Abstract
Staphylococcus aureus is a gram-positive bacterium. It is a foremost cause of skin and respiratory infections, endocarditis, osteomyelitis, Ritter’s disease, and bacteraemia. Topoisomerase enzyme is involved in preventing or correcting topological problems of overwinding or underwinding occurring in DNA before replication process. An exhaustive molecular modeling studies that includes pharmacophore modeling, ligand-based three-dimensional quantitative structure-activity relationship (3D-QSAR), molecular docking, molecular dynamics simulation, and ADME calculations were performed on isothiazoloquinolones derivatives which are reported as effective inhibitors against topoisomerase IV of wild type S. aureus. In pharmacophore modeling by using pharmacophore alignment and scoring engine (PHASE) a five-point model (AHHRR.3) was generated with existing compounds having statistical significant as correlation coefficient (R 2 = 0.954), cross-validation coefficient (Q 2 = 0.650), and F value of 130.5. Ligand-based 3D-QSAR study was applied using comparative molecular field analysis (CoMFA) with Q 2 = 0.616, R 2 = 0.989, and comparative molecular similarity indices analysis (CoMSIA) with Q 2 = 0.510, R 2 = 0.995. The predictive ability of this model was determined using a test set of molecules that gave acceptable predictive correlation (R 2 Pred) values 0.55 and 0.56 for CoMFA and CoMSIA, respectively. Docking and molecular dynamic simulations were employed to position the inhibitors into protein active site to find out the most probable binding mode and most reliable conformations. Developed pharmacophore models and docking methods provide guidance to design enhanced activity molecules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus aureus is a gram-positive bacterium. It is a leading cause of food poisoning, soft tissue infections, and skin infections like abscesses, respiratory infections like sinusitis, blood stream infections, endocarditis, osteomyelitis, Ritter’s disease, and bacteraemia in the developed world. 4-quinolones have antibacterial activity against DNA gyrase and topoisomerase IV. During translocation of transcription and replication arised topological stress relieves with gyrase, topoisomerase IV is a decatenating enzyme that resolves interlinked daughter chromosomes following DNA replication. Since both enzymes are required for cell growth and division. With trapping of gyrase and topoisomerase IV on DNA probably leads to the lethal release of double-strand DNA breaks. Both topoisomerase IV and gyrase use a double-strand passage mode of action [1]. However, the enzymes differ in a fundamental way, gyrase wraps DNA around itself, while topoisomerase IV does not [2]. Indeed, wrapping seems to be the principal difference between the enzymes, since removal of a portion of the gyrase A protein converts gyrase into an enzyme that has a strong decatenating activity, much like that of topoisomerase IV [3].
Computational drug design approaches are vastly employed in development and optimization of inhibitors. A detailed study of molecular interactions of topoisomerase IV inhibitors with the protein will help in design of novel molecules for better antibacterial activity. Our main objective was to obtain structural requirements for topoisomerase IV inhibitors and design novel molecules. In this present study, we have employed and constructed the pharmacophore model through pharmacophore alignment and scoring engine (PHASE) [4] and developed ligand-based 3D-QSAR [5, 6] models using comparative molecular field analysis (CoMFA) [7, 8] and comparative molecular similarity indices analysis (CoMSIA) [9], to assist in the further discovery and design of potent topoisomerase inhibitors. Partial least square (PLS) [10] based statistical analysis was carried out on 42 molecules to identify the correlation. The contour maps generated enabled us to explain the observed variation in activity and guided us to design new molecules. In addition, a combined computational approach, including the docking analysis and molecular dynamics (MD) simulations were also employed to elucidate the probable binding modes of these antagonists at the binding site.
Materials and methods
All the molecular modeling calculations were performed on IBM MX1300 server with Linux operating system. Pharmacophore modeling was performed using Phase V3.2, and molecular docking was performed using Glide 5.6 in Schrödinger suite 2010. 3D-QSAR studies based on CoMFA and CoMSIA were performed using SYBYL X 2.1. Molecular dynamics simulations were carried out using Desmond 3.8.
Detail protocol for PHASE, docking, CoMFA, CoMSIA, and molecular dynamics studies are well-documented in many of the previous reports [11, 12]. Here, we give the brief description of the protocol followed for the present study. A set comprising of 42 compounds having biological activity against TOPO-IV was selected from the available literatures [13,14,15]. Inhibitory potencies of the compounds included in data set reported as IC50 values varied from 0.1 to 64 μM and were converted into pIC50. The structure of ligands were built using build panel in Schrödinger suite and optimized using Ligprep applying OPLS 2005 force field (Table 1).
The low-energy conformer were selected and imported into PHASE module; these were classified into active and inactive based on threshold “Active > (5.5) pIC50 < (5.0) inactive.” Conformers for all the molecules were generated by applying default parameters. Pharmacophoric feature cites were created for the set of molecules. The chemical pharmacophore features include two acceptors [A], two donors [D], and two aromatic rings [R]. Common pharmacophoric features were selected from active molecules, at most five variants and least three variants were generated with a tolerance of 1 Å. It resulted in a possible combination of features which were screened through shape and volume scoring for both active and inactive molecules. The molecule set was divided into training set of 30 and test set of 12 molecules. QSAR model was generated using PLS with three factors and best model with good R 2; Q 2 were selected.
Molecules were docked into Topo–IV active site using Glide 5.6; catalytic domain of TOPO-IV enzyme in complex with kibdelomycin (pdb id: 4URN) was downloaded from the protein data bank [16] for docking studies. Prior to docking, the protein was prepared using protein preparation wizard; a grid was generated around the active site by selecting co-crystallized ligand. Van der Waals scaling for receptor atoms was set to 0.9 [17]. Extra-precision XP docking mode was used for molecular docking. The docked conformers were used for CoMFA and CoMSIA studies. CoMFA and CoMSIA studies were carried out as described by Carmer et al. [4, 7] and Ramesh et al. [18]; the process is briefly described here. Dock pose of each ligand was imported into SYBYLX-2.1 molecular modeling program package [19], and Gasteiger–Huckel [20] charges were assigned. The active conformations were then aligned on docked pose of most active molecule 41 in the series, using ALIGN DATABASE command in SYBYL X 2.1 taking the substructure that is common to all (Fig. 1). The resulting alignment is shown in Fig. 2. Standard Tripos force fields were employed for CoMFA and CoMSIA analysis. A 3D cubic lattice of dimension 4 Å in each direction with each lattice intersection of a regularly spaced grid of 2.0 Å was created. Steric and electrostatic parameters were calculated in CoMFA fields, while hydrophobic, acceptor, and donor parameters in addition to steric and electrostatic were calculated in CoMSIA fields at each lattice. The sp3 carbon atom was used as variables. To improve the signal-to-noise ratio column filtering value (s) was set to 2.0 Kcal/mol by omitting lattice points whose energy variations were below the threshold. Leave-one-out (LOO) cross-validations were performed to determine the optimum number of components (ONC) and coefficient Q 2 LOO. The obtained ONC was applied to derive the final QSAR model. Validation of CoMFA and CoMSIA derived model was performed by predicting the activity of test set compounds.
To estimate the stability and binding interactions, MD simulations were performed using Desmond 3.8 [21] applying OPLS 2005 force field [22]. The crystallized protein along with ligand and docked complex of molecule S-1 were used for the MD simulations. The protein-ligand complex was solvated using SPC [23] water molecules in an orthorhombic box of 15 Å × 15 Å × 15 Å size. Counter ions were added to balance the net charge of the system; salt of concentration was maintained at 0.15 mol/l during the entire course of simulation. The system was minimized initially with restraints on solvate followed by without restrains using steepest descent and LBFGs algorithm until value converged to 5 kcal/mol/Å. Further, the system was relaxed with 12 and 24 ps MD simulation before extensive MD simulation of 5 ns with 2 fs time step at NTP canonical ensemble within periodic boundary conditions. Nose-Hower thermostat approach with a relaxation time of 1 ps was used to maintain the temperature during MD simulation, and the pressure was maintained at 1.0135 bars by applying Martyanu-Tobaisklin barostat approach using coupling-style isotropic with a relaxation time of 2 ps. Non-bonded interaction was computed at a truncated distances of 9 Å, and long-range electrostatic interactions were calculated using PME method with Ewald tolerance of 1e-09. The SHAKE algorithm [24] is applied to all bonds including hydrogen bond. MD simulation trajectories were recorded at 4.8 ps, and the recording interval energy was set at 1.2 ps. The RMSD with respect to simulation time was calculated for the protein-ligand complex.
Result and discussion
To understand and elucidate the mode of interaction and required pharmacophoric features for S. aureus TOPO-IV enzyme, a combination of ligand-based quantitative structure activity studies were performed on a set of 42 molecules. PHASE-QSAR approach was applied for ligand-based studies; the molecular set of 42 molecules were classified into active and inactives based on their activity that ranged between 4.19 and 7.00. The threshold for active was set to active > (5.5) pIC50 < (5.0) inactive. Pharmacophoric sites were identified for each molecule in the set; common pharmacophore hypothesis was selected from conformation of the active molecules using a tree-based partitioning technique. Obtained common pharmacophore hypothesis were scored and ranked based on contributions from the alignment of site points and vectors, volume overlap, selectivity, relative conformational energy, and activity of active molecules followed by scoring in terms of inactive molecules. The hypothesis was rescored to identify a better hypothesis that can differentiate between active and inactives. Pharmacophoric features thus obtained were AHHRR.3. The survival scores of the hypothesis is provided in Table 2; distance and angles of the sites are provided in supplementary file (Fig. S1).
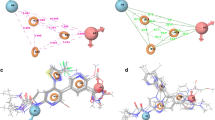
Figure 3a shows the pharmacophore sites aligned on the most active molecule; the features include a hydrogen bond acceptor (A3, pink sphere with two arrows), two ring system (R12, R14, orange ring), and two hydrophobic regions (H7, H9, green spheres). The hydrogen bond acceptor A3 is aligned to the carbonyl oxygen present on six member ring; the ring features are aligned onto fused six membered rings. Hydrophobic feature is seen on the cyclo propyl ring nitrogen (H9) and on the fluorine attached to fused ring (H7). An atom-based QSAR model was generated with respect to these pharmacophoric features, where in the complete data set of molecules was aligned to hypothesis AHHRR. Four factor PLS analyses was carried out using a training set of 30 molecules that yielded a R 2 of 0.954 with SD value of 0.176, Fisher test value of 130.5, and RMSE of 0.388. The predictive ability of the model was assessed using a test set of 12 molecules that gave a regression coefficient (Q 2) of 0.65. All the statistical parameters for the model are provided in Table 3.
a PHASE-generated pharmacophore model AHHRR.3 illustrating hydrogen bond acceptor (A3 red), hydrogen bond donor (H7,H9 green) and aromatic ring (R12,R14 red) features and b pictorial representation of the cubes generated using the QSAR model, blue cubes indicate favorable regions, while red cubes indicate unfavorable region for the activity of most active molecule 41
Figure 3b shows the combined coefficients of QSAR model for possible substitution on the molecules, the blue cubes represent favorable region for substitution, and at region of red cubes substitutions are disfavored. To further ascertain the structural features required for receptor binding and inhibition, receptor-based QSAR analysis was performed on the molecular set. All the 42 molecules were docked into active site of TOPO-IV using GLIDE 5.6 by applying XP docking protocol. Dock pose analysis of the molecules corroborate with PHASE results, where in we can see a hydrogen bond interaction between carbonyl oxygen on six membered ring (A3) and ALA 212. Dock pose of most active molecule 41 is shown in Fig. 4; the molecules show hydrogen bond interaction with ALA 212 and GLY 80.
Dock pose of all molecules thus obtained were subjected to standard CoMFA and CoMSIA analysis. Statistically, the results obtained for CoMFA and CoMSIA indicate the stability of 3D QSAR model, CoMFA model generated from 30 molecule training set gave R 2 ncv of 0.981 and Q 2 loo of 0.616. In the case of CoMSIA model, R 2 ncv of 0.995 and Q 2 loo of 0.510 were obtained. These models were externally validated using 12 test set molecules that yielded R 2 pred of 0.55 and 0.56 for CoMFA and CoMSIA, respectively. All the values obtained are in acceptable range; these are tabulated in Table 4. Scatter plot of experimental and predicted activity is shown in Fig. 5, and values are provided in Table 5. The prediction error in the model is measured in terms of MAE and RMSE for test, based on this, it was evident; the reason for low R 2 pred values, MAE of 0.449, and 0.448 for CoMFA, and CoMSIA is high in terms of standard requirement of MAE (MAE ≤ 0.1 × training set range AND MAE + 3 × σ ≤ 0.2 × training set range) [25]. Analysis of test set in the case of both CoMFA and CoMSIA clearly indicated large deviation in prediction for molecules 24 and 33. Considering these molecules as outliers, the resulting R 2 pred values improved to 0.700 and 0.644 for CoMFA and CoMSIA, respectively. Similar observations were made in the case of PHASE model, wherein R 2 pred value improved to 0.729. The MAE values of 0.253, 0.256, and 0.192 for CoMFA, CoMSIA, and PHASE respectively were obtained which are in acceptable range. Another important parameter to be considered for QSAR models is the applicability domain, representing the chemical space from which a model is derived that gives a reliable prediction [26, 27]. Applicability domain of the training set was calculated as reported by Zhang et al. and Kaur M [28, 29], training set applicability domain (APD) was found to be about 3.789, and all the nearest distance of test molecules were with this APD value (Table 5).
The most promising factor of CoMFA and CoMSIA analysis is the elucidation of structural parameter required for biological activity, which can be obtained from contour map analysis. CoMFA provides with steric (favored—green; disfavored—yellow) and electrostatic (red for electronegative favored and blue for electronegative disfavored) contour. CoMSIA provides with hydrogen bond acceptor (favored—magenta; disfavored—red), hydrogen bond donor (favored—cyan, disfavored—purple), and hydrophobic (favored—yellow; disfavored—white).
All of the contours represented the default 80 and 20% level contributions for favored and disfavored regions, respectively. Dihydroisoindole of highest active molecule was orients toward sterically favored green region indicate that substitution on it with steric group increases activity. Hydrogen of isothiozole ring orients toward electronegative region, replacement of this hydrogen by electronegative favored group or atom increases the activity. CoMSIA steric and electrostatic map is similar to the CoMFA steric and electrostatic map showing favored and disfavored regions besides this methoxy of dihydroquinoline group orients toward electronegative group. Instead of methoxy, substitution of more electronegative group will increase activity (Fig. 6).
a CoMFA steric standard deviation (S.D.* coefficient) contour maps illustrating steric and electrostatic features in combination with molecule 41.CoMSIA S.D.* coefficient contour maps illustrating b steric and c electrostatic features in combination with molecule 41. The green contour indicates a sterically favored region; yellow maps calls for a reduction of this potential to improve activity. Blue indicates a positive charge preferred region, and red contour indicated an electronegative group substitution to improve activity
Figure 7a–c shows the contour maps derived from the CoMSIA PLS model. To demonstrate its affinity for the hydrophobic, H-bond acceptor and H-bond donor regions of inhibitors. Dihydroisoindole orients toward hydrophobic region and methoxy of dihydroquinolline toward hydrophilic region indicate substitution of dihydroisoindole with bulky group, and substitution instead of methoxy with hydrophilic group will increase activity.
CoMSIA S.D.* coefficient contour maps illustrating hydrophobic, acceptor, and donor features in combination with 41 molecule. a The yellow contour for hydrophobic favored region, white indicates the hydrophilic favored region. b The magenta contour for H-bond acceptor group increase activity, red indicates the disfavored region. c The cyan contour for H-bond donor group increase activity, purple indicates the disfavored region
Structural requirements for inhibitory activity of TOPO-IV were elucidated based on the contour map analysis of both CoMFA and CoMSIA (Fig. 8). The molecules were modified to further improve the inhibition activity toward topoisomerase. Compound 41 was chosen as a reference structure to design new molecules that are within the applicability domain range of 3.789. The newly designed molecules (Table 6) were docked into the protein active site. Dock poses were used to predict the activity by applying the 3D-QSAR model (Table 7) (Fig. 9).
The pharmacokinetic parameters (ADME) were calculated for the designed molecules using QikProp 3.4. The compounds were assessed for their basic parameters of Lipinski’s rule of 5 and other pharmacokinetic parameters. Supplementary file (Table S1) shows the results obtained from QikProp with their permissible range. The important parameters with their permissible ranges are tabulated in Table S1 (Supplementary Material). The optimum value of rotatable bonds (0–15) and polar surface area (7–200 Angstroms) holds a great importance on the oral bioavailability of the drug molecules. The active test isothiazoloquinolones derivatives demonstrated results of the descriptors to be in the prescribed range thus owing good bioavailability. Intestinal absorption or permeation is also one of the important factors to be studied in concern with the absorption of the drug molecule, which was further confirmed by predicted Caco-2 cell permeability (QPPCaco), used as model for gut–blood barrier. Caco-2 cell permeability prediction of the test compounds indicates excellent results predicting good intestinal absorption. Also, the QikProp descriptor for blood/brain partition coefficient QPlogBB showed reliable prediction for all the test compounds and reference drugs. The cell permeability of the blood brain barrier mimic MDCK cells (QPPMDCK) also displayed reliable results. The aqueous solubility parameter (QPlog S) of the test entities was assessed, and the compounds were also found to be in the permissible range (< 0.5).
Molecular dynamics simulations
Stability, inter-intramolecular interaction and conformational changes in a protein-ligand complex can be assessed insilco using molecular dynamics simulations. Hence, 5 ns MD simulation was performed to compare newly designed molecules based on ligand-based QSAR and the crystal structure ligand. Figure 10 shows RMSD of protein backbone as a function of simulation time for each complex with respect to initial structure. Crystal structure complex showed RMSD change from 0.9 to 1.5 Å during first 200 ps and during the course of simulation; RMSD ranged between 1.5 Å and 1.9 Å. In the case of designed molecule initially, the RMSD ranged from 0.8 to 1.5 Å in first 200 ps and between 1.5 to 2.1 Å for the rest of the course. The averaged RMSD of the crystal structure complex was found to be around 1.7 Å, whereas for S-1 protein complex, it was around 1.8 Å indicating similar stability for both the complexes. To understand the mode of interaction of the ligand with protein, ligand interaction was analyzed throughout the course of dynamics run. Figure 11 shows ligand interaction diagram of crystal structure and designed molecule complex during the course of the simulation. The binding site of TOPO-IV protein has the following amino acid residues GLU 45, ASP 48, ASN 49,ASP 52,GLU 53, ASN 56, TYR 58, ASP 76, ARG 79,GLY 80, MET 81, PRO 82, THR 83, GLY 84, ILE 85, ILE 96, LEU 100, ARG 138, ASP 139, and THR 168. The designed molecule showed 96% hydrogen bond interaction with ASP 76. The crystal ligand showed 57, 35, and 30% hydrogen bond interaction with ARG 138 ASP 52 and GLU 53 respectively in simulation time (Fig. 12). The interaction diagrams clearly indicate that the designed molecule has similar mode of interaction with the receptor.
Conclusion
Combined computational approach was applied to give an insight into the structural basis and inhibition mechanism for a series of isothiazoloquinolone derivatives as TOPO-IV inhibitors. 3D-QSAR studies were performed to provide a structural framework for understanding the structure activity relationship of these compounds. The generated pharmacophore model AHHRR.3 exhibited good correlation and predictive power and satisfactory agreement between experiment and theory. Receptor-based 3D-QSAR by using CoMFA and CoMSIA methodologies were used to build models for topoisomerase IV inhibitory activity of the isothiazoloquinolone derivatives. Based on the detailed contour map analysis, improvement in topoisomerase IV binding affinity can be achieved through conformationally restricted substitution at dihydroisoindole and at nitrogen of isothiozole in reference molecule. The 3D-QSAR model generated has a good predicative ability and can be used to design new molecules with better activity. Molecular docking and molecular dynamics studies were performed to understand possible binding poses and their stability for these compounds. The low value of RMSD between the initial complex structure and the energy minimized final average complex structure suggests that the derived docked complex is close to equilibrium. Further, ADME predictions were performed for these compounds to determine their drug likeness.
References
Roca J (1995) The mechanisms of DNA topoisomerases. Trends Biochem Sci 20:156–160
Peng H, Marians KJ (1995) The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem 270:25286–25290
Kampranis SC, Maxwell A (1996) Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci U S A 93:14416–14421
Shubham S, Bhanwar SC, Manish S, Ruchi M (2016) Pharmacophore modeling and 3D-QSAR studies of galloyl benzamides as potent P-gp inhibitors. Med Chem Res 25:1140–1147
Mohammed AA, Janarthanan T, Naga ST (2017) Molecular insights on analogs of imidazo[1,2-a] pyridine, azaindole, and pyridylurea towards ParE using pharmacophore modeling, molecular docking, and dynamic simulation. Struct Chem 28:1187–1200
Sree KS, Vijjulatha M (2010) Molecular docking and 3D-QSAR studies on triazolinone and pyridazinone, non-nucleoside inhibitor of HIV-1 reverse transcriptase. J Mol Model 16:1169–1178
Cramer III RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Cramer III RD, Patterson DE, Bunce JD (1988) Cross validation, bootstrapping, and partial least squares compared with multiple regression in conventional QSAR studies. Quant Struct Act Relat 7:18–25
Klebe G, Abraham U, Mietzer T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146
Wold S, Johansson A, Cochi M (1993) PLS-partial least squares projection to latent structures. In: Kubinyl H (ed) 3D–QSAR in drug design: theory, methods and application. ESCOM, Liedin, pp 523–550
Ujashkumar AS, Hemantkumar SD, Shivajirao SK, Vithal MK (2010) Pharmacophore generation and atom-based 3D-QSAR of novel 2-(4-methylsulfonylphenyl)pyrimidines as COX-2 inhibitors. Mol Divers 14:559–568
Ramesh I, Srilata B, Sree KS, Vijjulatha M (2017) Molecular modeling-driven approach for identification of Janus kinase 1 inhibitors through 3D-QSAR, docking and molecular dynamics simulations. J Recept Signal Transduc 37:453–469
Jason AW, Qiuping W, Edlaine L, Akihiro H, Yongsheng S, Jijun C, Christopher WM, Yangsi O, Steven DP, Jane AT, Christy LT, Milind D, Michael JP, Barton JB (2006) Isothiazoloquinolones containing functionalized aromatic hydrocarbons at the 7-position: synthesis and in vitro activity of a series of potent antibacterial agents with diminished cytotoxicity in human cells. Bioorg Med Chem Lett 16:1272–1276
Jason AW, Yongsheng S, Qiuping W, Edlaine L, Akihiro H, Jijun C, Christopher WM, Yangsi O, Steven DP, Jane AT, Christy LT, Milind D, Michael JP, Barton JB (2006) Biological evaluation of isothiazoloquinolones containing aromatic heterocycles at the 7-position: in vitro activity of a series of potent antibacterial agents that are effective against methicillin-resistant Staphylococcus aureus. Bioorg Med Chem Lett 16:1277–1281
Qiuping W, Edlaine L, Akihiro H, Godwin CGP, David MN, Yongsheng S, Jane AT, Christopher WM, Christy LT, Jijun C, Steven DP, Yangsi O, Milind D, Pucci MJ, Douglas DB, Barton JB, Wiles J (2007) Isothiazoloquinolones with enhanced antistaphylococcal activities against multidrug resistant strains: effects of structural modifications at the 6-, 7-, and 8-positions. J Med Chem 50:199–210
Jun L, Sangita P, Nandini S, Stephen MS, Ryuta K, Masaya T, Yasumichi F, Kevin JL, Sheo BS (2014) Structures of Kibdelomycin bound to Staphylococcus aureus GyrB and ParE showed a novel U-shaped binding mode. ACS Chem Biol 9:2023–2031
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Ramesh I, Srilata B, Sree KS, Vijjulatha M (2016) Molecular docking, 3D QSAR and dynamics simulation studies of imidazo-pyrrolopyridines as janus kinase 1 (JAK 1) inhibitors. Comp Bio Chem 64:33–46
Sybyl-X-2.1 version, (2013) Tripos Inc., St. Louis (MO)
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228
Kaminski GA, Friesner RA (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem 105:6474–6487
Strahan GD, Keniry MA, Shafer RH (1998) NMR structure refinement and dynamics of the K+-[d(G3T4G3)]2 quadruplex via particle mesh Ewald molecular dynamics simulations. Biophys J 75:968–981
Berendsen HJC, Grigera JR, Straatsma TP (1987) The missing term in effective pair potentials. J Phys Chem 91:6269–6271
Andersen HC (1983) Rattle: a “velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys 52:24–34
Roy K, Das RN, Ambure P, Aher RB (2016) Be aware of error measures. Further studies on validation of predictive QSAR models. Chemom Intell Lab Syst 152:18–33
Gadaleta D, Mangiatordi GF, Catto M, Carotti A, Nicolotti O (2016) Applicability domain for QSAR models: where theory meets reality. Int J Quant Struct Prop Relat 1(1):45–63
Roy K, Kar S, Ambure P (2015) On a simple approach for determining applicability domain of QSAR models. Chemom Intell Lab Sys 145:22–29
Zhang S, Golbraikh A, Oloff S, Kohn H, Tropsha A (2006) A novel automated lazy learning qsar (all-qsar) approach: method development, applications, and virtual screening of chemical databases using validated all-qsar models. J Chem Inf Model 46:1984–1995
Kaur M, Kumari A, Bahia MS, Silakari O (2013) Designing of new multi-targeted inhibitors of spleen tyrosine kinase (Syk) and zeta-associated protein of 70 kDa (ZAP-70) using hierarchical virtual screening protocol. J Mol Graph Model 39:165–175
Acknowledgments
We greatly acknowledge Tripos Inc., USA and Schrödinger LLC, New York, for providing the software. This research was made possible through grants from DST-SERB (SB/EMEQ-004/2013), CSIR (01/(2436)/10/EMR-II), and UGC (42-233/2013(SR)), New Delhi, India. The authors SB and RI would like to acknowledge the financial support from UGC for research fellowships. We wish to express our gratitude to the Department of Chemistry, Osmania University for providing facilities to carry out the research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 159 kb)
Rights and permissions
About this article
Cite this article
Ballu, S., Itteboina, R., Sivan, S.K. et al. Pharmacophore modeling, 3D-QSAR, docking, and molecular dynamics simulation on topoisomerase IV inhibitors of wild type Staphylococcus aureus . Struct Chem 29, 593–605 (2018). https://doi.org/10.1007/s11224-017-1056-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-017-1056-2