Abstract
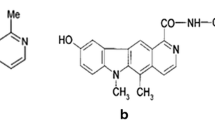
Certain DNA minor groove binders, especially bis-benzimdazole containing compounds, such as Hoechst 33258 and its derivatives, act as potent topoisomerase I inhibitors. The mechanism of action of these drugs is complex and involves hindering the breakage/reunion reaction of topoisomerase I. In the present work, molecular modeling studies have been performed to develop a pharmacophore and 3D-quantitative structure–activity relationship (QSAR) model based on bis- and ter-benzimidazoles, in an attempt to recognize the features that must be present in a molecule for it to behave as a topoisomerase I inhibitor. A data set comprising thirty bis-benzimidazoles and ter-benzimidazoles, known for their cytotoxicity against the RPMI-8402 lymphoblastoma cell line, has been chosen for this study. A five-point common pharmacophore hypothesis (CPH), with two acceptors, one donor and two aromatic features, has been derived for pharmacophore-based alignment of the molecules. The QSAR model, hence generated, shows a reasonable predictive Q2 value of 0.465. The CPH and contour map analyses display features that render antiproliferative properties to molecules against tumor cell lines, thereby ceasing cell growth. Further, the pharmacophore model has been utilized to develop lead molecules that can provide stability to the DNA–topoisomerase I cleavable complex, in turn inhibiting the activity of the enzyme. Virtual screening, followed by docking of obtained hits into the minor groove of B-DNA, gave three potent drugs, which are already approved drugs. The drug having the best fitness and binding score was further docked into the DNA–topoisomerase I cleavable complex. The present study opens up a new dimension in development of drugs for topoisomerase I inhibition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA topoisomerase I plays a significant role in DNA replication and transcription, as well as in chromosome segregation and condensation [1,2,3,4,5]. It introduces transient single strand breaks in the absence of an energy cofactor (ATP independent) and is therefore required during transcription, especially elongation [4, 6,7,8]. Topoisomerase I directs DNA cleavage and forms a covalent enzyme-DNA intermediate, called “cleavable complex.” This is followed by DNA relaxation and, finally, relegation of the phosphate backbone to restore the continuity of the DNA. Thus, topoisomerase I possesses immense importance in almost all stages of the cell cycle. As a result, the role of mammalian DNA topoisomerases as molecular targets for anticancer and antitumor drugs has been explored, and it was found that topoisomerase inhibition could curb cancerous cell growth across a variety of cell lines. However, the mechanism of action of topoisomerase I targeting anticancer drugs is very complex [9,10,11,12,13,14,15,16,17].
Certain DNA minor groove binders are known to act as topoisomerase I inhibitors. Hoechst 33342 (Fig. 1) and its analogues act as a structurally unique class of topoisomerase I poisons [18]. These drugs are known to hinder the breakage/ reunion reaction of topoisomerase I, in which the enzyme is reversibly trapped in a state where the DNA is cleaved [19]. The single-strand DNA breaks induced by minor groove binding drugs are highly site specific.
Studies with bis- and ter-benzimidazoles have shown that their interaction with the minor groove of DNA is essential for the poisoning of topoisomerase I. A study [20] established that a 3,4-dimethoxyphenyl bis-benzimidazole derivative of Hoechst acts as a novel DNA topoisomerase inhibitor with preference of targeting E. coli topoisomerase I. Recently, a series of Hoechst 33258 (Fig. 1) based mono- and bis-benzimidazoles has been synthesized and their E. coli DNA topoisomerase I inhibition, B-DNA binding, and antibacterial activity have been evaluated [21]. A 2D-quantitative structure–activity relationship (2D-QSAR) model, developed on the topoisomerase inhibitory potency of 5-substituted ter-benzimidazoles [22], indicated that the lipophilic activity of substituents at the fifth position of these ter-benzimidazoles can strongly influence cytotoxic activity. In another study [23], the topoisomerase I inhibition by bis- and ter-benzimidazoles was reviewed and individual 2D-QSAR models were generated for each class of inhibitors. A 3D-QSAR model was also generated using CoMFA [24] and CoMSIA [25] molecular field methods. The results were compared with multiple linear regression (MLR) models. The study reported that the hydrogen bond donor efficacy of the minor groove binders is a very important factor in enzyme inhibition [26].
However, minor groove binding alone is not a sufficient criterion for topoisomerase I trapping, as distamycin, berenil, and netropsin (which are good minor groove binders) do not poison topoisomerase I [27, 28]. It can thus be suggested that the stabilization of DNA–topoisomerase I covalent complexes may depend on the capacity of the drug to induce DNA bending or to stabilize a bent DNA conformation [17, 29].
After it was established that Hoechst 33258 and its derivatives can act as topoisomerase poisons, their cytotoxicity against the human lymphoblastoma cell line, RPMI-8402, was also tested by several groups [22, 30,31,32,33,34,35,36]. RPMI-8402, established from the peripheral blood of Homo sapiens with acute lymphoblastic leukemia (ALL), is a round single-cell suspension which grows partly in clumps and forms tumors [37]. The cytotoxicity of Hoechst derivatives towards these cancerous cell lines opens up their use as antitumor agents. The topoisomerase I extracted from these cell lines can be successfully inhibited, and further, cell growth can thus be hindered.
In the present study, we have developed pharmacophore and 3D-QSAR models based on bis- and ter-benzimidazoles, in an attempt to recognize the features that must be present in a molecule for it to behave as a topoisomerase I inhibitor. Further, virtual screening and molecular docking studies have been performed on external ligand sets extracted from the ZINC database. The obtained hits have been subjected to in silico pharmacokinetic studies and evaluated on Lipinski’s rule of five in order to pass the drug acceptability criteria. In all, our pharmacophore model will represent features that render antiproliferative properties to molecules against tumor cell lines.
Computational details
Pharmacophore building and atom-based 3D-QSAR
We utilized the PHASEFootnote 1 (Pharmacophore Alignment and Scoring Engine) module developed by Schrödinger, Inc. to perform both pharmacophore modeling and atom-based 3D-QSAR using the “develop pharmacophore model” workflow. PHASE thoroughly explores all possible conformations across rotatable bonds and retains only the most reasonable conformations. It finds possible pharmacophores using a high-dimensional, tree-based partitioning algorithm. After this, alignments of active ligands are done on generated pharmacophores according to an open, highly configurable scoring function. Partial least-squares (PLS) regression [38,39,40] is used for predicting a significant pharmacophore model [41, 42]. The alignment of molecules obtained from PHASE is used as the input for development of an atom-based 3D-QSAR model.
A data set comprising 30 bis-benzimidazole and ter-benzimidazole based Hoechst 33258 derivatives (Table S1) that are known for their cytotoxicity against the RPMI-8402 lymphoblastoma cell line was used in our study. The molecules were chosen keeping in view that they must cover maximum diversity under similar biological assay conditions [22, 30,31,32,33,34,35,36]. The molecules were geometrically refined retaining their specified chiralities, and all possible ionization states at the target pH 7.0 were generated to incorporate the states that have possibility of existence at the physiological pH. From the X-ray analysis, it is proven that in the Hoechst molecule, both –NHs of benzimidazole rings face the minor groove of DNA and adopt a slightly twisted planar structure in order to form intermolecular hydrogen bonds with DNA bases [43]. It has also been found that it requires very little energy (of the order of a few kJ mol−1) to flip the two benzimidazole rings along the central bond connecting them [44], and therefore it is not justified to carry a rigorous conformational search via PHASE.
After this, the activity threshold was set for these molecules. All IC50 values were converted into pIC50 for convenience. The pIC50 values range from 3.82 to 7.52, and these values were used to divide the molecules into three categories. Molecules with pIC50 above 6.20 were tagged active, while those with pIC50 below 5.60 were labeled inactive (Table S1). All inhibitors having pIC50 values between 5.60 and 6.20 were considered moderately active. With this threshold, we obtained 15 actives and 8 inactives, which were used for pharmacophore generation and scoring [42].
Common pharmacophoric hypotheses (CPHs) were generated for three to seven variant lists, comprising the pharmacophore features, hydrogen bond acceptor (A), hydrogen bond donor (D) (hydrogens bonded to N, O, P, S), hydrophobic group (H) (alkyl chains, Cl, Br, F, I), negatively ionizable (N), positively ionizable (P), and aromatic ring (R). These CPHs were then examined using a scoring function in the “score hypotheses” panel to obtain best alignment of the active ligands [42]. After this, a scoring procedure was applied to identify the pharmacophore from each surviving n-dimensional box that yields the best alignment of the active set ligands.
The predictive ability of a hypothesis which scored well with actives but less with inactives was evaluated by correlating the observed and estimated activity of the training and test set molecules using PLS analysis. For the QSAR studies, compounds showing high activities were included in both the training and test sets, especially in the training set, so as to provide important information on the pharmacophore requirements.
Virtual screening and molecular docking
After establishing a pharmacophore model, it is imperative to identify from a set of known drugs the structures which fit the model, and are hence likely to bind DNA, thereby stabilizing the DNA–topoisomerase I complex. A small library of commercially available approved drugs from the ZINC Drug Database (ZDD) (www.zincdocking.org), comprising 2924 drugs, was chosen for the study. These molecules were first energy minimized through multiple minimizations using the OPLS-2005 force field, keeping the force field defined electrostatic treatment tools available in MacroModel, Schrödinger, Inc.
The enumeration of tautomeric, ionization, and stereoisomeric states is an important step in virtual screening. These states were generated using LigPrep at the physiological pH 7 ± 2. A total of 42,071 states were thus generated. These states were then subjected to “Find Matches to Hypothesis Panel” in PHASE, and rigorous conformational search was performed during the process. The pharmacokinetic drug likeliness and Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) parameters [45, 46] were then evaluated for the obtained hits using the QikProp module available in Schrödinger, Inc.
For molecular docking, we chose the Dickerson Drew structure of B-DNA complexed with Hoechst 33258 (PDB ID: 1DNH) [43]. The minimization of the macromolecule and generation of the receptor grid were carried out on the same lines as reported in our earlier work [47]. Glide standard precision (SP) and extra precision (XP) methodologies were utilized to select the top lead molecules. The Prime MM/GBSA calculations were also performed using the Ligand and Structure-Based Descriptors (LSBD) application of the Schrödinger software package.
In a further step to validate the results, docking studies of the proposed molecules with the DNA–topoisomerase I cleavable complex were then performed. Due to unavailability of the crystal structure of the ternary complex of DNA–topoisomerase I with the minor grove binder, the binary complex was chosen instead. The starting coordinates of the human topoisomerase I in complex with a 22-base pair duplex oligonucleotide having the d(AAAAAGACTTAGAAAAATTTTT)-3′ sequence (PDB ID: 1A36) were imported from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (www.rcsb.org) and refined for further work using the protein preparation workflow in the Schrödinger suite.
Results and discussion
Survival scores—internal and external data set predictions for CPHs
Thirty Hoechst derivatives with varying activities against RPMI 8402 cell lines were selected for CPH generation. Two hundred and twenty three-featured, 337 four-featured, 267 five-featured, 107 six-featured, and 16 seven-featured probable CPHs were thus generated from the list of variants. The CPHs of three and four featured variants were rejected, as they were unable to define the complete binding space of the large Hoechst based molecules. On applying the scoring function for five featured CPHs, 75 CPHs belonging to five broad types AADRR, ADRRR, ADDRR, DRRRR, and ARRRR survived. The survival as well as survival minus inactive (“survival-inactive”) scores for each class of hypotheses were obtained and analyzed. Out of the 75 surviving hypotheses, the ones that were the best scorers in their respective category, and also those that aligned on the molecule in the most diverse ways, are tabulated in Table S2. Further details are provided in the supplementary information.
A good hypothesis is one that not only matches the actives significantly but also deviates appreciably from the inactives. The larger the value of the survival-inactive score, the better is the hypothesis in distinguishing the actives from the inactives [42]. From Table S2 for the five-featured CPHs, it is observed that the hypotheses AARRR.22 and AARRR.24 have the least survival scores (2.858 and 3.364, respectively) as compared to the other CPHs. Therefore, this feature family (AARRR) can be safely dropped from further statistical analysis. For the other hypotheses, some have exactly the same survival scores, with little or no difference in the survival-inactive score. The same is also true for the six- and seven-featured hypotheses (Tables S3 and S4). Since there is little to distinguish amongst the various hypotheses in each variant, we proceeded with 3D-QSAR generation using all five, six, and seven-featured CPHs.
The QSAR results for the six and seven featured CPHs are found to be unsatisfactory (Tables S3 and S4). There is great mismatch between the R2 and Q2 values for these variants, signifying that the QSAR model is not a good one. Negative Q2 values are obtained for these CPHs, indicating over-fitting of data. Despite good regression with the training set molecules (R2), most of the six and seven featured hypotheses show poor regression coefficient with the test set molecules (Q2). The Pearson-R values, which are a correlation between the predicted and observed activities for the test set, are also unsatisfactory. Though the results for seven featured CPHs are slightly better than those for six featured CPHs, both show large deviation in the test set activity predictions and were, therefore, rejected.
We now discuss the five featured CPHs. To select the best hypothesis amongst these, different combinations of the training and test set molecules were generated and analyzed using PHASE PLS analysis. All the 30 ligands were aligned on each of these five featured CPHs, and random training (50%) and test (50%) sets were thereby generated. The summary of the statistical data of the random set for the selected five featured CPHs with three PLS factors is listed in Table 1. It can be seen that all the hypotheses are statistically significant (p < 10−4).
A good QSAR model is one for which the Q2 values for the tested ligands are comparable to the R2 values obtained for the training set ligands. Also, the Pearson-R values should be greater than 0.5, and RMSE values should be low [41, 42, 48,49,50]. From Table 1, it is observed that the largest Q2 values are obtained for ADRRR.98 and AADRR.4. Their Pearson-R coefficients are also the highest and comparable. These hypotheses are therefore better than the others and are thus considered for further discussion.
A second combination of the training and test set molecules was generated by applying a 50% random training set selection. The training set was analyzed using three PLS factors, and the predictivity of the hypotheses was analyzed with the test set molecules (Table S6). Here again it is observed that for both AADRR.4 and ADRRR.98, the statistics are comparable in one respect or the other. For example, both the hypotheses have comparable Q2 values (0.3627 and 0.3647, respectively, for AADRR.4 and ADRRR.98), and it cannot be conclusively asserted which of the two hypotheses, AADRR.4 and ADRRR.98, is the better of the two.
In order to gain further clarity in this respect, the above two hypotheses were tested over an external test set of ligands. For testing the external test set predictivity of the CPHs, 277 known topoisomerase I inhibitors were imported from the Binding DB database (www.bindingdb.org), along with their IC50 values. The conformations for these 277 molecules were generated via the MacroModel conformation search panel using a mixed MCMM/LMOD (Mixed Monte Carlo Multiple Minimum/Low Mode) search with distance-dependant dielectric solvation treatment using the OPLS-2005 force field. For each molecule, all conformers with maximum energy difference of 10 kcal mol−1 relative to the global energy minimum conformer were retained. A total of 1929 conformations were thus obtained for these 277 ligands.
Using the “Find Matches to Hypothesis” panel of PHASE, it was found that the numbers of hits/ligands matching the two competing hypotheses, AADRR.4 and ADRRR.98, are 38 and 15, respectively. The activity for each of these molecules was then predicted. One point to be emphasized here is that the external data set comprised all the topoisomerase I poisons and did not specifically include DNA minor groove binders. Topoisomerase inhibition can be ascertained via many routes, and stabilization of the DNA–topoisomerase cleavable complex by the ligands is one of them. The lower number of hits obtained is justified, as the pharmacophore model is built on bis- and ter-benzimidazoles only, but we have scanned it over the entire library of topoisomerase poisons, of which Hoechst derivatives are just a small part. The number of hits also signifies that these ligands present in the topoisomerase inhibitor library can inhibit the enzyme in a similar manner as the DNA minor groove binders do. It will be of much interest to see the binding affinity of these drugs towards DNA. As the mechanism of topoisomerase I inhibition via DNA minor groove binders is still unclear, it will be interesting to study theoretically the mechanism of Hoechst and its bis- and ter-benzimidazole derivatives as blocking agents towards the DNA–topoisomerase cleavable complex.
A plot of the predicted activity for each of these molecules versus their experimental activity for each QSAR model, along with the corresponding regression coefficients, is presented in Fig. 2.
From the external data set predictions of the two pharmacophore models, it is observed that the regression coefficient between the predicted and experimental activity is better for hypothesis AADRR.4. The CPH ADRRR.98, although only a little behind AADRR.4 in terms of R2, was rejected, as the number of hits obtained for the former (15) is much less than that for the former (38). Since the internal data set predictivities (Table 1), as well as external data set predictions (Fig. 2), are both satisfactory for the CPH AADRR.4, it emerges as the “best” pharmacophore model for defining the features required for topoisomerase inhibition and cytotoxicity towards the RPMI 8402 cell lines. The details of this pharmacophore model are presented below.
AADRR.4—the pharmacophore model
The features represented in the best pharmacophore model are two acceptors, one donor, and two aromatic rings. The acceptors are the nitrogens of the bis-benzimidazole moiety that possess lone pairs, while the hydrogen bond donor group is represented by one of the –NH of the benzimidazole system (Fig. 3). The two aromatic features are also a part of the bis-benzimidazole unit. Any molecule with these features at the specified distance and angle (Tables S7 and S8) is expected to be a potent drug towards RPMI 8402 cell lines.
Model AADRR.4 representing features a distances and b angles between them. Acceptor is indicated as a light red sphere A1 and A3 with lone pair vectors; donor D5 is indicated by a light blue sphere centered on the H-atom with an arrow pointing in the direction of a potential H-bond. R12 and R13 represent the aromatic rings features denoted by orange rings
In Fig. 4, all the thirty molecules are superimposed on the model AADRR.4, and it is found that almost all molecules superimpose on the pharmacophore model with pretty good accuracy. As the Hoechst molecule contains a central core of the bis-benzimidazole moiety, the presence of two acceptors, one donor, and two aromatic ring features in our pharmacophore model is an indicator that these features could be important in deciding the biological activity of Hoechst derivatives.
The fitness score is a measure of how well a conformer matches a pharmacophore model [41, 42]. The fitness scores of all the molecules with respect to the model AADRR.4 and the predicted and experimental activities are listed in Table S9. Compound S# 24 with the maximum possible fitness score of 3.00 is presented in Fig. 5. As stated earlier, all the ligands fit on the pharmacophore model with sufficient accuracy (Fig. 4), which is also confirmed from the fact that even the lowest fitness score value (2.41; Table S9) is appreciably good (> 80%).
In Fig. 6 are presented the scatter plots of predicted versus experimental pIC50 for two different combinations of training and test set molecules—one with random 50% training set selection and the other with random 70% training set selection. A reasonable correlation is observed between the R2 and Q2 values within both the sets. Moreover, the validity of this model had already been confirmed by the good regression coefficient obtained for an external data set (Fig. 2a).
Interpretation of contour maps
In the QSAR visualization panel, contour maps were found for hydrogen bond donor, hydrophobic/non-polar part, and electron withdrawing groups, but no contour maps could be found for hydrogen bond acceptor groups. This implies that the presence of hydrogen bond acceptor groups in bis-benzimidazoles and ter-benzimidazoles hardly makes any contribution to the biological activity of these ligands. In other words, the position of the heterocyclic N atom in the benzimidazole lacks significance as far as cytotoxicity is concerned. These contour maps generated from pharmacophore model AADRR.4 give an idea regarding positioning of various groups in 3D space in the absence of the receptor.
Figures 7, 8, and 9 illustrate the hydrogen bond donor, hydrophobic, and electron withdrawing properties of the model superimposed on the most potent molecule S# 13 and the least potent molecule S# 20. The colored regions in the contour maps denote the placement of substituents/groups that would increase or decrease the activity of drug. The orange colored regions (Fig. 7) denote the 3D space where the presence of a hydrogen bond donor will cause increment in the activity of drugs against topoisomerase I inhibition, whereas the green colored regions indicate decrement in the same.
Atom-based 3D-QSAR model based on a most potent topoisomerase inhibitor and b least potent topoisomerase inhibitor, illustrating the hydrogen bond donor feature. The orange regions denote where hydrogen bond donor groups increase the cytotoxicity of molecules towards RPMI 8402 cell lines, and the green regions denote 3D space where hydrogen bond donor decreases the cytotoxicity of molecules towards RPMI 8402 cell lines
Atom-based 3D-QSAR model based on a most potent topoisomerase inhibitor and b weak topoisomerase inhibitor illustrating the hydrophobic/non-polar feature. The yellow regions denote where hydrophobic groups increase the cytotoxicity of molecules towards RPMI 8402 cell lines, and the pink regions denote where hydrophobic groups decrease the cytotoxicity of molecules towards RPMI 8402 cell lines
Atom-based 3D-QSAR model based on the most potent topoisomerase inhibitor (a) and weak topoisomerase inhibitor (b) illustrating the electron withdrawing feature. The cyan regions denote where electron withdrawing groups increase the cytotoxicity of molecules towards RPMI 8402 cell lines, and the purple regions denote where electron withdrawing groups decrease the cytotoxicity of molecules towards RPMI 8402 cell lines
It can be seen that both the ligands are ter-benzimidazoles, and only one benzimidazole ring, Bz1 (Table S1), is responsible for the enhancement of the biological activity of these ligands, as this is the only group that overlaps the orange colored regions (Fig. 7). No other position of hydrogen bond donor seems significant. In addition, the presence of a hydrogen bond donor at the third benzimidazole ring, Bz3 (Table S1) seems unfavorable for the activity (Fig. 7), as this overlaps the green region. This is in agreement with a proposal [26] that the central imidazole –NH moiety contributes less to the activity in comparison to the equivalent groups on the other two benzimidazoles.
Apart from the –NH of benzimidazole rings, the presence of a hydrogen bond donor at the fifth position of the benzimidazole Bz3 in ter-benzimidazoles also decreases the activity. The molecule S# 20 is the least potent topoisomerase inhibitor, as the presence of the –OH group at this position overlaps with the green regions of the QSAR model.
Figure 8a, b shows regions around the most and least potent topoisomerase I inhibitor with respect to hydrophobic/non-polar groups. The yellow regions reflect the positions where the presence of non-polar groups (such as aromatic rings/aliphatic chains) increases the topoisomerase activity. The presence of a –CH3 group on the aliphatic piperazinyl ring in Hoechst 33342 also falls in this yellow region. As far as the ter-benzimidazoles are concerned, the most potent molecule S# 13 contains the non-polar pyridine ring, which overlaps with the yellow region, thereby increasing the activity. Similarly, ligand S# 30 has two hydrophobic phenyl rings at the fourth and fifth positions of the benzimidazole ring Bz3 (Table S1), which overlap with the yellow region of the QSAR model, and hence, this molecule possesses significant activity.
The low topoisomerase I activity of the minor groove binder S# 20 (Fig. 8b) is due to its inability to interact with the hydrophobically favored yellow region. The compound is small in size and so does not contain large hydrophobic substituents at the specified yellow regions, causing poorer activity. On the other hand, the pink colored region falling on the –NH of the first benzimidazole ring (extreme left) indicates that the polar groups are favored at this position and any hydrophobic group would result in lowering of activity against topoisomerase inhibition.
Figure 9 denotes the effect of electron withdrawing groups on the cytotoxicity of these ligands towards RPMI 8402 cell lines. The electron withdrawing groups lying in the cyan region enhance molecular activity, while those present in the purple region result in lowering of activity. Amongst all the ligands considered in this study, the activity of molecule S# 4 is significantly good (Table S9) due to the presence of the electron withdrawing –NO2 group at the fourth position of bis-benzimidazole, which overlaps with the cyan region. In the most potent inhibitor S# 13, the hydrophobic group is present at the fifth position of the benzimidazole ring Bz3 (Table S1), which plays an important part in the enhancement of its activity. Hence, the presence of an electron withdrawing group at this position would decrease the cytotoxicity of the molecule due to its overlap with the purple colored region, as illustrated in Fig. 9a.
In Fig. 10 are presented the combined effects of H-bond donor, hydrophobic group, and electron withdrawing groups on the activity of the molecules—S# 13 and S# 20. The presence of these groups in the blue colored regions enhances the activity, while their presence in the red colored regions decreases activity.
From Fig. 10a, it is clear that, for the most active molecule, there are maximum blue colored regions overlapping the available features in the molecule, while for the least active ligand (Fig. 10b), the number of features overlapping with the unfavorable red regions is greater. We conclude that, for a molecule to be an active inhibitor of topoisomerase I, the H-bond acceptor, H-bond donor, and hydrophobic groups should preferably lie in the blue colored regions with minimal presence in the red region.
Following the alignment of the Hoechst derivatives on the pharmacophore and 3D-QSAR model, we proceeded towards virtual screening.
Virtual screening
Finding the matches to hypothesis AADRR.4
To further validate the pharmacophore model AADRR.4 and to find the lead molecules from the known drug database that can inhibit the topoisomerase I enzyme, we performed a query-based search on the 42,071 states generated of the 2924 drugs. A total of 1197 molecules were obtained as hits. The maximum value of the fitness score obtained was 1.92. The distribution of molecules in the various fitness score ranges is given in Table 2.
It is observed that, out of the 1197 molecules, there are 221 (~ 19%) that exhibit fitness scores ≥ 1.50 (i.e., 50%). The low percentage can be explained on the basis that we are in search of drugs that resemble DNA minor groove binders based on bis- and ter-benzimidazoles. The aim is to search for an alternative mechanism induced by DNA minor grove binders in the inhibition of the topoisomerase enzyme. Usually, topoisomerase I inhibitors attack the enzyme site; however, topoisomerase I poisoning can also be achieved by the drug attacking both DNA and the enzyme at the site of cleavage. Therefore, fitness cannot be the sole criterion in determining new leads, and hence docking plays a significant role in enumerating the results, and therefore, we proceeded with docking studies in three stages. The first part is the docking of the obtained hits within the minor groove of B-DNA (PDB ID: 1DNH). The second part is docking of bis- and ter-benzimidazoles (used to obtain the pharmacophore model) within the minor groove of B-DNA to correlate the binding pattern between the proposed drugs and bis- and ter-benzimidazoles (the known minor groove binders). The third part is an attempt to actually mimic the real system by docking one of the best proposed drugs within the minor groove of DNA complexed with topoisomerase I (PDB ID: 1A36).
ADMET properties and docking studies of obtained hits within the minor groove of B-DNA
We chose the crystal structure of the Dickerson–Drew DNA dodecamer d(CGCGAATTCGCG)2 in complexation with Hoechst 33258 (PDB ID: 1DNH) [43], obtained from the Protein Data Bank (www.rcsb.org), for docking analysis of the hits. Hoechst 33258 is known as a prominent minor groove binder that displays specific binding towards the adenine–thymine (AT)-rich region of the minor groove of B-DNA [43]. Therefore, the docking studies were carried out by replacing Hoechst 33258 with the obtained hits. This was done in order to understand the interaction modes of these ligands with the AATT rich sites of DNA. The preparation and minimization of the macromolecule were performed as described in our previous work [47]. The initial analysis and filtering were performed using the standard precision (SP) docking protocol [51]. Table S10 gives the distribution of ligands in the various glide-SP score ranges, along with the number of molecules (within each range) that have fitness scores ≥ 1.50. The glide scores are all negative, and the highest magnitude of the glide-SP score is − 11.24 and the least magnitude is − 0.69.
From Table S10, it is evident that the number of ligands with good fitness along with an appreciable docking score is small (only ~ 10% of the ligands lie in the range − 12.00 < G score ≤ − 5.00). Therefore, we subjected all the ligands having glide score ≤ − 5.00 and fitness score ≥ 1.50 to the extra precision (XP) docking methodology [52]. The complete data for glide-XP docking, along with fitness score, predicted activity and predicted IC50 in micromolars of the 116 ligands is tabulated in Table S11. It is observed that the docking score reduces drastically in XP docking. For example, for ligand Z1, the magnitude of the glide-XP score decreased to − 9.24 from − 11.24 in glide-SP.
Now, before proceeding to the discussion on binding, it is important to predict the pharmacokinetic profile of these ligands. We used the QikProp module available in Schrödinger to calculate the ADMET properties. Out of the 116 states of the ligands, QikProp successfully processed 103 (Table S12) and failed to predict descriptors for the remaining 13 states of ligands (Fig. S1). All these 13 states of ligands possess a negatively charged carbon atom, which is unusual for any drug molecule. Moreover, QikProp does not process ligands that are either odd electron systems or charged. Out of these 13 failed ligands, seven are different structural forms of the ligand dasatinib (ZINC21982951), three are different structural forms of ZINC19632618, two are different structural forms of ZINC12503187, and one belonged to ZINC13916432. Hence, we proceed with the discussion of 103 ligands. As most of the hits are commercially available drugs, they possess acceptable ADMET properties. Nevertheless, on screening these compounds, some of the drugs (26 out of 103) violated Lipinski’s rule of five. This could be due to the fact that some drugs do not have good oral bioavailability and permeability. Therefore, a suitable drug need not cross the threshold imposed by Lipinski’s rule of five and biomacromolecules can also be administered parenterally [53, 54].
The predicted activity IC50 of obtained hits that qualify in QikProp ranges from 0.36 to 35.48 μM. LogPo/w, the n-octanol-water partition coefficient, is a measure of the hydrophobicity/lipophilicity of a compound [55, 56]. From Table S12, we can see that almost all the drugs have high logPo/w values, which indicates that these compounds are lipophilic in nature. This is also supported by the low solubility of these drugs in aqueous solution (logS value). Out of 103, 22 drugs have logS values below the permissible range, i.e., − 6.5 to 0.5. Apparent Caco-2 cell permeability, which is a measure of the ability of a drug to cross the gut–blood barrier, and apparent MDCK cell permeability, which predicts the permeability for the blood/brain barrier, are high for the drugs having logPo/w values greater than 6. This suggests that lipophilicity enhances the chance of the drug to cross these cell barriers. Also, the predicted skin permeability, logKp, and predicted binding to human serum albumin, logKHSA, are also found to lie within the acceptable range for these drugs (Table S12).
A major concern arises on observing that the IC50 value for the blockage of HERG K+ channels for almost all the drugs (93 out of 103) is below − 5 (Table S12). The human ether-a-go-go related gene (HERG) K+ channel, best known for its involvement in the electrical activity of the heart that coordinates the heart’s beating, is a molecular target responsible for the cardiac toxicity of a wide variety of drugs [57]. A low value of logHERG indicates high cardiac toxicity of drugs [58]. Similarly, there are 21 molecules having low (< − 3) predicted brain/blood partition coefficient (logBB) (Table S12). This could be due to the fact that these drugs are too polar to cross the blood/brain barrier. A correlation of − 0.8348 between logPw and logBB for these 21 molecules validates the explanation. Therefore, a hydrophilic drug finds difficulty in crossing the lipophilic blood/brain barrier.
The top scorer ligand Z1 having the highest glide score and Emodel (− 124.50 kcal mol−1) could not be filtered in QikProp. However, ligand Z5, a state of this drug commercially known as dasatinib (ZINC21982951) qualifies as a drug with significant binding parameters.
Keeping all the parameters viz. fitness score, predicted IC50, binding energy, scoring functions and ADMET properties in mind, three drugs are proposed as DNA–topoisomerase I complex inhibitors. These are dasatinib (Z5), lapatinib (Z25), and novobiocin (Z22) (Fig. 11).
Dasatinib, sold under the brand name sprycel, is a tyrosine kinase inhibitor having immense applications in chemotherapy medication, especially chronic myelogenous leukemia (CML) and acute lymphoblastic leukemia (ALL) [59]. One of its states, Z5, with total charge + 1, is found to fit well on the AADRR.4 pharmacophore model with fitness score 1.88 (Fig. 12). Geometrically, it is observed that it complements the curvature of the DNA helix and hence can effectively bind in the minor groove.
Docking studies revealed that Z5 successfully fits in the minor groove of B-DNA with − 104.99 kcal mol−1 model energy score (Emodel) and − 8.10 glide score. It has been proved that, for DNA–ligand interactions, there exists a good correlation between the glide score, glide energy, and Emodel [47]. Since Emodel combines the glide score, the non-bonded interaction energy, and the excess internal energy of the generated ligand conformation, we will explain the results based on Emodel.
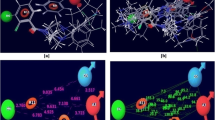
The complex formed between double stranded B-DNA and Z5 is quite stable. The Gibbs energy of binding of the ligand Z5 to DNA was found to be − 47.14 kcal mol−1 indicating maximum stabilization as compared to other proposed drugs (Table 3). The stabilization is due to various interactive forces such as electrostatic interactions and hydrogen bond formations. The N–H linker in between the thiazole and pyrimidine rings of the ligand forms bifurcated hydrogen bonds, one end of which is linked to O2 of the thymine (dT7) of one DNA strand and the other with O2 of the thymine (dT19) of the complementary DNA strand (Fig. 13a–c). The existence of bifurcated hydrogen bonds was confirmed by measuring the hydrogen bond angle between the hydrogen bond donor, hydrogen, and the hydrogen bond acceptor. The bond distances of NH…O(dT7) and NH…O(dT19) were found to be 2.572 Å and 2.423 Å, respectively (Fig. 13c). The bond angles between N…H…O(dT7) and N…H…O(dT19) were found to be 140.7° and 128.5°, respectively, affirming the formation of bifurcated hydrogen bonds, since hydrogen bond angles for bifurcated interactions typically lie between 120° and 160°. This feature of the drug forming bifurcated hydrogen bond with DNA strands resembles Hoechst 33258 [43].
a A two-dimensional interactive diagram of Z5 within the AATT pocket of DNA showing formation of hydrogen bonds between Z5 and DNA (purple colored solid lines). b A three-dimensional view of Z5 embedded in DNA. c Displaying interaction of Z5 with only dT7 and dT19 involved in bifurcated hydrogen bonds (yellow colored dashed lines)
The pharmacokinetic profile of Z5 displayed no violation of Lipinski’s rule of five and therefore it can be orally administered. Its percentage human oral absorption in the GI tract is 84%. All the predicted properties, except logHERG and logBB, were found to be in the permissible range (Table S12).
The second proposed drug, novobiocin (Z22), having minimal predicted IC50 of 0.77 μM, is a known inhibitor of DNA gyrase and topoisomerase IV [60, 61]. It is also known as albamycin or cathomycin, which is an aminocoumarin antibiotic that is produced by the actinomycete Streptomyces niveus [62]. This drug fails terribly in the pharmacokinetic profile (Table S12) with three violations of Lipinski’s rule of five but displays remarkable binding properties within the minor groove of B-DNA (Table 3). Hence, it is proposed that this drug may be administered intravenously and not orally. Next is lapatinib (Z25), an established orally active drug for breast cancer and other solid tumors [63]. It interrupts both epidermal growth factor receptor (EGFR) and HER-2 tyrosine kinases [64, 65]. None of the proposed drugs are known inhibitors of topoisomerase I. However, all three of them show anticancer properties against various cell lines and are shown to affect various DNA enzymes.
Docking of bis-benzimidazole and ter-benzimidazole containing molecules within the minor groove of B-DNA and comparison with the proposed drugs
After obtaining the three lead molecules which bind significantly within the DNA pocket, it is imperative to compare their binding interactions with the known DNA binders. These are the bis- and ter-benzimidazole-containing drugs that also display cytotoxicity against the RPMI-8402 cell line. It is well established that bis-benzimidazole and ter-benzimidazole containing drugs specifically trap DNA–topoisomerase I reversible cleavable complexes in various cancerous cell lines [21, 28, 33, 34, 36] and hence are potential candidates in antitumor chemotherapy. The analysis of the pharmacokinetic profile of these drugs revealed the same (Tables S13 and S14). Seventeen of the 30 ligands followed Lipinski’s rule of 5 with no violations, while the remaining ligands displayed up to 2 violations in the same. This is mainly due to poor aqueous solubility of these drugs. However, up to two violations of Lipinski’s rule are permitted.
In order to gain more insight, we performed glide-XP docking of these drugs (Table S1) within the prepared B-DNA. Docking results, along with the fitness score, experimental and predicted activities, are given in Table S15. Surprisingly, ligand S# 24, which has the highest fitness score of 3.00 on the pharmacophore model AADRR.4, did not turn out to be the best binder (glide score − 7.85 and Emodel − 110 kcal mol−1). On the other hand, ligand S# 3, having fitness score 2.53, has the second lowest predicted activity of 0.03 μM (Table S15) and appears to be the most potent amongst all 30 ligands, with appreciable binding parameters (Table S15). This is a derivative of Hoechst 33342, where the piperazine ring is replaced by a piperdinyl ring (Fig. S2). It has been experimentally shown to report good topoisomerase mediated DNA cleavage. The ratio of [Hoechst 33342]/[Drug] was 0.5, where [Hoechst 33342] and [Drug] are the concentrations that cause 50% cleavage of DNA in the presence of calf thymus topoisomerase I [35]. Table 3 shows comparative docking results for the three proposed drugs and a known inhibitor S# 3 within the AATT rich region of B-DNA.
It can be seen that the Gibbs energies of binding for the three proposed drugs are better than that for S# 3. Dasatinib stands out amongst all as far as the glide score, Emodel, and Gibbs energy of binding parameters are concerned (Table 3). Therefore, dasatinib (Z5) is proposed to be the most potent drug for DNA–topoisomerase I enzyme inhibition via binding through the DNA minor groove, and hence, further studies are performed with this drug.
Docking of dasatinib (Z5) into the human DNA–topoisomerase I cleavable complex
Aiming towards finding a drug that can catalytically inhibit the topoisomerase I enzyme via a lesser known mechanism, we performed molecular modeling studies on the ternary complex. The ternary complexes available in the protein data bank comprise of enzyme, nucleic acid, and an intercalator binding nucleic acid (PDB IDs: 1TL8 and 1K4T). Due to unavailability of the crystal structure of any ternary complex having a ligand binding the minor groove of DNA, we chose a crystal structure of a binary complex for our work. The crystal structure comprises 22 dodecamer sequenced DNA (5′-AAAAAGACTTAGAAAAATTTTT-3′)2 and human topoisomerase I (PDB ID: 1A36). The active site of the cleavage reaction consists of the DNA bases thymine (dT10 or dT−1) and adenine (dA11 or dA+1) and residues Arg488, Arg590, His632, and Tyr723 of the enzyme topoisomerase I. The phosphodiester bond between dT−1 and dA+1 is cleaved and relegated with the help of His632 and Tyr723 [66].

Further, it has been shown that, unlike other agents such as camptothecins and indolocarbazoles that interact at the cleavage site via intercalation between − 1 and + 1 base-pairs, bis-benzimidazoles derivatives display inhibition by attacking the minor groove. The binding of the drug occurs at a distal position, i.e., + 4 to + 8 base pairs downstream from the cleavage site [67].
These drugs, and especially Hoechst derivatives, prefer the AATT rich region of the minor groove. Since our entire focus was on developing the lead molecules that bind within DNA similar to Hoechst derivatives, we chose the AATT rich site within a 22-base pair duplex oligonucleotide ((5′-A−10A−9A−8A−7A−6G−5A−4C−3T−2T−1A+1G+2A+3A+4A+5A+6A+7T+8T+9T+10T+11T+12-3′)2). Making use of the results obtained in [67], we prepared the receptor grid by choosing the base pairs starting from + 6 to + 9 positions from the cleavage site. The box was made at the centroid of the residues dA16 to dT19 of one strand and dA104 to dT107 of the complementary strand (Fig. S3).
Glide-XP docking with a van der Waals radius scaling factor of 1.0 and charge scale factor of 1.0 was performed. This was done in order to soften the potential of the non-polar parts of the receptor. The glide score was found to be − 7.78 with Emodel − 82.64 kcal mol−1. The ligand seems to prefer the AAAA/TTTT sequence over the AATT sequence, as depicted by the four hydrogen bonds it forms with the adenines and thymines of both strands (Fig. 14; Fig. S3). Table S16 gives the hydrogen bond distances formed between Z5 and the DNA bases. Also, there exist some π–π stacking interactions between the adenine (dA14) and pyrimidinamine groups of Z5 (shown with green line in Fig. 14).
The Gibbs energy of binding for the complex was found to be − 33.65 kcal mol−1, suggesting stable complex formation. This was also verified by calculating the OPLS-2005 energies of the minimized enzyme–DNA binary complex (PDB: 1A36), dasatinib (Z5), and the ternary complex of Z5-DNA–topoisomerase I obtained after XP-docking, using the following Eq. (1).
The energies of the binary complex, ligand Z5, and ternary complex were found to be − 9581.6, − 52.7, and − 10,173.6 kcal mol−1, respectively. The stabilization energy was thus calculated as − 539.3 kcal mol−1. Though the complex formed is stable, many aspects on the interaction still need to be explored. Therefore, further analysis on the binding mode and mechanism of inhibitory action of the drug Z5 on the DNA–topoisomerase I complex is the purview of our next work.
Conclusions
Molecular modeling studies were performed to develop a predictive CPH and used for alignment in atom-based 3D-QSAR studies. A five-point CPH AADRR.4, with two hydrogen bond acceptors, one hydrogen bond donor, and two aromatic features, for pharmacophore-based alignment of molecules was derived using PHASE. This hypothesis was selected from a pool by correlating the observed and estimated activity for the training and test set molecules using PLS analysis. The QSAR model, so generated, showed a reasonable predictive Q2 value of 0.465. The contour maps of the models were analyzed to give structural insight for activity improvement of future novel topoisomerase I inhibitors. The CPH also provides a powerful template for virtual screening and design of new DNA directed topoisomerase poisons. Virtual screening and docking methodologies were utilized to find lead molecules that bind in a manner similar to bis- and ter-benzimidazoles. Three drugs, namely dasatinib, lapatinib, and novobiocin, are proposed to have the best DNA binding properties. Dasatinib, having the optimum fitness and binding score with predicted activity of 2.52 μM, shows stabilization towards the DNA–topoisomerase I cleavable complex. Unlike Hoechst derivatives, dasatinib prefers AAAA/TTTT over the AATT region of double stranded DNA in complexation with the enzyme. The mechanism of action of the drug is complex and requires various methodologies for complete understanding. However, this study enables us to explore a new path in the development of novel anticancer agents.
Notes
PHASE, Version 3.0, Schrödinger, LLC, NY2009
References
Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF (1996) Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev 10:2564–2576
Lee MP, Brown SD, Chen A, Hsieh TS (1993) DNA topoisomerase I is essential in Drosophila melanogaster. Proc Natl Acad Sci U S A 90:6656–6660
Morham SG, Kluckman KD, Voulomanos N, Smithies O (1996) Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol Cell Biol 16:6804–6809
Wang JC (1996) DNA topoisomerases. Annu Rev Biochem 65:635–692
Zhang CX, Chen AD, Gettel NJ, Hsieh TS (2000) Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev Biol 222:27–40
Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413
Champoux JJ (1990) Mechanistic aspects of type-I topoisomerases. In: Cozzarelli NR, Wang JC (eds) DNA topology and its biological effects, vol 20. Cold Spring Harbor Laboratory Press, Plainview, New York, pp 217–242
Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3:430–440
Hertzberg RP, Hecht SM, Caranfa MJ (1989) On the mechanism of topoisomerase I inhibition by camptothecin: evidence for binding to an enzyme-DNA complex. Biochemistry 28:4629–4638
Hertzberg RP, Busby RW, Caranfa MJ, Holden KG, Johnson RK, Hecht SM, Kingsbury WD (1990) Irreversible trapping of the DNA-topoisomerase I covalent complex. Affinity labeling of the camptothecin binding site. J Biol Chem 265:19287–19295
Hsiang YH, Hertzberg RP, Hecht SM, Liu LF (1985) Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem 260:14873–14878
Liu LF (1989) DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem 58:351–375
Liu LF, D’Arpa P (1992) Topoisomerase-targeting antitumor drugs: mechanisms of cytotoxicity and resistance. Important Adv Oncol 79–89
Pommier Y, Kohlhagen G, Kohn KW, Leteurtre F, Wani MC, Wall ME (1995) Interaction of an alkylating camptothecin derivative with a DNA base at topoisomerase I-DNA cleavage sites. Proc Natl Acad Sci U S A 92:8861–8865
Porter SE, Champoux JJ (1989) The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res 17:8521–8532
Potmesil M, Giovanella BC, Wall ME, Liu LF, Silber R, Stehlin JS, Wani MC, Hochster H (1993) Preclinical and clinical development of DNA topoisomerase I inhibitors in the United States. In: Andoh T, Ikeda H, Oguro M (eds) Molecular biology of DNA topoisomerases and its application to chemotherapy. CRC Press, Boca Raton, pp 301–311
Xu Z, Li T-K, Kim JS, LaVoie EJ, Breslauer KJ, Liu LF, Pilch DS (1998) DNA minor groove binding-directed poisoning of human DNA topoisomerase I by terbenzimidazoles. Biochemistry 37:3558–3566
Bailly C (2000) Topoisomerase I poisons and suppressors as anticancer drugs. Curr Med Chem 7:39–58
Pommier Y (1993) DNA topoisomerase I and II in cancer chemotherapy: update and perspectives. Cancer Chemother Pharmacol 32:103–108
Bansal S, Sinha D, Singh M, Cheng B, Tse-Dinh Y-C, Tandon V (2012) 3,4-Dimethoxyphenyl bis-benzimidazole, a novel DNA topoisomerase inhibitor that preferentially targets Escherichia coli topoisomerase I. J Antimicrob Chemother 67:2882–2891
Ranjan N, Story S, Fulcrand G, Leng F, Ahmad M, King A, Sur S, Wang W, Tse-Dinh Y-C, Arya DP (2017) Selective inhibition of Escherichia coli RNA and DNA topoisomerase I by Hoechst 33258 derived mono- and bisbenzimidazoles. J Med Chem 60:4904–4922
Kim JS, Sun Q, Yu C, Liu A, Liu LF, LaVoie EJ (1998) Quantitative structure-activity relationships on 5-substituted terbenzimidazoles as topoisomerase I poisons and antitumor agents. Bioorg Med Chem 6:163–172
Mekapati SB, Hansch C (2001) Comparative QSAR studies on bibenzimidazoles and terbenzimidazoles inhibiting topoisomerase I. Bioorg Med Chem 9:2885–2893
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Klebe G (1998) Comparative molecular similarity indices analysis, CoMSIA. Perspect Drug Disc Des 12:87–104
Winkler D (2011) Modelling topoisomerase I inhibition by minor groove binders. Bioorg Med Chem 19:1450–1457
Beerman TA, McHugh MM, Sigmund R, Lown JW, Rao KE, Bathini Y (1992) Effects of analogs of the DNA minor groove binder Hoechst 33258 on topoisomerase II and I mediated activities. Biochim Biophys Acta 1131:53–61
Chen AY, Chiang Y, Gatto B, Liu LF (1993) DNA minor groove-binding ligands: a different class of mammalian DNA topoisomerase I inhibitors. Proc Natl Acad Sci U S A 90:8131–8135
Pilch DS, Xu Z, Sun Q, LaVoie EJ, Liu LF, Breslauer KJ (1997) A terbenzimidazole that preferentially binds and conformationally alters structurally distinct DNA duplex domains: a potential mechanism for topoisomerase I poisoning. Proc Natl Acad Sci U S A 94:13565–13570
Jin S, Kim JS, Sim S-P, Liu A, Pilch DS, Liu LF, LaVoie EJ (2000) Heterocyclic bibenzimidazole derivatives as topoisomerase I inhibitors. Bioorg Med Chem Lett 10:719–723
Kim JS, Gatto B, Yu C, Liu A, Liu LF, LaVoie EJ (1996) Substituted 2,5’-bi-1H-benzimidazoles: topoisomerase I inhibition and cytotoxicity. J Med Chem 39:992–998
Kim JS, Yu C, Liu A, Liu LF, LaVoie EJ (1997) Terbenzimidazoles: influence of 2″-, 4-, and 5-substituents on cytotoxicity and relative potency as topoisomerase I poisons. J Med Chem 40:2818–2824
Rangarajan M, Kim JS, Jin S, Sim S-P, Liu A, Pilch DS, Liu LF, LaVoie EJ (2000) 2″-Substituted 5-phenylterbenzimidazoles as topoisomerase I poisons. Bioorg Med Chem 8:1371–1382
Rangarajan M, Kim JS, Sim S-P, Liu A, Liu LF, LaVoie EJ (2000) Topoisomerase I inhibition and cytotoxicity of 5-bromo-and 5-phenylterbenzimidazoles. Bioorg Med Chem 8:2591–2600
Sun Q, Gatto B, Yu C, Liu A, Liu LF, LaVoie EJ (1994) Structure activity of topoisomerase I poisons related to Hoechst 33342. Bioorg Med Chem Lett 24:2871–2876
Sun Q, Gatto B, Yu C, Liu A, Liu LF, LaVoie EJ (1995) Synthesis and evaluation of terbenzimidazoles as topoisomerase I inhibitors. J Med Chem 38:3638–3644
Huang CC (1974) Cytogenetic study of human lymphoid T-cell lines derived from lymphocytic leukemia. J Natl Cancer Inst 53:655–660
Wold S, Ruhe A, Wold H, Dunn III WJ (1984) The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J Sci Stat Comp 5:735–743
Wold H (1985) Partial least squares. In: Kotz S, Johnson NL (eds) Encyclopedia of statistical sciences, vol 6. Wiley, New York, pp 581–591
Wold S, Johansson E, Cocchi M (1993) PLS-partial least squares projections to latent structures. In: Kubinyi H (ed) 3D QSAR in drug design: theory, method and applications. ESCOM Science Publishers, Leiden, pp 523–550
Dixon SL, Smondyrev AM, Rao SN (2006) PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 67:370–372
Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Freisner RA (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671
Teng M-K, Usman N, Frederick CA, Wang AH-J (1988) The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res 16:2671–2690
Issar U, Kumari T, Arora R, Kakkar R (2017) Conformational properties of DNA minor groove binder Hoechst 33258 in gas phase and in aqueous solution. Comp Theo Chem 1113:32–41
Duffy EM, Jorgensen WL (2000) Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J Am Chem Soc 122:2878–2888
Jorgensen WL, Duffy EM (2000) Prediction of drug solubility from Monte Carlo simulations. Bioorg Med Chem Lett 10:1155–1158
Issar U, Kumari T, Kakkar R (2015) Assessment of molecular binding of Hoechst 33258 analogues into DNA using docking and MM/GBSA approach. J Comp Sci 10:166–177
Kubinyi H (2008) Comparative molecular field analysis (CoMFA). In: Gasteiger J (ed) Handbook of chemoinformatics—from data to knowledge. Wiley-VCH Verlag GmbH, Weinheim, pp 1555–1574
Ray S (2012) QSAR modeling of antitmycobacterial activities of N-benzylsalicylamides and N-benzylsalicylthioamides derivatives against Mycobacterium kansasii CNCTC My (6509/96) using stepwise and PLS method. Int J ChemTech Res 4:41–47
Tenenhaus M, Vinzi VE, Chatelin Y-M, Lauro C (2005) PLS path modeling. Comp Stat Data Anal 48:159–205
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shaw DE, Shelley M, Perry JK, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 49:6177–6196
Giménez BG, Santos MS, Ferrarini M, Fernandes JP (2010) Evaluation of blockbuster drugs under the rule-of-five. Pharmazie 65:148–152
Zhang MQ, Wilkinson B (2007) Drug discovery beyond the ‘rule-of-five’. Curr Opin Biotechnol 18:478–488
Matijević-Sosa J, Cvetnić Z (2005) Antimicrobial activity of N-phthaloylamino acid hydroxamates. Acta Pharma 55:387–399
Michot J-M, Seral C, van Bambeke F, Mingeot-Leclercq M-P, Tulkens PM (2005) Influence of efflux transporters on the accumulation and efflux of four quinolones (ciprofloxacin, levofloxacin, garenoxacin, and moxifloxacin) in J774 macrophages. Antimicrob Agents Chemother 49:2429–2437
Vandenberg JI, Walker BD, Campbell TJ (2001) HERG K+ channels: friend and foe. Trends Pharmacol Sci 22:240–246
Aronov AM (2005) Predictive in silico modeling for hERG channel blockers. Drug Discov Today 10:149–155
Steinberg M (2007) Dasatinib: a tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. Clin Ther 29:2289–2308
Bellon S, Parsons JD, Wei Y, Hayakawa K, Swenson LL, Charifson PS, Lippke JA, Aldape R, Gross CH (2004) Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob Agents Chemother 48:1856–1864
Burlison JA, Neckers L, Smith AB, Maxwell A, Blagg BS (2006) Novobiocin: redesigning a DNA gyrase inhibitor for selective inhibition of hsp90. J Am Chem Soc 128:15529–15536
Lanoot B, Vancanneyt M, Cleenwerck I, Wang L, Li W, Liu Z, Swings J (2002) The search for synonyms among streptomycetes by using SDS-PAGE of whole-cell proteins. Emendation of the species Streptomyces aurantiacus, Streptomyces cacaoi subsp. cacaoi, Streptomyces caeruleus and Streptomyces violaceus. Int J SysT Evol Microbiol 52:823–829
Burris IIIHA (2004) Dual kinase inhibition in the treatment of breast cancer: initial experience with the EGFR/ErbB-2 inhibitor lapatinib. Oncologist 3:10–15
Higa GM, Abraham J (2007) Lapatinib in the treatment of breast cancer. Future Drugs 7:1183–1192
Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, Keith BR, Murray DM, Knight WB, Mullin RJ, Gilmer TM (2001) The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther 1:85–94
Stewart LM, Redinbo MR, Qiu XY, Hol WG, Champoux JJ (1998) A model for the mechanism of human topoisomerase I. Science 279(5356):1534–1541
Khan QA, Pilch DS (2007) Topoisomerase I-mediated DNA cleavage induced by the minor groove-directed binding of bibenzimidazoles to a distal site. J Mol Biol 365:561–569
Acknowledgments
U.I. thanks the University Grants Commission (UGC), and R.A. and T.K. thank the Council of Scientific and Industrial Research (CSIR) for junior and senior research fellowships.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 1284 kb)
Rights and permissions
About this article
Cite this article
Issar, U., Arora, R., Kumari, T. et al. Combined pharmacophore-guided 3D-QSAR, molecular docking, and virtual screening on bis-benzimidazoles and ter-benzimidazoles as DNA–topoisomerase I poisons. Struct Chem 30, 1185–1201 (2019). https://doi.org/10.1007/s11224-018-1257-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-018-1257-3