Abstract
This study monitored the transcriptional response of OsDHODH1 under nitrosative stress conditions relative to the transcripts accumulations for the core mitochondrial cytochrome c oxidase1 (CcOX1) subunit, nuclear CcOX subunits 5b and 5c, two rice nitrate reductases (OsNIA1 and OsNIA2), and nitric oxide excess 1 (OsNOE1) genes. Our findings reveal that short-term exposure of rice seedlings to 1 mM SNP (Nitric oxide donor) applied exogenously for 1 h resulted in significant down-regulation of OsDHODH1 expression in all rice cultivars. In addition, the transcriptional patterns for the CcOX subunits, which are known to have a high affinity for nitric oxide, showed that the core catalytic subunit (OsCcOX1) and the nuclear subunit (OsCcOX5b) were up-regulated, while the nuclear subunit (OsCcOX5c) gene expression was suppressed. OsGSNOR1 expression was enhanced or decreased concomitant with a decrease or increase in SNO accumulation, particularly at the basal level. Moreover, high OsNIA1 expression was consistent with impaired root development, whereas low transcript accumulation matched a balanced root-growth pattern. This suggests that OsNIA1 expression would prevail over OsNIA2 expression under nitrosative stress response in rice. The level of malondialdehyde (MDA) content increased with the increase in SNP concentration, translating enhanced oxidative damage to the cell. We also observed increased catalase activity in response to 5 mM SNP suggesting that potential cross-talk exist between nitrosative and oxidative stress. These results collectively suggest a possible role of OsDHODH1 and OsCcOX5b role in plant root growth during nitrosative stress responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitric oxide (NO), a versatile bioactive gaseous molecule, has the potential to diffuse through biological membranes owing to its physicochemical properties (Khan et al. 2014). NO plays an important role in diverse physiological processes, such as plant metabolism, defense systems, enzyme activity, stomatal closure, photosynthesis, expression of cell cycle genes, as well as other major metabolic processes (del Rio et al. 2014). As documented in several studies, many pathways for NO synthesis have been suggested in plants, operating either through oxidation or reduction of certain biochemical products (Gupta et al. 2011). NO is also known for its role in stimulating plant growth (Kopyra 2004), promoting auxin-mediated activation of cell division (Ötvös et al. 2005), regulating plant root growth, etc. The promotion or inhibition of root growth depends on NO concentrations (Sun et al. 2017). In addition, excessive NO accumulation has been shown to induce stressful conditions in plants (Oz et al. 2015). NO signaling may involve a large and very interactive web, with Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS), functioning through crosstalk in a competitive way with one another, that may leading to a beneficial outcome for plant cells (Wang et al. 2013a). Since NO has been recognized as an important modulator of biological processes (Khan et al. 2014), increasing attention has been paid to the mechanism underlying the synthesis of NO in plants. Many studies have investigated the plants mechanisms of NO synthesis similar to those seen in animals.
Because of their sessile nature, plants are usually exposed to various stresses, which include oxidative stress caused by either abiotic or biotic stimuli (Allen 1995). During the stress period, Reactive Oxygen Species (ROS) accumulate to the extent of being detrimental to plant fitness (Borsani et al. 2001). To respond to the stressful environmental conditions, plants activate antioxidant systems, which include Catalase (CAT) activity among others. CAT helps maintain a balanced reduction–oxidation state and decrease oxidative stress caused by H2O2 generation (Foyer and Noctor 2003). The enzymatic and non-enzymatic systems in plants are assumed indispensable to maintain the ROS accumulation under control in adverse conditions. Moreover, these ROS scavenging systems do not act alone but have an interactive regulation mechanism alongside ROS–antioxidants. From this perspective, NO and its derived molecules are regarded as being part of the complex signaling network defense mechanisms against changed environmental conditions (Chaki et al. 2015).
The oxidative stress-induced ROS accumulation has been shown to result in increased lipid peroxidation and cell membrane degradation. Malondialdehyde (MDA), one of the organic compounds that accumulate in the cell experiencing excess ROS production, is widely used to investigate the oxidative damage to the cell membrane in plants (Hasanuzzaman et al. 2012; Parankusam et al. 2017).
Plant mitochondria are the major sites for NO production (Vanlerberghe 2013) through the action of cytochrome c oxidase (CcOX) under hypoxic conditions via nitrite reduction. Cytochrome c oxidase (CcOX) is also known as the terminal component (complex IV) of the mitochondrial electron transport chain (ETC) (Miki 2002). The CcOX contains the closely associated hemea3 (Fe 3+a3 ) and copper (Cu 2+B ) that are referred to as the binuclear center where NO and oxygen (O2) competitively bind (Shiva 2010). The binding activity of NO to the binuclear site present in CcOX1 is well described (Sarti et al. 2012). Once bound, NO has the ability to modulate the activity or function of the hemeproteins, affording signal transduction (Reedy et al. 2008).
The mitochondrial electron transport chain (ETC) is known as the major site for the production of Reactive Oxygen Species (ROS) (Møller 2001). Inhibition of the ETC results in a significant decrease in NO accumulation and delay of cell death. The same phenomenon allows an increase in ROS levels. However, NO production deficiency under the same conditions avoids cell death by reducing peroxynitrite (ONOO−) formation, as reported previously (Igamberdiev et al. 2014). Besides its above-mentioned role, NO acts as a signaling molecule in both the biotic and abiotic stress signal transduction pathways in plants (Khan et al. 2014).
De novo pyrimidine biosynthesis occurs in plants via an oxidation–reduction reaction pathway, whereby dihydroorotate (DHO) is oxidized by dihydroorotate dehydrogenase (DHODH) at the fourth step in a Nicotinamide dinucleotide (Kim et al. 2017) dependent redox reaction to yield orotic acid (OA) (Zrenner et al. 2006). The DHODH links the respiratory chain functionally and physically, delivering electrons to ubiquinone through the ETC (Fang et al. 2013). The respiratory ETC couples electron transfer from organic substrates to molecular oxygen together with proton translocation across the inner mitochondrial membrane (Schertl and Braun 2014). Some enzymatic assays and related studies in mammalian cells have suggested that the animal DHODH is indirectly inhibited when respiration is disturbed by diverse respiratory chain inhibitors (Beuneu et al. 2000), such as cyanide (CN) and NO, as previously reported by Sarti et al. (2012).
In the course of their life cycle, plants may be subjected to various unfavorable situations, such as low availability of oxygen for their respiration (for instance, caused by drought stress), whereby plant tissues are required to cope with restricted aerobic metabolic activity (Oliveira and Salgado 2014).
The rooting system of plants has a wide range of functions for plant survival throughout its life cycle, including uptake of water and essential nutrient elements and serving as structural support for the plant. Many studies aiming to dissect the molecular and genetic mechanisms modulating the development of the rice rooting system hypothesized that the roots are critical for rice adaptation to a variety of abiotic stress conditions, thereby contributing to the understanding of crop establishment, growth and viability, and productivity (Verstraeten et al. 2014).
Sodium nitroprusside (SNP) is a NO donor commonly used to induce nitrosative stress in plants (Antoniou et al. 2013; Singh et al. 2009). During dissociation into NO, SNP simultaneously releases Nitrosonium ion (NO+) and cyanide (Lewis et al. 2017a), which in turn affects photosynthetic electron transport, among other processes (Wodala et al. 2010). In addition, cyanide (CN) acts as a potent inhibitor of nitrate reductase (NR) and CcOX, both of which are reported to be active in various in vivo NO production pathways. Furthermore, dissociation of NO from SNP in vivo was reported to be mediated by sulfhydryl groups of protein thiols (Grossi and D’Angelo 2005). NO generation through SNP occurs at a low but sustained rate (lasting several hours), though it also depends on the intensity and quality of light, to some extent. Another important NO donor is S-nitrosoglutathione (GSNO) that is more stable and acts both as NO donor and NO reservoir (Corpas et al. 2013; Feechan et al. 2005).
This study explored a number of undescribed pathways to investigate the transcriptional regulation of the plant OsDHODH1 gene under nitrosative stress induced by SNP (as NO donor). Here, we discussed about the regulatory network of the OsDHODH1 gene under nitrosative stress by investigating a few of the possible interactions with high potential for regulating the target gene transcripts levels in six rice (subspecies indica and japonica) cultivars, in relation to NR encoded genes expression and cytochrome c oxidase core mitochondrial membrane and nuclear subunits. Three indica cultivars (Lioto, Irat112 and Sipi) selected for their wide cultivation in different agro-ecological zones in the Democratic Republic of Congo, with Irat112 used for its known resistance to water deficit and agronomic performances (Aboukhadrah et al. 2015). Three japonica cultivars (Jinbu, Odae) previously reported for cold tolerance (Kim et al. 2007), and Tunnae were included as genetic materials to conduct the experiments.
We monitored gene expression from samples harvested 1 h after exposure to SNP and GSNO (the bioactive form of NO donor present in living cells) while phenotypic data were recorded 10 days after nitrosative stress was imposed. The results improved our understanding of the factors involved in the modulation of OsDHODH1 gene expression in rice associated with drought tolerance. This study also investigated the dose-dependent effects of two widely used NO donors on plant growth, as well as catalase enzyme activity and the level of lipid peroxidation by malondialdehyde (MDA) content.
Methods
Experimental design and plants materials
A completely randomized design was used as the experimental design for the current study with at least three replications. Seedlings were grown in the growth chamber under controlled conditions. Three indica cultivars (Lioto, Irat112 and Sipi) selected for their wide cultivation in different agro-ecological zones in the Democratic Republic of Congo. Irat112 was used for its known tolerance to water deficit and its agronomic performances (Aboukhadrah et al. 2015). This study included three japonica cultivars (Jinbu, Odae) previously reported for cold tolerance (Kim et al. 2007), and Tunnae as genetic materials to conduct the experiments.
Rice seeds were sterilized, in 25% (v/v) Prochloraz solution (62.5 µl/125 ml distilled water), for 24 h at 37 °C in an incubator, followed by rinsing three times for 3 h with distilled water. Sterilized rice seeds were then germinated and grown, in Hoagland solution at 25 °C and a 16/8 h light/dark cycle, for up to 14 days.
Nitrosative stress was induced by either GSNO, known as the bioactive form of NO donor present in living cells (Broniowska et al. 2013) or SNP to 14-day old rice seedlings. These two NO donors were used based on their wide use in a number of studies (Antoniou et al. 2013; Begara-Morales et al. 2014; Imran et al. 2018; Seabra and Oliveira 2016). SNP during dissociation into NO simultaneously releases cyanide (Lewis et al. 2017b), which particularly drawn our attention in this study for it is also reported to interact with the cytochrome c oxidase (Complex IV of the mitochondrial electron transport chain). Moreover, NO donation by SNP is assumed to be continuous and long-lasting as compared to other NO donors (Zandonadi et al. 2010). Plants were exposed to NO donors through irrigation methods as described earlier (Mun et al. 2018). Different concentrations of both NO donors were used to identify the most effective. Two NO donors were applied in multiple doses (Antoniou et al. 2013). Root samples were collected at one-time point for all NO sources and concentrations.
Preparation and optimization of NO donors
The GSNO solutions, with two concentration gradients 1 mM and 5 mM, were prepared using Sodium nitrite (NaNO2, SIGMA-ALDRICH Lot No MKBH2411 V S-2252, Japan) and Glutathione (Sigma Lot No 1734C061). Similarly, SNP [(Na2(Fe(CN)5NO)2H2O) Sigma cat# S-3421)], another NO donor, was also used with two concentration gradients, of 1 mM and 5 mM. Initially, two concentrations of both NO donors were screened to identify the one that gives significant effects in different rice cultivars. For this, different sets of rice seedlings grown in the same conditions were exposed to either 1 mM and 5 mM GSNO or 1 mM and 5 mM SNP (Antoniou et al. 2013) through irrigation method as described earlier by Mun et al. (2018), in which we used CySNO (another NO donor) gradient concentration. The phenotype of the six rice genotypes was observed 10 days after their exposure to nitrosative stress. Because of the necrotic phenotype observed in all rice cultivars particularly in response to 5 mM SNP (Fig.S2), we opted to use 1 mM concentration for both GSNO and SNP as the NO donors for further sub-screening. Since both 1 mM GSNO and 1 mM SNP induced similar phenotypic responses, we selected 1 mM SNP as NO donor to use in downstream experiments. For gene expression study, rice seedlings were exposed to 1 mM SNP and samples were collected 1 h post-stress and the transcript accumulation for different genes was analyzed.
Total RNA isolation, cDNA synthesis, and qPCR analysis
Total RNA was isolated from approximatively 100 mg of roots from control and stressed plants using the TRI-Solution™ Reagent (Cat. No.: TS200-001, Virginia Tech Bio-Technology, Lot: 337871401001) as described by the manufacturer company. Thereafter, the complementary DNA (cDNA) was synthesized as described earlier by Imran et al. (2018). Briefly, RNA with good integrity was used to synthesize cDNA using BioFACT™ RT-Kit (BioFACT™ Republic of Korea). qRT-PCR was performed as described earlier (Sharma et al. 2016). Briefly, qRT-PCR was performed for selected genes (Table S1) using Real-time PCR master mix including SYBR green (BioFact Korea) along with 100 ng of template DNA and 10 nM of each forward and reverse primers in a final volume of 20 µL. A no template control was used as control. For transcript accumulation, a 2-step reaction including initial denaturation at 95 °C for 15 min, followed by denaturation at 95 °C for 5 s, then annealing and extension at 65 °C for 30 s, was performed in a Real-time PCR machine (Eco™ Illumina). The data from 40 total reaction cycles were normalized with the relative expression of rice Actin1.
Catalase activity assay
The spectrophotometric method was used for CAT enzyme activity assay as previously described (Adamu et al. 2018). About 100 mg of roots samples were ground to fine pounder in liquid nitrogen and immediately homogenized in phosphate buffer (pH 7.5). The homogenate was kept on ice for about 10 min followed by centrifugation at 12,000 rpm for 10 min at 4 °C. The supernatant was then transferred to fresh Eppendorf tubes and used as crude enzyme source for CAT activity. 50 µL of H2O2 was added to the 50 µL crude extract, and the absorbance of the reaction was read at 240 nm wavelength. The catalase activity was expressed in Unit per milligram of the sample (Aebi 1984).
Lipid peroxidation assay
Lipid peroxidation level was studied as the malondialdehyde (MDA) content, determined by the thiobarbituric acid (TBA) also called thiobarbituric acid-reactive substances (TBARS) reaction (Jambunathan 2010). Approximatively 200 mg of control and treated samples were homogenized in 4 mL of 0.1% Trichloro acetic acid (TCA) with a ceramic mortar and pestle. The homogenate was centrifuged at 10,000 rpm for 15 min. The supernatant (1 mL) was collected and 2 mL of 20% TCA containing 0.5% TBA was added. The mixture was then incubated at 95 °C in a water bath for 30 min and immediately cooled on ice. The mixture was centrifuged at 10,000 rpm for 10 min. The absorbance of the supernatant was read at 532 nm and 600 nm wavelength (A600 as the non-specific absorbance, which is subtracted from the A532 reading). MDA content was calculated using the MDA extinction coefficient of 155 mM−1 cm−1.
SNO level measurement
Cellular SNO levels were quantified using methods described by Imran et al. (2018). Briefly, the leaf samples from control seedlings (0 mM SNP) and SNP treated seedlings were ground in KPI buffer (pH 5.5) and centrifuged at 13,000 rpm for 10 min, the supernatant was measured in a NO analyzer NOA280i (Sievers, USA) according to the manufacturer’s instructions. Protein content was measured using a Bradford assay (Kruger 2002) and SNO contents were calculated.
Statistical analysis
Data from samples collected in triplicate from all experiments underwent statistical analysis using GraphPad Prism software (Version 7.00, 1992-2016 GraphPad). The analysis of variance (ANOVA) for Completely Randomized Design was performed and the LSD calculated where needed at a significance level of 0.05.
Results
Screening of different NO donors for optimal response
Prior to challenging the rice cultivars with nitrosative stress, we screened two different NO donors for identification of the NO source with optimal distinctive responses in various genotypes. Rice seedlings (approximately 14-days old) were exposed to SNP and GSNO, each at concentrations of 1 mM and 5 mM. Based on the phenotypic response of the six rice cultivars (Fig. S1 and Fig. S6), we selected 1 mM SNP for subsequent experiments, as widely explained in the subsection “Preparation and optimization of NO donors” of the “Methods” section.
NO is essential for controlling the growth of rice seedlings under SNP-induced nitrosative stress in a varietal-dependent manner
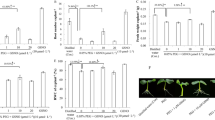
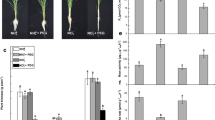
By visual observation, we observed changes in the phenotype of six different rice cultivars after their exposure to SNP as a NO donor at two different concentrations for 10 days. In the subsequent phenotypic analysis, significant results were obtained when seedlings were exposed to 1 mM SNP as compared to the control (0 mM). The general trend in the response of six rice cultivars shows that 1 mM SNP exerts a negative impact on rice seedlings growth. A priori, we could observe that 1 mM SNP inhibited shoot growth in Tunnae, Lioto, Irat112, while Jinbu and Odae showed increased shoot growth. However, the Sipi cultivar exhibited slightly reduced shoot growth, but not statistically significant. In contrast, 5 mM SNP did not result in a significant change of shoot growth (Fig.S1b) and root growth (Fig. 1) phenotypes, except in Irat112 (inhibited).
SNP-induced nitrosative stress modulates root development in rice. a The changes observed in root development of rice seedlings exposed to 1 mM and 5 mM SNP for 10 days. b The phenotypic response to nitrosative stress is shown to be genotype-dependent. White bars represent the seedlings grown solely in Hoagland nutrient solution; the grey bars indicate rice seedlings exposed to 1 mM SNP and black bars show seedlings under 5 mM SNP. LSD test revealed a highly significant inhibitory effect of 1 mM SNP compared to 5 mM. Error bars represent ± SD of the means. *P < 0.05, **P < 0.01, ***P < 0.001 and ns non-significant
We also observed a necrotic phenotype, particularly between 5-10 days after 5 mM SNP was applied (Fig. 1 and Fig. S6), making this an inappropriate SNP concentration for assessing the nitrosative response of rice seedlings. It is important to note that when SNP is applied at concentrations greater than 1 mM it may induce both nitrosative stress and oxidative damage to rice plant tissues.
Exogenous application of NO modulates root growth in rice seedlings
We also measured root growth after NO treatment in all the cultivars and found that rice cultivars Odae, Tunnae, Lioto, and Irat112 showed root inhibition under 1 mM SNP, while Jinbu and Sipi exhibited a balanced root growth (Fig.S1c and S1d). The inhibition of root growth ranged between 18.6% (lowest) to 66.2% (highest) under GSNO and 18.6% (lowest) to 85.4% (highest) under SNP-mediated nitrosative stress (Fig.S1c and Fig.S1d). Out of six different genotypes, SNP had less effect on the root length of Sipi (the indica rice cultivar) compared to well-grown seedlings. In general, the Sipi phenotype did not change significantly after exposure to the nitrosative stress induced either by 1 mM GSNO or 1 mM SNP, therefore, translating strong plasticity against modified environmental conditions.
Of greater interest was an increase in total root weight, recorded as follows (Fig. 2): 177% (GSNO) and 327% (SNP) for Jinbu; 13% (GSNO) and 55% (SNP) in Irat112. However, root growth was inhibited in Odae and Tunnae, ranging from 37.1% to 66% under 1 mM GSNO and 76.5–85.4% under 1 mM SNP.
Root: Shoot ratio suggests enhanced biomass under SNP. a The pattern of shoot weight and root weight b measured on 14-day old rice seedlings from six cultivars 10 days after 1 mM SNP application. c The ratio between the root and shoot weight refers to the root system development in relation to the shoot growth pattern in response to SNP-induced nitrosative stress. White bars represent unstressed seedlings, while black bars show SNP-treated seedlings. All bars are mean values of three replications ± SD and means are compared between untreated and treated samples. Asterisks and ns marks on top of bars show the significance level. *P < 0.05, **P < 0.01, ***P < 0.001 and ns non-significant
Quantitative real-time RT-PCR Analysis
Nitric Oxide down-regulates OsDHODH1 while inducing NR genes and CcOX subunits
Our study first observed that rice seedlings exposed to 1 mM SNP as the NO source showed highly significant suppression of OsDHODH1 transcript abundance (Fig. 3a) in roots of different japonica and indica rice cultivars 1 h after their exposure to nitrosative stress. On the contrary, SNP concentrations above 1 mM resulted in tissue necrosis and plant death (Fig. S1d).
The transcriptional nitrosative response of OsDHODH1 in relation to CcOX and NR genes. a The transcriptional nitrosative response of OsDHODH1, b the core mitochondrial CcOX encoding subunit 1, c the nuclear subunit (OsCcOX5b), d the nuclear subunit (OsCcOX5c). e Nitrate reductase genes, OsNIA1, f OsNIA2 and g the rice catalase encoded by OsNOE1 in the roots of 14-day old rice seedlings exposed to 1 mM SNP for 1 h. The expression values were normalized to rice Actin1. Student’s t tests revealed highly significant effects of SNP on the relative gene expression. All means are compared with their corresponding controls. ns non-significant and *** P < 0.001
SNP induced expression of both NR genes, OsNIA1 (Fig. 3e) and OsNIA2 (Fig. 3f). However, the relative expression of OsNIA1 was significantly higher than the one of OsNIA2. The highest OsNIA1 expression level matched the highest SNO level (in Tunnae). In addition, OsNOE1 encoding the rice catalase OsCATC was similarly induced by SNP (Fig. 3g).
Furthermore, we were interested to know how the activity of the CcOX subunits could contribute to NO-mediated suppression of OsDHODH1 expression. We, therefore, performed qPCR to monitor the transcript levels of three important CcOX subunits: the mitochondrial-encoded CcOX1 and the nuclear-encoded CcOX5b and CcOX5c. Treatment with SNP significantly induced expression of the CcOX1 (core catalytic for enzyme activity) and CcOX5b (nuclear) subunits. However, the transcript of the nuclear-encoded CcOX5c was dramatically down-regulated as a result of NO source treatment.
OsGSNOR1 gene expression and intracellular SNO accumulation in the roots of rice seedlings
In this study, OsNIA1 and OsNIA2 are predicted to have a functional protein–protein interaction with OsDHODH1 (Fig. S2). The rice cultivar Tunnae (japonica) exhibited the highest OsNIA1 transcript level while having the lowest OsNIA2 expression. This cultivar also had the most severe inhibition of root growth by SNP. In contrast, Lioto cultivar (indica) showed the lowest OsNIA1 expression pattern and the highest OsNIA2 transcript accumulation. However, Irat112 (indica) revealed an intermediate expression pattern of OsNIA1 and OsNIA2 when compared to other rice genotypes used in this study.
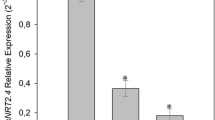
The specificity of the transcripts levels of OsNIA1 and OsNIA2 and the expression pattern of rice catalase encoded by OsNOE1 gene influenced our choice to measure the basal SNO levels relative to the expression of OsGSNOR1 gene in three rice genotypes. As presented in Fig. 4a, the accumulation pattern of basal SNO levels was concomitant with the decrease/increase in the basal OsGSNOR1 gene expression (Fig. 4b).
Intracellular SNO levels and OsGSNOR1 expression in roots 1 h after SNP application. a The increased basal SNO accumulation of level in three different rice genotypes (white bars) concomitant with the reduced pattern of OsGSNOR1 expression at basal level (white bar) b. The bars in both panels (a) and (b), respectively represent the SNO levels and OsGSNOR1 gene expression in the roots of seedlings exposed to 1 mM SNP for 1 h. White bars are means of non-treated samples and black bars represent SNO levels in treated samples ± SD. Asterisks indicate the significant level based on Student’s t test
An in silico analysis-based approach facilitated by computer simulations using the GPS-SNO algorithm by Ren et al. (2009) allowed us to predict that the Cysteine residue 196 (Cys-196) may be exposed with a high probability of being S-nitrosylated (Fig. S3, Fig. 5).
Proposed model of the functional interaction of NO with the CcOX subunits. The above figure proposes a model of nitric oxide (NO) interaction with the key core cytochrome c oxidase subunits (CcOX1) harboring the binuclear center, the active site where NO and cyanide (CN) are reported to competitively bind. Continuous lines show the recorded expression of target genes in the current study, while the discontinuous dotted lines indicate reported evidences by other research groups showing inhibition of the CcOX either by NO or CN by binding of NO or CN, not observed in the present study. The lower horizontal bar with different compartments identified by their colors represent the four complexes of the mitochondrial electron transport chain
Catalase activity and lipid peroxidation under nitrosative stress
To investigate the antioxidant systems activation and tackle the extent of lipid peroxidation in rice seedlings subjected to nitrosative stress, we measured the activity of CAT, and our results show that CAT activity was detected in Odae, Lioto Irat112 and Sipi cultivars soon after (1 h) 1 mM SNP was supplemented to the modified hydroponic culture medium. However, Jinbu and Tunnae showed reduced CAT activity (Fig. 6). In contrast, seedlings exposed to 5 mM SNP exhibited a highly significant increase in CAT activity in all cultivars (Fig. S4). To further confirm the possible oxidative damage we measured MDA contents and found that in SNP treated samples MDA contents were slightly increased (Fig. 7), but not significantly as compared to their corresponding controls. However, a highly significant increase in MDA content was recorded in all rice cultivars at early nitrosative stress induction (1 h) (Fig. S5). Both catalase activity and MDA content indicate the oxidative stress perception and lipid peroxidation (cell membrane degradation level) occurrence during the treatment period.
Induction of catalase enzyme activity induced under nitrosative stress. The above figure shows the catalase enzyme activity induced 1 h after SNP-induced nitrosative stress was imposed to rice seedlings. White and black bars are. Respectively, means from control and SNP-treated seedlings ± SD. Means from treated seedlings are compared to their corresponding controls and the statistical significance at P < 0.05 is indicated on top of each bar. ns non-significant and ***highly significance
Change in malondialdehyde (MDA) content 1 h after 1 mM SNP treatment. The extinct of lipid peroxidation was investigated by measuring the malondialdehyde (MDA) content at early time point of 1 mM SNP application. White and black bars are respectively means from control and SNP-treated seedlings ± SD. Means from trea66ted seedlings are compared to their corresponding controls and the statistical significance at P < 0.05 is indicated on top of each bar. ns non-significant
Discussion
Exogenous application of NO differentially affect root development in rice
NO is produced in nearly all organs of plants and plays a pivotal role in a number of physiological processes and molecular functions (Małolepsza 2007). As for the root, its implication is presumed in the coordination of growth for primary, secondary, and adventitious roots, which in turn are regulated by various factors at the genetic and environmental levels as reported by Corpas and Barroso (2015).
In the present study, 14-day old rice seedlings subjected to 10 days of direct exposure to NO donors resulted in significant inhibition of root growth in rice cultivars Jinbu, Odae, Tunnae, and Irat112, while Lioto and Sipi exhibited a balanced root growth pattern (Fig. 1). This response to nitrosative stress was shown to vary depending on the NO source concentration and the rice cultivar. According to Pagnussat et al. (2003), when NO and its derived molecules accumulate in the roots they contribute to root development. Moreover, the production of NO in plant roots under stressful conditions was shown to depend on a number of factors, such as the development stage of the plant, as well as the intensity and duration of exposure (Corpas and Barroso 2015).
Furthermore, NO was reported to favor proliferation and survival of the cell at relatively low concentrations (picomolar to nanomolar), while higher concentrations (micromolar to millimolar) favor cell cycle arrest, apoptosis, and senescence (Thomas et al. 2008). Despite its role as defined in the previous paragraphs, high levels of NO and NO-derived products accumulate under exogenous NO application causing nitrosative and oxidative damage (Valderrama et al. 2007).
Our results support that exogenous application of NO can act to control rice root development 10 days post-stress induction. The different inhibition patterns of root growth observed in different rice cultivars in this study are attributed to genotype-dependent NO inhibition. Therefore, the genetic background and diversity within rice cultivars could be one of the factors with significant influence over the response of genotypes to nitrosative stress. Other possible mechanisms are conceivable, including protein–protein interactions, as well as crosstalk between different signaling, biosynthetic, and metabolic pathways.
Nitric Oxide inhibits OsDHODH1 while inducing NR genes and differentially expressing CcOX subunits
Findings from the current study reveal that the expression of the gene encoding DHODH1 was significantly down-regulated in rice after treatment with SNP (Nitric Oxide donor). Prior to investigating the response of OsDHODH1 to nitrosative stress, we examined the transcripts levels of OsNIA1 and OsNIA2. Upon SNP treatment, expression of the two Nitrate reductase encoded genes were dramatically increased (Fig. 3e, f). This observation was confirmed by the increased SNO level (Fig. 4a) and the expression pattern of OsGSNOR1 (Fig. 4b), particularly at basal levels, confirming the effectiveness of nitrosative stress occurrence.
NO is reported to mimic hypoxia signaling. In accordance with Brown (2001), NO can antagonize hypoxia by inhibiting the mitochondrial CcOX, resulting in an increase in cellular O2 availability. Thus, high levels of O2 speed up the reaction with NO forming N2O3, which is a potent nitrosating agent (Nedospasov 2002).
Our results show that CcOX1 (Fig. 3b), the key catalytic core subunit of the cytochrome c oxidase embedded in the inner mitochondrial membrane and harboring twelve membrane-spanning domains, and the nuclear subunit CcOX5b (Fig. 3c) are both significantly up-regulated under nitrosative stress induced by SNP (Nitric Oxide donor). As stated by Mansilla et al. (2018), the inhibition of CcOX by NO is reversible. Unlike other ligands, NO binds to this binuclear center (hemea3-CuB) (Sarti et al. 2003), where it acts as an efficient reactant for the reduced heme–iron, thereby stabilizing the hemea3 Fe2+ while competing with O2, resulting in a reversible, transient inhibition of CcOX (Brunori et al. 1997).
Through conserved domain analysis from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov), this study has identified an interface on the subunit Vb/I in the conserved domain of CcOX5b, which would functionally interact with the core subunit CcOX1. Earlier, Poyton and McEwen (1996) reported that when the CcOX5b subunit was deleted, loss of cytochrome c oxidase activity was observed. From this perspective and owing to the transcriptional patterns of the two genes recorded in the current study, it is thought that the gene encoding CcOX5b could play an essential role in the activity of the core subunit 1. Additionally, this raises the possibility of signal transduction existing between the mitochondrial membrane and the nucleus.
Furthermore, Giegé et al. (2005) recorded a decreased expression of the genes encoding the CcOX5b and 5c subunits in rice seedlings exposed to hypoxic conditions. In our case, we report inhibition of the nuclear subunit CcOX5c (but not subunit 5b) by NO as a result of SNP supplementation (Fig. 3d). Besides, the findings of Curi et al. (2002) revealed that genes encoding the CcOX5c subunit were regulated by nitrate and oxygen availability in the Sunflower. Higher expression was recorded with increased nitrate concentrations, while a reduction in the transcript levels was observed under hypoxic conditions.
Therefore, it is believed that the activity of the nuclear CcOX5c subunit could play an important role in NO-mediated inhibition of OsDHODH1 gene expression in rice through the mitochondrial respiratory chain, partially explaining the suppression of OsDHODH1 expression with excessive NO accumulation upon SNP supplementation. From previous expression patterns, it is conceivable that CcOX5b could be necessary for the core CcOX1 subunit to be functionally active, which has a high affinity for NO.
It was interesting to observe that OsDHODH1 expression was significantly down-regulated upon application of SNP as the NO source to rice seedlings. In contrast, Liu et al. (2009) reported that OsDHODH1 was significantly enhanced by drought stress in rice, where its loss of function resulted in sensitivity to drought and its overexpression improved drought tolerance. Consequently, the authors suggested that OsDHODH1 could be a candidate gene playing a prevalent role in drought tolerance in rice. These results are similar to those recorded in our previous work (unpublished) showing OsDHODH1 is significantly up-regulated, in the same six cultivars used as genetic materials in the present study, upon drought stress induced by withholding water. Therefore, the nitrosative response pattern of OsDHODH1 recorded in the current study suggests that this gene could be involved in producing NO through the electron transport complexes (ETC) of the mitochondrial respiratory chain. Functional analysis or studies are needed to depict the role of OsDHODH1 in NO production in rice. This would include the use of knockouts and/or overexpressors lines either in rice or in the Arabidopsis dicot model plant where diverse mutant lines are available at the Arabidopsis Biological Resource Center (ABRC) or the Nottingham Arabidopsis Stock Center (NARSC). Moreover, up-regulation of OsNIA1 (6.0–8.1 Log2 FC, as shown in Fig. 3e) coupling with SNO levels concomitantly to the OsGSNOR1 gene expression pattern, would be explained by the suggestion made by Feechan et al. (2005) that GSNOR negatively regulates the process of protein S-nitrosylation, thus controlling endogenous NO levels.
The rice OsNOE1/ OsCATC gene expression is induced by SNP
OsNOE1 codes for rice catalase (OsCATC) (Wang et al. 2013b). Wendehenne (2016) reported that the rice neo1 (nitric oxide excess1) mutant accumulates high levels of NO in rice leaves. Emerging evidence indicates that loss of OsNOE1 function results in increased H2O2 levels in rice leaves (Lin et al. 2012). Catalase is known for its role in degrading H2O2, which would alleviate oxidative damage to cells. The CAT activity detected soon after SNP application is here perceived as an indication of the activation of the antioxidant (enzymatic) system in response to oxidative stress occurrence. On one hand, in 1 mM SNP treated seedlings, a slight increase of CAT activity was detected (Fig. 6). At the same SNP concentration, MDA content slightly increased but not significantly (Fig. 7). However, there was not a clear correlation between CAT activity and OsNOE1/OsCATC expression in 1 mM SNP treated seedlings, except, in Jinbu where CAT activity decreased. This decrease was in accordance with OsNOE1 transcriptional level in SNP treated seedlings. The same pattern was observed in 5 mM SNP treated Jinbu seedlings. On the other hand, enhanced CAT activity was recorded in several folds approximatively 1 h after 5 mM SNP treatment. The increase of CAT activity can be associated with the enhanced antioxidant system, as a defense mechanism activated in plants to cope with imbalance oxidation–reduction state caused by the increase in ROS production. The temporal assessment of cellular damage levels was performed by the determination of lipid peroxidation (MDA content), one of the markers associated in several studies with oxidative damage and the degradation of the cell membrane. Increasing concentration of SNP is shown to result in increasing cell membrane damage estimated by MDA content in each cultivar (Fig. S5). The phenotypic observation under 5 mM SNP favors the occurrence of cell membrane damage later recorded by the necrotic symptoms as presented in Fig. S6.
Under nitrosative stress, endogenous NO levels increase as a result of NR genes activation (NIA1 and NIA2) (Wendehenne 2016). A similar observation was made in the current study (Fig. 3e, f) when SNP was supplemented to the hydroponic solution to induce nitrosative stress to rice seedlings. Thus, the up-regulation of OsNOE1 (Fig. 3b) observed under a 1 h application of SNP is here perceived as a signal towards the maintenance of a balanced cellular redox status during the exposure period.
Putative interaction of OsDHODH1, NO, and the mitochondrial ETC
The enzyme encoded by OsDHODH1 gene is physically linked to the mitochondrial electron transport chain (ETC), which may cause its activity to also be functionally affected by the activity of ETC complexes. NO is reported to tightly interact with the respiratory chain at complex IV (CcOX) as mentioned in the introduction. The CcOX was previously reported as a key regulator of the overall mitochondrial respiratory chain/ETC activity. This implies that a significant reduction of CcOX activity would result in a decrease in ATP production (Pierron et al. 2012). The Eukaryotic CcOX contains about ten to fourteen subunits, representing proteins of unknown function. The CcOX is reported to be the electron acceptor from the reduced form of cytochrome c; and the CuB maintains a permanent equilibrium with hemea3, which allows the electron to be quickly transferred to the active binuclear center (Lamas 2005). The CcOX subunits CcOX1, the key catalytic core subunit located in the inner mitochondrial membrane, and the nuclear subunits CcOX5b and CcOX5c (Mansilla et al. 2018). CcOX1, CcOX2, and CcOX3 are the three core functional subunits of CcOX embedded in the inner mitochondrial membrane. However, in accordance with Pierron et al. (2012), some of the nuclear-encoded CcOX subunits play an active role in regulating the activity of the enzyme and are considered as central in the complex regulatory machinery underlying the activity of CcOX. The core subunit 1 functionally interacts with CcOX2, CcOX3, and the other nuclear subunits (including CcOX5b) through interface features in their respective conserved domains.
The transcript abundance of the three important CcOX subunits monitored through qPCR in this study revealed up-regulation of the OsCcOX1 and OsCcOX5b transcripts by NO exogenously applied (Fig. 3). In Fig. 5, this study proposed a model that illustrates how Nitric Oxide would interact with the OsDHODH1 and the three core CcOX subunits known to have a Copper bimetallic site whose role is as the electron-entry door of the enzyme. We used biological data generated in the current study by monitoring the transcriptional levels of the genes of interest by qPCR. To construct this model, we put to contribution in silico analysis using available public bioinformatics tools through conserved domain analysis (https://www.ncbi.nlm.nih.gov), GPS-SNO 1.0 software and available literature.
Conclusion
Emerging interests in Nitric Oxide-related studies have put this molecule at the forefront of research in plant biosciences. NO is known to mediate gene expression under multiple stress conditions. The output from years of NO-related experiments identified a series of NO-responsive genes in different plant species.
This study provides an insight into the transcriptional response of OsDHODH1 under nitrosative stress conditions to investigate the regulatory machinery underlying its expression against environmental cues. The phenotypic observations revealed that NO applied exogenously inhibits root growth in a genotype-dependent manner. Furthermore, the increase in SNP concentration (5 mM) resulted in increased lipid peroxidation level (high MDA content).
References
Aboukhadrah S, Abd Allah A, Gharib H, Sakran RM (2015) Effect of soil water deficit on yield and its components at the different growth stages in rice (Oryza sativa). Egypt J Agron 37:79–92
Adamu T, Mun B-G, Lee S-U, Hussain A, Yun B-W (2018) Exogenously applied nitric oxide enhances salt tolerance in rice (Oryza sativa L.) at seedling stage. Agronomy 8:276
Aebi H (1984) Catalase in vitro. Methods in enzymology, vol 105. Elsevier, Netherlands, pp 121–126
Allen RD (1995) Dissection of oxidative stress tolerance using transgenic plants. Plant Physiol 107:1049
Antoniou C, Filippou P, Mylona P, Fasoula D, Ioannides I, Polidoros A, Fotopoulos V (2013) Developmental stage-and concentration-specific sodium nitroprusside application results in nitrate reductase regulation and the modification of nitrate metabolism in leaves of Medicago truncatula plants. Plant signal Behav 8:e25479
Begara-Morales JC, Sánchez-Calvo B, Luque F, Leyva-Perez MO, Leterrier M, Corpas FJ, Barroso JB (2014) Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol 55:1080–1095
Beuneu C, Auger R, Löffler M, Guissani A, Lemaire G, Lepoivre M (2000) Indirect inhibition of mitochondrial dihydroorotate dehydrogenase activity by nitric oxide. Free Radical Biol Med 28:1206–1213
Borsani O, Dıaz P, Agius MF, Valpuesta V, Monza J (2001) Water stress generates an oxidative stress through the induction of a specific Cu/Zn superoxide dismutase in Lotus corniculatus leaves. Plant Sci 161:757–763
Broniowska KA, Diers AR, Hogg N (2013) S-nitrosoglutathione Biochimica et Biophysica Acta (BBA)-General Subjects 1830:3173-3181
Brown GC (2001) Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochimica Biophysica Acta (BBA) 1504:46–57 (Bioenergetics)
Brunori M, Giuffrè A, D’Itri E, Sarti P (1997) Internal electron transfer in cu-heme oxidases thermodynamic or kinetic control? J Biol Chem 272:19870–19874
Chaki M, Álvarez de Morales P, Ruiz C, Begara-Morales JC, Barroso JB, Corpas FJ, Palma JM (2015) Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Ann Bot 116:637–647
Corpas FJ, Barroso JB (2015) Functions of nitric oxide (NO) in roots during development and under adverse stress conditions. Plants 4:240–252
Corpas FJ, Alche JD, Barroso JB (2013) Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front Plant Sci. https://doi.org/10.3389/fpls.2013.00126
Curi GC, Chan RL, Gonzalez DH (2002) Genes encoding cytochrome c oxidase subunit 5c from sunflower (Helianthus annuus L.) are regulated by nitrate and oxygen availability. Plant Sci 163:897–905
del Rio LA, Corpas FJ, Barroso JB, López-Huertas E, Palma JM (2014) Function of peroxisomes as a cellular source of nitric oxide and other reactive nitrogen species. In: Nitric oxide in plants: metabolism and role in stress physiology. Springer, Switzerland, pp 33–55
Fang J et al (2013) Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci Rep 33:e00021
Feechan A, Kwon E, Yun B-W, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci 102:8054–8059
Foyer CH, Noctor G (2003) Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant 119:355–364
Giegé P, Sweetlove LJ, Cognat V, Leaver CJ (2005) Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell 17:1497–1512
Grossi L, D’Angelo S (2005) Sodium nitroprusside: mechanism of NO release mediated by sulfhydryl-containing molecules. J Med Chem 48:2622–2626
Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT (2011) On the origins of nitric oxide. Trends Plant Sci 16:160–168
Hasanuzzaman M, Nahar K, Alam MM, Fujita M (2012) Exogenous nitric oxide alleviates high temperature induced oxidative stress in wheat (‘Triticum aestivum’ L.) seedlings by modulating the antioxidant defense and glyoxalase system. Aust J Crop Sci 6:1314
Igamberdiev AU, Ratcliffe RG, Gupta KJ (2014) Plant mitochondria: source and target for nitric oxide. Mitochondrion 19:329–333
Imran QM et al (2018) Transcriptome wide identification and characterization of NO-responsive WRKY transcription factors in Arabidopsis thaliana L. Environ Exp Bot 148:128–143. https://doi.org/10.1016/j.envexpbot.2018.01.010
Jambunathan N (2010) Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Plant stress tolerance. Springer, Berlin, pp 291–297
Khan MN, Mobin M, Mohammad F, Corpas FJ (2014) Nitric oxide in plants: metabolism and role in stress physiology. Springer, Berlin
Kim S-H, Kim J-Y, Kim S-J, An K-S, An G, Kim S-R (2007) Isolation of cold stress-responsive genes in the reproductive organs, and characterization of the OsLti6b gene from rice (Oryza sativa L.). Plant Cell Rep 6:1097–1110
Kim YJ, Sukweenadhi J, Seok JW, Kang CH, Choi ES, Subramaniyam S, Yang DC (2017) Complete genome sequence of Paenibacillus yonginensis DCY84(T), a novel plant Symbiont that promotes growth via induced systemic resistance. Stand Genom Sci 15:5–9
Kopyra M (2004) The role of nitric oxide in plant growth regulation and responses to abiotic stresses. Acta Physiol Plant 26:459–473
Kruger N (2002) The Bradford method for protein quantitation. In: The protein protocols handbook, 2nd edn. Humana Press, Totowa, NJ, pp 15–21
Lamas S (2005) Nitric oxide, cell signaling, and gene expression. CRC Press, Boca Rato
Lewis RW, Unrine J, Bertsch PM, McNear DH (2017a) Silver engineered nanomaterials and ions elicit species-specific O-2 consumption responses in plant growth promoting rhizobacteria. Biointerphases. https://doi.org/10.1116/1.4995605
Lewis RW, Unrine J, Bertsch PM, McNear DH Jr (2017b) Silver engineered nanomaterials and ions elicit species-specific O2 consumption responses in plant growth promoting rhizobacteria. Biointerphases 12:05G604
Lin A et al (2012) Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol 158:451–464
Liu WY, Wang MM, Huang J, Tang HJ, Lan HX, Zhang HS (2009) The OsDHODH1 gene is involved in salt and drought tolerance in rice. J Integrat Plant Biol 51:825–833
Małolepsza U (2007) Nitric oxide production in plants. Postepy Biochemii 53:263–271
Mansilla N, Racca S, Gras DE, Gonzalez DH, Welchen E (2018) The complexity of mitochondrial complex iv: an update of cytochrome c oxidase biogenesis in plants. Int J Mol Sci 19:662
Miki T (2002) Mitochondrial complex IV (cytochrome c oxidase). Nihon Rinsho 60:149 (Japanese Journal of Clinical Medicine)
Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Biol 52:561–591
Mun B-G, Lee S-U, Hussain A, Kim H-H, Rolly NK, Jung K-H, Yun B-W (2018) S-nitrosocysteine-responsive genes modulate diverse regulatory pathways in Oryza sativa: a transcriptome profiling study. Funct Plant Biol 45:630–644
Nedospasov AA (2002) Is N2O3 the main nitrosating intermediate in aerated nitric oxide (NO) solutions in vivo? If so, where, when, and which one? J Biochem Mol Toxicol 16:109–120
Oliveira HC, Salgado I (2014) Role of plant mitochondria in nitric oxide homeostasis during oxygen deficiency. Nitric oxide in plants: metabolism and role in stress physiology. Springer, Berlin, pp 57–74
Ötvös K et al (2005) Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. Plant J 43:849–860
Oz MT, Eyidogan F, Yucel M, Öktem HA (2015) Functional role of nitric oxide under abiotic stress conditions. Nitric oxide action in abiotic stress responses in plants. Springer, Berlin, pp 21–41
Pagnussat GC, Lanteri ML, Lamattina L (2003) Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol 132:1241–1248
Parankusam S, Adimulam SS, Bhatnagar-Mathur P, Sharma KK (2017) Nitric oxide (NO) in plant heat stress tolerance: current knowledge and perspectives. Front Plant Sci 8:1582
Pierron D, Wildman DE, Hüttemann M, Markondapatnaikuni GC, Aras S, Grossman LI (2012) Cytochrome c oxidase: evolution of control via nuclear subunit addition. Biochimica Biophysica Acta (BBA) 1817:590–597 (Bioenergetics)
Poyton RO, McEwen JE (1996) Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem 65:563–607
Reedy CJ, Elvekrog MM, Gibney BR (2008) Development of a heme protein structure–electrochemical function database. Nucleic Acids Res 36:D307–D313
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19:271
Sarti P, Giuffrè A, Barone MC, Forte E, Mastronicola D, Brunori M (2003) Nitric oxide and cytochrome oxidase: reaction mechanisms from the enzyme to the cell. Free Radical Biol Med 34:509–520
Sarti P, Forte E, Mastronicola D, Giuffrè A, Arese M (2012) Cytochrome c oxidase and nitric oxide in action: molecular mechanisms and pathophysiological implications. Biochimica Biophysica Acta (BBA) 1817:610–619 (Bioenergetics)
Schertl P, Braun H-P (2014) Respiratory electron transfer pathways in plant mitochondria. Front Plant Sci 5:163
Seabra AB, Oliveira HC (2016) How nitric oxide donors can protect plants in a changing environment: what we know so far and perspectives. AIMS Mol Sci 3:692–718. https://doi.org/10.3934/molsci.2016.4.692
Sharma A et al (2016) Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol Bioch 106:82–90
Shiva S (2010) Mitochondria as metabolizers and targets of nitrite. Nitric Oxide 22:64–74
Singh HP, Kaur S, Batish DR, Sharma VP, Sharma N, Kohli RK (2009) Nitric oxide alleviates arsenic toxicity by reducing oxidative damage in the roots of Oryza sativa (rice). Nitric Oxide 20:289–297
Sun H, Tao J, Zhao Q, Xu G, Zhang Y (2017) Multiple roles of nitric oxide in root development and nitrogen uptake. Plant Signal Behav 12:e1274480
Thomas DD et al (2008) The chemical biology of nitric oxide: implications in cellular signaling. Free Radical Biol Med 45:18–31
Valderrama R et al (2007) Nitrosative stress in plants. Febs Lett 581:453–461
Vanlerberghe GC (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14:6805–6847
Verstraeten I, Schotte S, Geelen D (2014) Hypocotyl adventitious root organogenesis differs from lateral root development. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00495
Wang Y, Loake GJ, Chu C (2013a) Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death Frontiers in Plant Science 4
Wang Y, Loake GJ, Chu C (2013b) Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front Plant Sci 4:314
Wendehenne D (2016) Nitric oxide and signaling in plants, vol 77. Academic Press, pp 165–192
Wodala B, Ördög A, Horváth F (2010) The cost and risk of using sodium nitroprusside as a NO donor in chlorophyll fluorescence experiments. J Plant Physiol 167:1109–1111
Zandonadi DB et al (2010) Nitric oxide mediates humic acids-induced root development and plasma membrane H + -ATPase activation. Planta 231:1025–1036
Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57:805–836
Acknowledgements
This work was supported by a grant from the Next-Generation BioGreen 21 Program Rural Development Administration, (SSAC, Grant NO. PJ01242501), Republic of Korea.
Author information
Authors and Affiliations
Contributions
NKR executed the experiments and wrote the manuscript, SUL and BGM helped in experiments, QMI helped in writing the manuscript and reviewed it for technical contents, AH helped in designing the study, KMK, and BWK designed and supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no potential conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rolly, N.K., Lee, SU., Imran, Q.M. et al. Nitrosative stress-mediated inhibition of OsDHODH1 gene expression suggests roots growth reduction in rice (Oryza sativa L.). 3 Biotech 9, 273 (2019). https://doi.org/10.1007/s13205-019-1800-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1800-y