide oxidoreductases present in all living cells, reduces thioredoxin (Trx). The roles of NTR B under stress conditions have not been well characterized. In this study, a tomato NTR B (SlNTRB) gene was cloned and functionally characterized. The mRNA transcript level of SlNTRB was induced significantly after nitrate treatment. The seed germination rate of SlNTRB overexpressed transgenic tobacco was higher than wild type (WT) under nitrate stress. The growth of SlNTRB overexpressed plants was better than WT and the reactive oxygen species (ROS) accumulation were lower than WT under nitrate stress. The NTR activity, superoxide dismutase (SOD) activity, the GSH/GSSG ratio, and the expression of Trx were significantly higher than WT after nitrate stress treatment. The SlNTRB overexpressed transgenic tobacco seeds showed higher tolerance to H2O2 and methyl viologen (MV) treatments than WT. The above results indicated that the SlNTRB gene improved the plant nitrate stress tolerance by alleviating the oxidative damage.
Key message
Overexpression of tomato SlNTRB gene in tobacco enhances the excess nitrate stress tolerance with less oxidative damage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen (N), which is a component of biological macromolecules such as protein and DNA, is an important element for the normal growth and maintenance of plant development (Ortigosa et al. 2019). N participates in various important physiological or metabolic processes in plants, such as photosynthesis, photorespiration, respiration, amino acid synthesis, tricarboxylic acid (TCA) cycle, and chlorophyll synthesis (Makino 2011). In order to get better growth and higher returns, growers apply too much N fertilizer to the crops they grow. In fact, the absorption rate of N fertilizer in fruits and vegetables is very low (Qu et al. 2020). Less than 50% of applied N fertilizer is used by plants and the rest is either leached out in soil or exhausted as agricultural runoffs (Qiao et al. 2017). Excessive N fertilizer inputs lead to a waste of fertilizer, which is reflected by a decline in nitrogen use efficiency and secondary salinization (Zhang et al. 2022). NO3− and Ca2+ are the main anion and cation of soil secondary salinization during greenhouse cultivation (Wang et al. 2016). Ca(NO3)2 alone significantly induced changes in the components of cell wall, anatomical structure, and expression profiles of several lignin biosynthetic genes (An et al. 2018). The growth of cucumber seedlings was significantly inhibited after the treatment of Ca(NO3)2 stress (Xing et al. 2015).
Plants produce excessive reactive oxygen species (ROS) under stress, resulting in the destruction of biological macromolecules such as proteins, nucleic acids and lipid membranes in the body, increasing the leakage of solute membranes (Cerqueira et al. 2023). Maintenance of ROS is an important event for whole lifespan of plants, however, in special cases, toxic ROS molecules are largely accumulated under excess stresses and diverse enzymes played as ROS scavengers (Cha et al. 2015). Superoxide dismutase (SOD), as the first line of defense in the system, can convert O2.− into hydrogen peroxide and oxygen, effectively eliminating the damage caused by superoxide anions. After that, peroxidase (POD) (Jacquot et al. 2009) and catalase (Abenavoli et al. 2016) continue to break down the products of SOD metabolism, and these antioxidant enzymes form a tight defense line to eliminate the harm caused by ROS (Wang et al. 2023). The thioredoxin (Trx) system has a vital function in cellular antioxidative defense via eliminating the excessive generation of ROS. It can be reduced by ferridoxin dependent Trx reductase (FTR) or niacinamide adenine dinucleotide phosphate (NADPH) dependent Trx reductase (NTR) (Jacquot et al. 2009). NTR is a flavin protein family of disulfide oxidoreductase present in all living cells (Dietz 2007). Each NTR subunit has two cofactor binding domains, one FAD-binding domain and one NADPH binding domain (Hagglund et al. 2016). NTR uses NADPH to specifically reduce Trx-S2 to Trx-(SH)2, the form of which is a powerful protein disulfide reductase (Serrato et al. 2004).
Plants contain three NADPH-thioredoxin reductases (NTR), NTRA located in the cytosol, NTRB located in the mitochondria and the NTRC in plastid, with important metabolic functions (Souza et al. 2023a). NTRC is a kind of NTR unique to photosynthetic autotrophs. It mainly exists in chlorophylls. NTRC is a single peptide bifunctional enzyme composed of N-terminal NTR domain and C-terminal Trx domain, and has high affinity to NADPH (Najera et al. 2017). The lack of NTRC affects the expression of proteins involved in stress and REDOX reactions (Ojeda et al. 2017). While NTRA is found in the cytoplasm, NTRB is mainly found in mitochondria and interferes with developmental processes by regulating auxin signaling (Cejudo et al. 2012). The combination of NTRA and NTRB with glutathione in Arabidopsis alters auxin transport and metabolism (Bashandy et al. 2010). In plants, NTRB is the main mitochondrial thioredoxin reductase, capable of reducing thioredoxin in the cytoplasm (Reichheld et al. 2005). The mitochondrial NTRB reduction system is an important metabolic factor in the TCA cycle, and the contents of various metabolic substances in the TCA cycle will be significantly changed after mutation of NTRB gene. At the same time, the NTR system lacking NTRB also has an impact on amino acid metabolism, inhibiting glycine/serine metabolism (Porto et al. 2022).In the previous experiment, transcriptome analysis was performed on tomatoes treated with nitrates. Compared with the control group, the expression of NTRB under nitrate treatment was higher than that of NTRC. In addition, no significant increase or decrease of NTRA expression was found (Supplemental Table S2).
Tomato (Solanum lycopersicum) is one of the most consumed fruit and vegetable crops in the world. In our research of RNA-seq of tomato seedling under nitrate stress, we found SlNTRB was significantly induced after nitrate stress treatment in tomato root. To further investigate the potential roles of SlNTRB, we amplified the SlNTRB gene from tomato and analyzed the function of SlNTRB in transgenic tobacco plants under excess nitrate stress.
Materials and methods
Plant materials and stress treatment
Tomato seedlings were grown hydroponically in plastic tank containing aerated modified nutrient solution (Zhai et al. 2022). After 4 weeks, excess nitrate provided by KNO3 and Ca(NO3)2 were added into the nutrient solution to reach a final NO3− ion concentration of 100 mM. The normal NO3− ion concentration of 9 mM was used as a control. After excess nitrate treatment for 24 h, the roots were harvest and put at − 80 °C for later analysis.
Gene cloning and phylogenetic analysis of SlNTRB
Tomato SlNTRB gene (GenBank No. XM_004232546. 4) was cloned by PCR with the primer of ATGACCGGCAACTGTT and ATCACTCTTTCCCACTT. To further understand the relationship between SlNTRB and other plant NTRB, we compared the derived SlNTRB amino acid sequence with other protein sequences and build phylogenetic trees using MEGA X software.
Plasmid construction and identification of overexpressed SlNTRB tobacco
The cDNA sequence of the target gene SlNTRB was cloned with the primer, F: TACGAACGATACTCGACCCCATGACCGGCAACTGTT and R: CTAGAGTCGA CGGATCCCCCATCACTCTTTCCCACTT. The SlNTRB gene was amplified using DNA polymerase (Vazyme, Nanjing, China). The plant expression vector of pRI101-SlNTRB-GFP was constructed using ClonExpress II one-step cloning kit (Vazyme, Nanjing, China). The constructed plant expression vector was transformed into Agrobacterium tumefaciens (LB4404) and transgenic tobacco was obtained by leaf disk method (Horsch et al. 1985).
To verify that the SlNTRB gene had been successfully integrated into tobacco, the genomic DNA of tobacco was extracted using the CTAB method. The PCR was done with the plasmid of pRI101-SlNTRB-GFP vector used as positive control and wild-type (WT) genomic DNA as negative control. Then the transgenic plants were identified by qRT-PCR and western blot analysis.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from tobacco using TRIzol reagent (Dalian Takara, China) and detected by gel electrophoresis. SYBR PrimeScript™ RT-PCR Kit II (Takara, Dalian, China) was used for reverse transcription of RNA into cDNA. Real-time quantitative PCR was performed using HieffTM qPCR SYBR® Green Master Mix kit (Shanghai Yishen Biotechnology Co., LTD.) and BioRad CFX 96™ instrument of real-time quantitative system. NtActin was used as an internal reference gene in tobacco. Tomato Actin was used as inner control for qRT-PCR analysis in tomato. The relative expression of specific genes was quantified using the 2−∆∆Ct method. Three replicates were set up in each group to detect the expression of SlNTRB gene and related gene. These primer sequences are listed in Supplemental Table S1.
Western blot assay
Western blot analysis was performed according to Leverone et al. (1991). The proteins were first transferred to PVDF membrane by SDS-PAGE gel electrophoresis and then transferred to PVDF membrane by micro-trans-blotting electrophoresis. After blocking with skim milk, the primary antibody is used with antibodies containing the target gene. The second antibody is horseradish peroxidase labeled anti-mouse IGG. After washing, ECL developer was used for color development (Beijing Conway Century Biotechnology Co., LTD.), and chemiluminescence imaging in dark room.
Seeds germination and seedlings growth analysis under excess nitrate stress
Transgenic tobacco seeds were placed in a 1.5 mL centrifuge tube and soaked in sterile water for 3 h, sterilized with 60% sodium hypochlorite solution for 3 min, and washed with sterile water for 30 min. Spot seeds evenly on MS medium containing 0 mM or 150 mM nitrate. The seeds were cultured in a tissue culture room with a constant temperature of 25 °C and photocycle of 16 h/8 h. The germination data were recorded daily.
In order to determine the tolerance of tobacco seedlings to nitrate stress, transgenic tobacco and WT tobacco of four weeks in peat and perlite (1:1) matrix were used for stress treatment. The experimental group was treated with 150 mM nitrate (50 mL) every day, while the control group was treated with water. The experiment was treated for three weeks, and then the samples were taken.
Seeds germination analysis of SlNTRB overexpressed transgenic tobacco under oxidative treatment
In order to observe the tolerance of SlNTRB overexpressed tobacco to oxidative stress, the overexpressed tobacco seeds were treated with H2O2 and methyl viologen (MV). After the tobacco seeds were disinfected as described above, they were uniformly located in normal MS and MS medium containing 60 μM MV and 20 mM H2O2. The germination data were recorded every day.
Fluorescence analysis of endogenous ROS and NO
ROS in root tips was observed using 2, 7-dichlorodihydrofluorescein diacetate (H2DCF-DA). The washed root tips were placed in a dye containing 2 μM H2DCF-DA, dyed for 30 min, and then washed three times with 20 mM HEPES–KOH (pH 7.8) buffer for 45 min, and photographed with a Inverted fluorescence microscope of Leica (DMI 3000B model) (excited at 488 nm; Emission 525 nm) (Mazel et al. 2004).
The root tips were rinsed with clean water and placed in an EP tube containing 5 μM DAF-FMDA dye and left in darkness for 30 min. The root tips were removed and placed in 20 mM HEPES–KOH (pH 7.7) buffer. The root tips were repeatedly washed 3 times for 15 min each time. The cleaned samples were placed under an inverted fluorescence microscope to observe the content of NO in the root tips and photographed (excitation 495 nm; Emission 515 nm) (Zhao et al. 2009).
The fluorescence images taken were analyzed by Image j software (https://imagej.en.softonic.com/).
Determination of NTR enzyme activities
The NTR activity was determined according to Ye et al. (1997). The 0.5 g sample was ground into powder in a mortar with liquid nitrogen, and 3 mL 25 mM PBS buffer (1 M DTNB, 0.2 mM EDTA, 1% PVP) was added. After grinding, it was thoroughly mixed in a centrifuge tube and centrifuged at 4 °C and 12,000 rpm for 15 min. After centrifugation, 1 mL of supernatant was taken into a water bath at 55 °C for 10 min, then left on ice for 5 min and centrifuged at 4 °C and 12,000 rpm for 10 min. Take 1 mL of reaction solution with 25 mM PBS buffer (0.2 mM EDTA, 1 M DTNB), 50 µL reduced Coenzyme II (NADPH), 100 µL bovine Serum protein (BSA), add 200 µL sample, and determine the absorption value at OD412 with BSA as the control.
Analysis of H2O2 content
Xylenol orange method was used to determine the H2O2 content, and its light absorption value at OD560 was determined according to Gay and Gebicki (2003).
Determination of antioxidant activities and antioxidant substance contents
The activity of superoxide dismutase (SOD) was determined by Wang et al. (2021). After adding the reaction solution, the determination was performed at 560 nm. The determination of GSH and GSSG was referred to the method of Jiang and Zhang (2001).
Phylogenetic analysis of NTRB
The NTRB protein sequences of different species were downloaded from the National Center for Biotechnology Information (NCBI) database. Phylogenetic analysis of SlNTRB with nsNTRB from other plants. The deduced amino acid sequences were aligned multiple times to analyze their same conserved structures using Jalview software. In addition, the phylogenetic trees of NTRBs in different plants were constructed using the NJ method in MEGA 11 software. The accession number of the database is shown in parentheses after the plant name.
Data processing and analysis
Graphpad Prism 7.0 (GraphPad Software, La Jolla California USA, www.graphpad.com) was used for data statistics and processing. Student's t-test was used to determine the significance of the difference (* denotes P < 0.05; ** indicates P < 0.01; **** indicates P < 0.0001). The experimental data were composed of the mean ± standard deviation (SD) values of three repeated experiments.
Results
Bioinformatics analysis of SlNTRB
The identified full-length cDNA of SlNTRB was 1104 bp, which encodes a 368-amino-acid protein with a molecular mass of 40 kDa. The SlNTRB conservative sequence belongs to Trx reductive superfamily (Fig. 1A). Conservative domains are included TRX (thioredoxin-disulfide reductase) and Pyr (Pyridine nucleotide-disulphide oxidoreductase).
A phylogenetic tree was constructed using SlNTRB proteins with other proteins of tobacco, Arabidopsis, potato and pepper, Zea maize, cotton, rice (Fig. 1B). The result showed that SlNTRB was closely related to NTRB of potato.
The expression of SlNTRB in tomato seedlings under nitrate stress
The tomato seedlings were treated with 100 mM nitrate for 24 h, and the gene expression of SlNTRB was then measured by qRT-PCR. Under nitrate stress, the expression of SlNTRB was up-regulated by 4.9-fold of the control (Fig. 2). These results indicated that SlNTRB gene was involved in the regulation of nitrate stress in tomato.
Effects of excess nitrate stress on the expression of SlNTRB in tomato seedlings. The expression of SlNTRB in tomato roots was analyzed by qRT-PCR. Tomato plants were treated with 100 mM nitrate (KNO3, 50 mM and Ca(NO3)2, 25 mM) for 24 h. The normal NO3− ion concentration of 9 mM was used as a control (CK). The results represented mean ± standard deviation (SD) of three biological replicates. Data was analyzed with Student's t-test compared with CK under similar conditions and indicated by *P < 0.1; **P < 0.05
Identification of SlNTRB overexpressed tobacco
To further investigate the function of SlNTRB under nitrate stress, overexpression transgenic plants were obtained. Three SlNTRB overexpressed tobacco lines were selected for the identification by PCR and Western blot analysis. The genomic PCR showed three specific bands, all of which were consistent with the size of the plasmid PCR bands (Fig. 3A). Western blot assay showed that NTRB protein expression was significantly increased in OE-1, OE-2, and OE-3 in the transgenic plants compared with WT (Fig. 3B). The above results proved that the transgenic tobacco of these three lines were SlNTRB overexpressed tobacco.
Identification and molecular characteristics of SlNTRB overexpressed transgenic tobacco. A Genomic PCR analysis of SlNTRB in transgenic plants with specific primers. M: marker; P: positive control of the plasmid; WT: wild type; OE-1, 2, 3: different transgenic lines. B Western blot analysis of the SlNTRB transgenic plants. All the results represented mean ± standard deviation (SD) of three biological replicates. Data was analysed with Student's t-test compared with control (CK) under similar conditions and indicated by *P < 0.1; **P < 0.05
Effects of nitrate stress on germination of SlNTRB overexpressed tobacco seeds
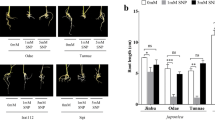
In order to verify the nitrate resistance of SlNTRB overexpressed tobacco seeds, tobacco germination test was conducted. As shown in the Fig. 4, there is no significant difference of the germination rate of WT and overexpression of SlNTRB transgenic tobacco seeds in MS medium (Fig. 4A). The germination rate of WT seeds in nitrate medium was 12.0% on the 8th day. The germination rates of SlNTRB overexpressed tobacco seeds of OE-1, OE-2, OE-3 were 92.0%, 84.0% and 80.0%, respectively on the 8th day after nitrate treatment (Fig. 4B).
Effects of nitrate stress on the growth, oxidative damage and NO accumulation in SlNTRB overexpressed tobacco seedlings
In the control, there is no difference of the growth size of overexpression tobacco and WT seedlings. However, after nitrate treatment, it was observed that the growth of SlNTRB overexpressed tobacco was better than that of WT plants (Fig. 5A). The H2O2 contents were then analyzed. As shown in Fig. 5B, there is no difference of the H2O2 contents between WT and overexpression plants. After nitrate treatment, the H2O2 contents were significantly lower than WT. The fluorescence intensity of ROS in lateral roots of WT plants was more than that of SlNTRB overexpressed tobacco, indicated that there is less oxidative damage to overexpression transgenic plants than WT plants (Fig. 5C, E). In the NO detection experiment, we found that after nitrate stress treatment, the fluorescence intensity of NO in the lateral roots of the three overexpressed tobacco was higher than that of WT tobacco (Fig. 5D, F). These results suggested that overexpression of SlNTRB increased the nitrate stress tolerance of tobacco by alleviating the oxidative damage.
Effects of excess nitrate stress on the phenotype (A), H2O2 contents (B), ROS accumulation (C, E) and NO accumulation (D;F) of SlNTRB overexpressed tobacco and WT seedlings. SlNTRB transgenic tobacco and WT plants were treated with 0 or 150 mM nitrate solution for 3 weeks. All the results represented mean ± standard deviation (SD) of three biological replicates. Data was analysed with Student's t-test compared with control (CK) under similar conditions and indicated by *P < 0.1; **P < 0.05; **** indicates P < 0.0001
Effect of nitrate stress on NTR and SOD activities, GSH/GSSG ratio and NtTrx expression in SlNTRB overexpressed tobacco
The NTR activity in SlNTRB overexpressed tobacco was significantly higher than that in WT tobacco under normal treatment. After nitrate stress, NTR activity in WT and overexpression transgenic plants were all increased, and NTR activities in OE-1, OE-2, and OE-3 were 137.2%, 111.5% and 118.1% of WT plants (Fig. 6A). In the control group, there was no significant difference in SOD activity between WT and overexpressed tobacco. After nitrate stress, SOD activity of overexpressed tobacco increased significantly (Fig. 6B). There was no significant difference of the GSH/GSSG ratio under normal treatment. After nitrate stress treatment, the ratio in overexpression tobacco was significantly increased, compared with WT (Fig. 6C). As shown in Fig. 6D, under normal treatment, the expression level of NtTrx genes in overexpressed tobacco had little difference with those in WT plants. However, under nitrate treatment, the expression of NtTrx genes in OE-1, OE-2 and OE-3 was 2.3-, 2.2- and 2.8-fold of WT.
Effects of excess nitrate stress on the enzyme activities of NTR and SOD, ratio of GSH/GSSG and expression of NtTrx of SlNTRB overexpressed tobacco and WT seedlings. SlNTRB transgenic tobacco and WT were treated with 0 or 150 mM nitrate solution for 3 weeks, the data was analyzed. All the results represented mean ± standard deviation (SD) of three biological replicates. Data was analysed with Student's t-test compared with control (CK) under similar conditions and indicated by *P < 0.1; **P < 0.05
Analysis of oxidative stress tolerance of SlNTRB overexpressed tobacco
In order to test the tolerance of SlNTRB overexpressed tobacco to oxidative stress, the overexpressed tobacco seeds were treated with H2O2 and MV. As shown in Fig. 7A, D, the germination rate of overexpression and WT tobacco in MS was basically similar. In the germination test treated with H2O2, the germination rate of WT tobacco was 32.2%, while the germination rate of SlNTRB overexpressed tobacco was 64.3%, 68.2%, and 68.2%, respectively (Fig. 7B, E). After MV treatment, the germination rate of WT was only 20.4%, while the germination rate of overexpressed tobacco was 40.4%, 36.3%, and 48.6%, respectively (Fig. 7C, F). These results showed that SlNTRB overexpressed tobacco enhanced the tolerance to oxidative damage.
Germination rate of SlNTRB overexpressed tobacco under H2O2 and MV stress conditions. A, B, C: Phenotypes of tobacco seeds grown on MS medium containing 0 (A), 20 mM H2O2 (B) or 60 μM of MV (C) for 8 days. D, E, F: Germination rate of tobacco seeds grown on MS medium containing 0 (D), 20 mM H2O2 (E) or 60 μM of MV (F) for 8 days
Discussion
Nitrate content is an essential indicator of the quality of vegetables but can cause stress at high levels (Abd-Elrahman et al. 2022). Redox regulation in heterotrophic organisms relies on NADPH, thioredoxins (Trxs), and an NADPH-dependent Trx reductase (NTR) (Gonzalez et al. 2019). Tx reductases control the redox state of Trx ubiquitous proteins that regulate a spectrum of enzymes by dithiol-disulfide exchange reactions (Buey et al. 2021). Arabidopsis and rice contain 3 NTRs which transfer reducing power to Thioredoxin/Peroxiredoxin system for scavenging ROS (Cha et al. 2015). In this study, a tomato NTR was isolated which belong to Trx reductive superfamily. Overexpression of Trx CDSP32 gene can alleviate oxidative damage in tobacco leaves under Cd exposure by regulating antioxidant defense system (Zhang et al. 2023). Ca and EGTA stimulated chickpea thioredoxin and NTR activities when compared to Cd-stressed (Sakouhi et al. 2021). In our study, tomato SlNTRB was significantly induced after nitrate treatment.
Plants contain three NTR located in the cytosol/mitochondria (NTRA/B) and the plastid (NTRC) with important metabolic functions (Souza et al. 2023b). Of the NTRA expressed by adjusting the quantity of ROS oxidation and drought tolerance in arabidopsis thaliana (Cha et al. 2014). Due to this functional redundancy between NTRA and NTRB, their physiological functions under environmental stresses are not clearly classified and remain elusive to date. Both ntrc and ntrabc mutants showed reduced growth and substantial metabolic alterations as compared to the wild type (Souza et al. 2023b). The ntra ntrb plants were characterized as producing wrinkled seeds, exhibiting slow rates of plant growth and accumulating high levels of anthocyanins (Reichheld et al. 2007; Bashandy et al. 2009), which could be expected to increase tolerance to abiotic stresses such as drought (Kovinich et al. 2014). It has been reported that ntra ntrb double mutant plant exhibits UV-C tolerance due to high accumulation of anthocyanin (Bashandy et al. 2009). In our study, the germination rate and seedling growth of SlNTRB transgenic tobacco were better than WT, suggesting that overexpression of SlNTRB enhanced the nitrate stress tolerance of tobacco.
Under oxidative stress, toxic ROS are largely accumulated in plant cells lead to cell damage and cause damage of DNA, RNA, protein and lipid (Rasheed et al. 2022). Thioredoxins (Trx) implicated redox reactions, conferred stress tolerance by enhancing the ROS scavenging ability of plants (Considine and Foyer 2021). To date, function of three NTRs in Arabidopsis was association with plant protection against oxidative stress (Lepisto et al. 2013; Cejudo et al. 2021). Under nitrate stress, the ROS fluorescence accumulation increased, while the contents in the overexpression SlNTRB plants were lower than WT plants. Besides, in our study, the SlNRTB overexpression plants showed high survival rates than WT under MV and H2O2 stress, which are well known to induce large accumulations of ROS and cause oxidative damage in plant cells. These results suggested that that SlNTRB overexpression improved the tolerance to the oxidative stress. The activity of NTR and expression of Trx were increased under nitrate stress in SlNTRB overexpression transgenic plants, suggesting that NTRB regulated cellular ROS levels via activated Trx systems in plant cells and protects the plants against stress.
Data availability
The data presented in this study are available on request from the corresponding author.
References
Abd-Elrahman SH, Saudy HS, Abd El-Fattah DA, Hashem FAE (2022) Effect of irrigation water and organic fertilizer on reducing nitrate accumulation and boosting lettuce productivity. J Soil Sci Plant Nutr 22(2):2144–2155. https://doi.org/10.1007/s42729-022-00799-8
Abenavoli MR, Longo C, Lupini A, Miller AJ, Araniti F, Mercati F, Princi MP, Sunseri F (2016) Phenotyping two tomato genotypes with different nitrogen use efficiency. Plant Physiol Biochem 107:21–32. https://doi.org/10.1016/j.plaphy.2016.04.021
An YH, Zhou H, Yuan YH, Li L, Sun J, Shu S, Guo SR (2018) 24-Epibrassinolide-induced alterations in the root cell walls of Cucumis sativus L. under Ca(NO(3))(2) stress. Protoplasma 255(3):841–850. https://doi.org/10.1007/s00709-017-1187-8
Bashandy T, Taconnat L, Renou JP, Meyer Y, Reichheld JP (2009) Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Mol Plant 2(2):249–258. https://doi.org/10.1093/mp/ssn065
Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22(2):376–391. https://doi.org/10.1105/tpc.109.071225
Buey RM, Fernandez-Justel D, Gonzalez-Holgado G, Martinez-Julvez M, Gonzalez-Lopez A, Velazquez-Campoy A, Medina M, Buchanan BB, Balsera M (2021) Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria. Plant Physiol 186(1):285–296. https://doi.org/10.1093/plphys/kiab072
Cejudo FJ, Ferrandez J, Cano B, Puerto-Galan L, Guinea M (2012) The function of the NADPH thioredoxin reductase C-2-Cys peroxiredoxin system in plastid redox regulation and signalling. FEBS Lett 586(18):2974–2980. https://doi.org/10.1016/j.febslet.2012.07.003
Cejudo FJ, González MC, Pérez-Ruiz JM (2021) Redox regulation of chloroplast metabolism. Plant Physiol 186(1):9–21. https://doi.org/10.1093/plphys/kiaa062
Cerqueira JVA, de Andrade MT, Rafael DD, Zhu F, Martins SVC, Nunes-Nesi A, Benedito V, Fernie AR, Zsögön A (2023) Anthocyanins and reactive oxygen species: a team of rivals regulating plant development? Plant Mol Biol 112(4–5):213–223. https://doi.org/10.1007/s11103-023-01362-4
Cha JY, Kim JY, Jung IJ, Kim MR, Melencion A, Alam SS, Yun DJ, Lee SY, Kim MG, Kim WY (2014) NADPH-dependent thioredoxin reductase A (NTRA) confers elevated tolerance to oxidative stress and drought. Plant Physiol Biochem 80:184–191. https://doi.org/10.1016/j.plaphy.2014.04.008
Cha JY, Barman DN, Kim MG, Kim WY (2015) Stress defense mechanisms of NADPH-dependent thioredoxin reductases (NTRs) in plants. Plant Signal Behav 10(5):e1017698. https://doi.org/10.1080/15592324.2015.1017698
Considine MJ, Foyer CH (2021) Oxygen and reactive oxygen species-dependent regulation of plant growth and development. Plant Physiol 186(1):79–92. https://doi.org/10.1093/plphys/kiaa077
Dietz KJ (2007) The dual function of plant peroxiredoxins in antioxidant defence and redox signaling. Subcell Biochem 44:267–294. https://doi.org/10.1007/978-1-4020-6051-9_13
Gay CA, Gebicki JM (2003) Measurement of protein and lipid hydroperoxides in biological systems by the ferric-xylenol orange method. Anal Biochem 315(1):29–35. https://doi.org/10.1016/s0003-2697(02)00606-1
Gonzalez M, Delgado-Requerey V, Ferrandez J, Serna A, Cejudo FJ (2019) Insights into the function of NADPH thioredoxin reductase C (NTRC) based on identification of NTRC-interacting proteins in vivo. J Exp Bot 70(20):5787–5798. https://doi.org/10.1093/jxb/erz326
Hagglund P, Finnie C, Yano H, Shahpiri A, Buchanan BB, Henriksen A, Svensson B (2016) Seed thioredoxin h. Biochim Biophys Acta 1864(8):974–982. https://doi.org/10.1016/j.bbapap.2016.02.014
Jacquot JP, Eklund H, Rouhier N, Schurmann P (2009) Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci 14(6):336–343. https://doi.org/10.1016/j.tplants.2009.03.005
Jiang M, Zhang J (2001) Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol 42(11):1265–1273. https://doi.org/10.1093/pcp/pce162
Kovinich N, Kayanja G, Chanoca A, Riedl K, Otegui MS, Grotewold E (2014) Not all anthocyanins are born equal: distinct patterns induced by stress in Arabidopsis. Planta 240(5):931–940. https://doi.org/10.1007/s00425-014-2079-1
Lepisto A, Pakula E, Toivola J, Krieger-Liszkay A, Vignols F, Rintamaki E (2013) Deletion of chloroplast NADPH-dependent thioredoxin reductase results in inability to regulate starch synthesis and causes stunted growth under short-day photoperiods. J Exp Bot 64(12):3843–3854. https://doi.org/10.1093/jxb/ert216
Leverone LA, Stroup TL, Caruso JL (1991) Western blot analysis of cereal grain prolamins using an antibody to carboxyl-linked indoleacetic acid. Plant Physiol 96(4):1076–1078. https://doi.org/10.1104/pp.96.4.1076
Makino A (2011) Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol 155(1):125–129. https://doi.org/10.1104/pp.110.165076
Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134(1):118–128. https://doi.org/10.1104/pp.103.025379
Najera VA, Gonzalez MC, Perez-Ruiz JM, Cejudo FJ (2017) An event of alternative splicing affects the expression of the NTRC gene, encoding NADPH-thioredoxin reductase C, in seed plants. Plant Sci 258:21–28. https://doi.org/10.1016/j.plantsci.2017.02.001
Ojeda V, Pérez-Ruiz JM, González M, Nájera VA, Sahrawy M, Serrato AJ, Geigenberger P, Cejudo FJ (2017) NADPH thioredoxin reductase C and thioredoxins act concertedly in seedling development. Plant Physiol 174(3):1436–1448. https://doi.org/10.1104/pp.17.00481
Ortigosa F, Valderrama-Martin JM, Avila C, Canovas FM, Canas RA (2019) Understanding plant nitrogen nutrition through a laboratory experiment. Biochem Mol Biol Educ 47(4):450–458. https://doi.org/10.1002/bmb.21239
Porto NP, Bret RSC, Souza PVL, Candido-Sobrinho SA, Medeiros DB, Fernie AR, Daloso DM (2022) Thioredoxins regulate the metabolic fluxes throughout the tricarboxylic acid cycle and associated pathways in a light-independent manner. Plant Physiol Biochem 193:36–49. https://doi.org/10.1016/j.plaphy.2022.10.022
Qiao P, Zhao F, Liu M, Gao D, Zhang H, Yan Y (2017) Hydrogen sulfide inhibits mitochondrial fission in neuroblastoma N2a cells through the Drp1/ERK1/2 signaling pathway. Mol Med Rep 16(1):971–977. https://doi.org/10.3892/mmr.2017.6627
Qu ZM, Qi XC, Shi RG, Zhao YJ, Hu ZP, Chen Q, Li CL (2020) Reduced N fertilizer application with optimal blend of controlled-release urea and urea improves tomato yield and quality in greenhouse production system. J Soil Sci Plant Nut 20(4):1741–1750. https://doi.org/10.1007/s42729-020-00244-8
Rasheed R, Ashraf MA, Ahmad SJN, Parveen N, Hussain I, Bashir R (2022) Taurine regulates ROS metabolism, osmotic adjustment, and nutrient uptake to lessen the effects of alkaline stress on L. plants. S Afr J Bot 148:482–498. https://doi.org/10.1016/j.sajb.2022.05.023
Horsch RB, Fry JE, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227(4691):1229–1231. https://doi.org/10.1126/science.227.4691.1229
Reichheld JP, Meyer E, Khafif M, Bonnard G, Meyer Y (2005) AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett 579(2):337–342. https://doi.org/10.1016/j.febslet.2004.11.094
Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, Meyer Y (2007) Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19(6):1851–1865. https://doi.org/10.1105/tpc.107.050849
Sakouhi L, Kharbech O, Ben Massoud M, Gharsallah C, Ben Hassine S, Munemasa S, Murata Y, Chaoui A (2021) Calcium and ethylene glycol tetraacetic acid mitigate toxicity and alteration of gene expression associated with cadmium stress in chickpea (Cicer arietinum L.) shoots. Protoplasma 258(4):849–861. https://doi.org/10.1007/s00709-020-01605-x
Serrato AJ, Perez-Ruiz JM, Spinola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279(42):43821–43827. https://doi.org/10.1074/jbc.M404696200
Souza PVL, Hou LY, Sun H, Poeker L, Lehman M, Bahadar H, Domingues AP, Dard A, Bariat L, Reichheld JP, Silveira JAG, Fernie AR, Timm S, Geigenberger P, Daloso DM (2023a) Plant NADPH-dependent thioredoxin reductases are crucial for the metabolism of sink leaves and plant acclimation to elevated CO. Plant Cell Environ 46(8):2337–2357. https://doi.org/10.1111/pce.14631
Souza PVL, Hou LY, Sun H, Poeker L, Lehman M, Bahadar H, Domingues AP, Dard A, Bariat L, Reichheld JP, Silveira JAG, Fernie AR, Timm S, Geigenberger P, Daloso DM (2023b) Plant NADPH-dependent thioredoxin reductases are crucial for the metabolism of sink leaves and plant acclimation to elevated CO2. Plant Cell Environ 46(8):2337–2357. https://doi.org/10.1111/pce.14631
Wang Y, Xu L, Tang M, Jiang H, Chen W, Zhang W, Wang R, Liu L (2016) Functional and integrative analysis of the proteomic profile of radish root under Pb exposure. Front Plant Sci 7:1871. https://doi.org/10.3389/fpls.2016.01871
Wang XY, Jiang JX, Wu ZH, Li XM, Dong SK (2021) Relationship between soil moisture content and superoxide dismutase activity in soybean leaves in northeast China. J Biobased Mater Bioenergy 15(6):799–804. https://doi.org/10.1166/jbmb.2021.2148
Wang G, Zhang L, Zhang S, Li B, Li J, Wang X, Zhang J, Guan C, Ji J (2023) The combined use of a plant growth promoting Bacillus sp. strain and GABA promotes the growth of rice under salt stress by regulating antioxidant enzyme system, enhancing photosynthesis and improving soil enzyme activities. Microbiol Res 266:127225. https://doi.org/10.1016/j.micres.2022.127225
Xing WW, Li L, Gao P, Li H, Shao QS, Shu S, Sun J, Guo SR (2015) Effects of grafting with pumpkin rootstock on carbohydrate metabolism in cucumber seedlings under Ca(NO3)2 stress. Plant Physiol Biochem 87:124–132. https://doi.org/10.1016/j.plaphy.2014.12.011
Ye B, Gitler C, Gressel J (1997) A high-sensitivity, single-gel, polyacrylamide gel electrophoresis method for the quantitative determination of glutathione reductases. Anal Biochem 246(2):159–165. https://doi.org/10.1006/abio.1996.9985
Zhai J, Qi Q, Wang M, Yan J, Li K, Xu H (2022) Overexpression of tomato thioredoxin h (SlTrxh) enhances excess nitrate stress tolerance in transgenic tobacco interacting with SlPrx protein. Plant Sci 315:111137. https://doi.org/10.1016/j.plantsci.2021.111137
Zhang YJ, Yu SG, Li ZJ, Chang TT, Xu QC, Xu HL, Zhang J (2022) Effects of excessive nitrogen fertilizer and soil moisture deficiency on antioxidant enzyme system and osmotic adjustment in tomato seedlings. Int J Agr Biol Eng 15(2):127–134. https://doi.org/10.25165/j.ijabe.20221502.5555
Zhang HB, Yao TT, Wang Y, Wang JC, Song JQ, Cui CC, Ji GX, Cao JN, Muhammad S, Ao H, Zhang HH (2023) CDSP32-overexpressing tobacco plants improves cadmium tolerance by modulating antioxidant mechanism. Plant Physiol Biochem 194:524–532. https://doi.org/10.1016/j.plaphy.2022.11.036
Zhao MG, Chen L, Zhang LL, Zhang WH (2009) Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol 151(2):755–767. https://doi.org/10.1104/pp.109.140996
Acknowledgements
This research was funded by the National Natural Science Foundation of China (Grant No. 32260753) and the Yunnan Ten Thousand Talents Plan: Young & Elite Talents Project.
Author information
Authors and Affiliations
Contributions
HNX designed the project. YTN and STQ conducted the experiments and analyzed the data and wrote the manuscript. KZL helped in the writing of the article. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Joyce Van Eck.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nie, Y., Qiao, S., Li, K. et al. Tomato SlNTRB improved excess nitrate stress tolerance by alleviating the oxidative damage in transgenic tobacco. Plant Cell Tiss Organ Cult 156, 94 (2024). https://doi.org/10.1007/s11240-023-02661-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11240-023-02661-w