Abstract
Salinity stress is a major environmental factor impeding barley productivity on a global scale. Enhancing salt tolerance in barley is crucial for maintaining crop yield and agricultural sustainability. This study aimed to elucidate the genetic underpinnings of salt tolerance in barley, focusing on the identification of key genomic loci and quantitative trait nucleotides (QTNs) associated with stress resilience, particularly through the modulation of antioxidant defense mechanisms. A diverse set of barley lines was subjected to salinity stress conditions, and a genome-wide association study (GWAS) was conducted. Ten morphophysiological parameters related to salt stress tolerance, such as antioxidant enzyme activities, were evaluated. This study revealed distinct genomic loci and QTNs intricately linked with salt tolerance in barley. These genetic markers were found to influence the plant’s antioxidant defenses, including enzymes like superoxide dismutase, catalase, and ascorbate peroxidase, which play pivotal roles in mitigating oxidative damage under salinity stress after the application of Se NPs. For instance, significant SNP (G:A) on chromosome 7H at position (521,369,195- 521,370,360 bp). Within this region, a gene mapped as peroxidase, HORVU.MOREX.r3.7HG0718950, was detected for SS_Se, SL_C, and SL_Se, suggesting the essential role of this gene in regulating the resilience of plant responses to salinity stress after application of selenium nanoparticles. Our findings provide a deeper understanding of the complex genetic architecture of salt tolerance in barley, highlighting the critical role of specific genomic regions in controlling antioxidant responses under salinity stress. The identified loci and QTNs serve as valuable genetic resources for the development of salt-tolerant barley cultivars, contributing to global food security and agricultural resilience in saline-prone environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity stress is a major environmental factor that adversely affects plant growth and productivity. Understanding the impact of salinity on barley is crucial for developing effective management strategies (Kamran et al. 2020; Thabet et al. 2021b; Tobe et al. 2003). Elevated levels of salt in the soil can greatly diminish the speed and consistency of seed germination in barley (Rasouli et al. 2024). During the initial stages of growth, plants are highly vulnerable to salinity, which can result in inhibited growth and inadequate root formation (Nikolić et al. 2023). High levels of sodium and chloride ions can be harmful to plant cells, as they interfere with cellular functions and upset the balance of ions. Salinity can impede the absorption of vital elements such as potassium and calcium, hence hindering plant growth even more (Atta et al. 2023). Plants can display diminished leaf area and decreased plant height in saline environments. Increased salinity can cause alterations in the structure of the root system, which can impact the plant’s capacity to uptake water and essential nutrients (Thabet and Alqudah 2023). Furthermore, the impact of salinity stress typically leads to a substantial decrease in grain output as a result of the combined influence on both growth and reproductive development. Munns et al. (2020) documented that salt stress hampers the growth and development of plants through the induction of osmotic and ion stress. Osmotic stress arises from an elevation in soluble salts, while ion toxicity results from the presence of exchangeable sodium (Na+) during salinity stress (Thabet et al. 2021a). The effects of salt stress on plants exhibit significant variation, contingent upon the specific type and concentration of salt employed, ambient conditions, plant species, different cultivars within a species, and the developmental phases of the plant (Tabur et al. 2021). Plants evolve intricate ways to endure salt. Plant resistance to salinity can be categorized into three primary types: osmotic stress tolerance, exclusion of Na+ or Cl−, and tissue tolerance to accumulated Na+ or Cl− (Munns and Tester 2008). Abiotic stressful circumstances, such as salt, lead to the generation of harmful free radicals, specifically reactive oxygen species (ROS), which harm crop physiological processes, growth, and yield, resulting in reduced yield (Thabet et al. 2021a). In addition, harmful free radicals induce harm to cell membranes, oxidize proteins, and cause DNA damage, ultimately leading to cell death (Feng et al. 2015). Plants have developed regulatory mechanisms to maintain the stability of their life processes in response to damage caused by ROS. Hence, the involvement of active oxygen-scavenging enzymes, such as superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and other crucial enzymes, is crucial in this process (Zhang et al. 2021). To address the issue of salt stress, it is crucial to evaluate different crop types for their ability to tolerate salinity and enhance their levels of salinity tolerance (Wu et al. 2015). While the creation of crop varieties that can tolerate salt is advantageous in certain situations, the process of cultivating and evaluating these varieties is laborious typically taking around 8–10 years (Noreen et al. 2021). On the other hand, the use of certain organic or inorganic compounds, known as the shot-gun technique, could be a viable strategy to mitigate the effects of salinity stress and safeguard various crops like wheat and barley (Agbolade et al. 2019).
Selenium (Se) nanoparticles have become prominent in the agricultural industry due to their distinct characteristics and advantages (Song et al. 2023). Selenium nanoparticles (Se NPs) have the potential to be utilized for crop fortification, suggesting increases in the selenium level in crops, which is crucial for human well-being (Thabet and Alqudah 2023). They assist plants in managing diverse stressors such as water scarcity, high salt levels, and pollution from heavy metals. This is a result of their antioxidant capabilities, which counteract the damage caused by oxidative stress in plants (Bano et al. 2021). Nanoparticles can affect the absorption of nutrients and the chemical processes in plants, resulting in enhanced growth and increased crop production (Samynathan et al. 2023). They can enhance the dietary value and shelf life of agricultural produce, hence improving its quality. Selenium nanoparticles can transform hazardous forms of elements in the soil into less damaging forms, hence decreasing soil toxicity (Song et al. 2023). They provide an environmentally conscious approach to agriculture as a substitute for artificial pesticides and fertilizers. It is essential to comprehend the natural and physiological consequences of selenium nanoparticles (Gudkov et al. 2020). Prudent regulation is necessary to ensure their safe utilization. Continued research is necessary to comprehensively comprehend the processes by which they operate and to enhance their utilization in agriculture (Devi et al. 2023). Se NPs have significant potential to revolutionize agricultural methods. Due to their capacity to promote vegetative growth, provide resistance to infections, and increase crop productivity, while also being renewable, they are considered a great asset in the field of sustainability in the agricultural sector (Pyrzynska and Sentkowska 2022). Nevertheless, continuous study and meticulous control are needed to fully exploit their complete capabilities while guaranteeing well-being (Zohra et al. 2021).
Barley (Hordeum vulgare L.) is a widely grown cereal crop that ranks fourth in global popularity. It is planted for multiple reasons, including consumption as food, livestock sustenance, and numerous brewing and malting processes globally (Meints and Hayes 2019; Thabet et al. 2020). Although barley is a biotic and abiotic stress-tolerant crop, its yield suffers greatly in saline environments (Noreen et al. 2021; Thabet et al. 2021a). Barley, despite its ability to withstand both biotic and abiotic stress, experiences significant yield reduction in saline settings (Thabet et al. 2023). Barley cultivars display various degrees of salinity tolerance at the molecular level, which is a crucial factor to consider when growing them in saline regions. The primary objective of breeding and genetic engineering endeavors is to cultivate barley cultivars that possess heightened resistance to salt (Thabet et al. 2021b). On the whole, salinity presents a substantial obstacle to the cultivation of barley, impacting all aspects of plant development, ranging from germination to crop production. Salt stress can significantly reduce the productivity of barley crops by causing physiological, biochemical, and morphological alterations (Thabet et al. 2022b). Gaining a comprehensive understanding of these effects is crucial for the development of management strategies and breeding initiatives focused on enhancing barley’s ability to withstand salinity (Thabet et al. 2018). Given the significance of barley as a nutritional crop, we propose that applying selenium nanoparticles (Se NPs) to the leaves can enhance growth and metabolic processes, leading to an overall increase in barley output when exposed to salinity stress.
Quantitative traits exhibit variation at specific locations on chromosomes known as quantitative trait loci (QTLs). Al-Tamimi et al. (2016) discovered many QTLs associated with shoot and root growth in the presence of salt stress. These QTLs indicate the presence of genetic loci that control water absorption and nutrient transportation, enabling plants to withstand high salinity levels. Within saline environments, there are significant QTLs that have an impact on the absorption, movement, and storage of ions such as sodium (Na+), potassium (K+), and chloride (Cl−). QTLs can influence the expression or function of ion transporters and channels. During periods of stress, the quantitative trait loci (QTLs) responsible for growth duration have an impact on the processes of tillering, flowering, and maturity, as stated by Alqudah et al. (2014). QTL mapping in barley has discovered specific regions in the genome that influence several characteristics, such as salt tolerance (Thabet et al. 2021a). Recently, Thabet et al. (2022a) have discovered quantitative trait loci (QTLs) associated with antioxidant components in barley when subjected to salt stress. For instance, there is a notable single-nucleotide polymorphism (SNP) A:C located on chromosome 7H that controls the expression of SOD_S and APX_S. Interestingly, Thabet and Alqudah (2023) discovered agricultural quantitative trait loci (QTNs) following the application of selenium nanoparticles to the leaves under slightly saline conditions. Hence, our work sought to examine the genetic connections associated with the foliar application of selenium nanoparticles (Se NPs) during the vegetative stage, which improves stress tolerance in barley under salt stress. Furthermore, it investigates the crucial genomic areas linked to antioxidant chemicals as well as agronomical and yield-related characteristics.
Materials and Methods
Plant Material
In the present investigation, a total of 138 spring barley accessions from various geographical origins were examined, encompassing 54 cultivars and 84 landraces. The collection was divided into 63 two-rowed and 75 six-rowed. The genotyping-by-sequencing (GBS) method was employed to genotype all the accessions, resulting in the identification of 19,276 single-nucleotide polymorphisms (SNPs), as outlined by Milner et al. (2019). The diverse collection utilized in this investigation was obtained from the German ex situ IPK-Gatersleben GeneBank (Table S1).
Experimental Setup
Over 2 years, 138 spring barley accessions were grown at the University of Fayoum Experimental Station (29°11′20.36″N, 30°10′06.45″E). Experimental planting began on November 25 and ended on April 20 (2022/2023). We collected soil samples from the experimental farm site before each trial to analyze physicochemical and fertility status (Table S2) using (Jackson 1958) techniques. As indicated in Dahnke and Whitney (1988), these findings revealed that the experimental soil had a high salt content, as shown by an ECe value of 8.51–8.61 dS m−1 for both seasons (Table S2). The barley seeds underwent surface sterilization using sodium hypochlorite for a duration of 10 min. Subsequently, they were rinsed with distilled water and left to dry in the air for a period of 2 h. A total of five rows, each measuring 3 m in length, were planted with a spacing of 0.23 m between the rows. Each row was divided into hills, with four seeds put in each hill. The distance between the hills was maintained at 15–20 cm. There were two robust seedlings on each hill prior to the initial watering. Prior to field planting, the soil was enriched with 280 kg per hectare of (NH4)2SO4 (20% nitrogen) and 350 kg per hectare of Ca2+ superphosphate (15.5% phosphorus pentoxide). In a fully randomized design, a concentration of 1-mM selenium nanoparticles (Se NPs) was administered as a foliar treatment, while distilled water was used as a control. Three replicas were utilized. Sigma Aldrich Co. offered spherical selenium nanoparticles (Se NPs) with a diameter ranging from 10 to 45 nm, a surface area of 30 to 50 m2/g, a density of 3.89 g/cm3, and a purity of 99.5%. Following a period of 25 days, the plants were subjected to two rounds of spraying with distilled water (as a control) and 1-mM Se NPs (the second and third sprays) when they reached the ages of 35 and 45 days, respectively.
Morphological Traits
Spike length (SL), measured in centimeters from the spike’s base to its tip, excluding the awns, was one of five phenotypic features analyzed. The number of spikelets per spike (SS) counted each spike’s produced spikelets. Grain count (GS) requires counting each spike’s grains. The weight of grains per spike (WGS) was recorded in grams, and thousand kernel weight (TKW) was determined by counting 100 seeds from the collective seed batch of each genotype using a high-speed automatic seed counter (Model-IC-VA, Aidex-Japan) and amplifying the subsequent grain weights by 10.
Antioxidant Enzymes Determination
The enzymes were extracted using the method outlined by Garratt et al. (2002). The enzymes ascorbate peroxidase (APX), glutathione reductase (GR), catalase (CAT), and superoxide dismutase (SOD) were evaluated, as stated by Maehly and Chance (1954), Sairam et al. (1997), and Rao et al. (1998). Nonenzymatic antioxidants ascorbic acid and glutathione were assayed as described in methods by De Kok et al. (1986).
Genome-Wide Association Scan and Candidate Genes
The barley population underwent genotyping utilizing genotyping by sequencing (GBS). The GWAS study was conducted using the FarmCPU technique, which assessed the genome containing 19,000 single-nucleotide polymorphisms (SNPs) (Milner et al. 2019). The analysis also included the evaluation of the best linear unbiased estimators (BLUEs) for the evaluated variables. The FarmCPU statistical model is a robust approach for GWAS analysis that allows us to effectively manage both false-positive and false-negative correlations (Alqudah et al. 2020). The physical positions of SNP markers were determined using the Morex genome sequence v2 (Monat et al. 2019). The genome-wide significance criterion for the GWAS was established as P < 0.0001 (equivalent to − log10 (P) ≥ 3) for all traits. The genome-wide pairwise estimates of linkage disequilibrium (LD) were computed as the squared correlation between pairs of SNPs (r2). The study conducted by Alqudah et al. (2020) involved calculating the LD block for a given physical distance, which included the strongest linked markers. This was done to determine the physical interval of the related quantitative nucleotide region (QNR). The identified physical interval was then used in subsequent analysis to identify candidate genes.
Using the BARLEX IPK database, we identified the candidate genes and their gene annotations based on the physical positions of SNP markers (https://apex.ipk-gatersleben.de/apex/f?p=284:10). Moreover, expression profiles for our potential candidates were measured using the RNA-Seq expression database at different developmental stages in barley according to the OPEN-ACCESS version of the Genevestigator program. The program includes the transcriptome and expression of barley genes from different tissues and organs such as anatomy and development from controlled and stressed conditions.
Results
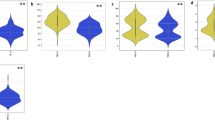
The average genotypic values for SOD, CAT, APX, GR, AsA, and GSH were measured as 0.21, 0.058, 0.97, 0.38, 29.16, and 34.86 μmol mg−1 protein, respectively (Table S3). The average genotypic values for SOD, CAT, APX, GR, AsA, and GSH were measured as 0.67, 0.20, 3.76, 1.18, 101.10, and 104.4 μmol mg−1 protein, respectively, after the application of Se NPs to the leaves. These data may be found in Table S4. Enzymatic antioxidants, specifically SOD, CAT, APX, and GR, exhibited a significant increase of 69%, 71%, 74%, and 67%, respectively, when treated with Se NPs relative to the control, as shown in Fig. 1. Nonenzymatic antioxidants ascorbic acid (AsA) and glutathione (GSH) increased by 71% and 67%, respectively, as illustrated in Fig. 1. All enzymatic and nonenzymatic antioxidants showed substantial natural variation in both treatments, as indicated by the data in Table S5 and Table S6. The heritability values varied from 69.22 for AsA to 96.93 for SOD in normal settings and from 90.92 for AsA to 98 for GSH following Se NPs treatment (Table S4). The minimum, maximum, and mean values for each agronomic trait across all genotypes can be found in Table S2 and Table S3. The mean performance of all yield features was significantly higher under Se NPs compared to the control circumstances, as shown in Table S5 and Table S6. The average values for the yield attributes SL, SS, GS, WGS, and TKW were 8.21, 9.49, 43.33, 1.37, and 39.43, respectively, as indicated in Figure and Table S1. The average genotypic values for SL, SS, GS, WGS, and TKW were measured as 12.52, 14.82, 52.93, 3.01, and 57.32, respectively, under Se NPs treatment. These results may be found in Table S5 and Table S6. Significantly, all morphological and yield parameters exhibited increment by 34%, 36%, 18%, 54%, and 31% for SL, SS, GS, WGS, and TKW, respectively, after Se NPs treatment in contrast to the control (salt stress), as depicted in Fig. 2. The broad-sense heritability (H2) values varied from 94.62 for TKW_C to 99.3 for GS_C, as shown in Table S6. The H2 was observed in GS (99.55), followed by SL (98.81), while the lowest H2 was found in TKW (95.17) (Table S6).
Correlation Analyses
Pearson’s correlations across attributes, determined by the mean of all accessions for each treatment condition, showed a significant association among all examined traits for both treatments. Strong negative correlations were found between nonenzymatic antioxidants (AsA and GSH) and enzymatic antioxidants (SOD, CAT, APX, and GR) in both control and Se NPs conditions. This indicates that nanoparticles play a crucial role in improving antioxidant defenses, which in turn promotes plant resilience in environments with high salt levels (Fig. 3). Simultaneously, there were notable connections observed among all the examined agronomic characteristics (SL, SS, GS, WGS, and TKW) that exhibited positive correlations in the two scenarios. This indicates the existence of shared genetic factors that influence all agricultural features and the process of grain filling in response to salt stress (Fig. 3).
Genome-Wide Association Scan (GWAS)
The GWAS analysis revealed 653 SNPs that showed significant associations with 10 characteristics and met the criteria of − Log10 (p) ≥ 3.0 under both control and Se NPs treatments (Table S7). Among the 653 SNPs, we detected 77, 51, 36, and 28 SNPs each connected with antioxidant components, specifically CAT_C, GSH_C, SOD_C, CAT_Se, SOD_Se, and GSH_Se. In contrast, less than 10 SNPs were linked to each of the other attributes mentioned (Table S6 and Figure S1). A total of 86 single-nucleotide polymorphisms (SNPs) with a p-value of -log10 ≥ 3 were found to be substantially associated with all agronomic characteristics across all chromosomes (Table S7). The largest number of markers was observed for SL_C (101 SNPs), after SL_C and SL_Se (64 SNPs each), while 40 SNPs were linked to each of the other variables (Table S7 and Figure S2). Highly significant SNP markers (− log10 (p-value) > 11) have been identified and correlated with GS_Se (p = 2.22E-17) on chr 2H at position 572,438,008 bp; SOD_C (p = 2.29E-15) on chr 5H at position 536,154,191bp; and GS_C (p = 2.040E-13) on chr 2H at position 572,438,008 bp (Table S7). This research identified 30 genomic regions through marker-trait associations (MTAs) and LD. These regions, located on chromosomes 1H, 2H, 3H, 5H, 6H, and 7H, consist of 24 potential candidate genes. These genes were found to regulate antioxidant components in response to salt exposure as described in Table S8 and Table S9. At position (6,695,881–6,697,565 bp) on chromosome 1H, a gene called HORVU.MOREX.r3.1HG0003270 was discovered. This gene encodes a protein from the Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family. It was found at the specific position of 6,697,338 bp, which is associated with all antioxidant components in both treatments, as shown in Fig. 4. Moreover, there is a consistent region with a highly significant SNP (A:G) at position 8,959,918 base pairs. The gene HORVU.MOREX.r3.1HG0004230, which is designated as an elongation factor, has been found inside the aforementioned region (Fig. 4). Furthermore, genetic loci with substantial LD were identified on chromosome 1H. Specifically, SNP with a (T:C) allele was found at position 58,238,555–58,239,948 bp. The gene HORVU.MOREX.r3.1HG0018170, which is known as polyadenylate-binding protein 1-B-binding protein, was identified in the GS_Se, WGS_C, and WGS_Se (Fig. 4). Additionally, the receptor-like protein kinase plays a role in salt tolerance. The HORVU.MOREX.r3.6HG0596150 gene was identified at position (374,041,230–374,043,818 bp) of the (T:C) SNP on chromosome 6H. This gene is involved in antioxidants in both circumstances (Fig. 4), indicating its vital role in controlling plant responses and determining the final grain yield in barley. A significant SNP (G:A) was found on chromosome 7H at position 521,369,195–521,370,360 bp. Within this region, a gene called peroxidase, HORVU.MOREX.r3.7HG0718950, was identified for SS_Se, SL_C, and SL_Se (Fig. 4). This suggests that this gene plays a crucial role in restricting the plant’s ability to respond to salt stress after Se NPs treatment (Fig. 5).
Locus zoom on regional plot colocalization of highly associated QTNs that control the variation of the studied traits including ascorbate peroxidase (APX), glutathione reductase (GR), and ascorbic acid (AsA), glutathione (GSH), weight grains per spikes (WGS), and grains per spikes (GS) under both control (c) and selenium nanoparticles (Se NPs). The x-axis shows the chromosomes and the QTN order. The y-axis shows the − log10 (p-value) for each QTN marker
Differential gene expression of the HORVU.MOREX.r3.7HG0718950 gene that encodes peroxidase at different developmental stages in barley. (a) Up- and/or downregulation underlying the natural phenotypic variation in several organs of barley. (b) Up- and/or downregulation underlying the natural phenotypic variation under abiotic stress conditions, including salt stress
Discussion
Salinity stress is one of the most detrimental factors affecting the growth, development, and yield of barley, a major cereal crop. Under salinity stress, plants experience osmotic and ionic stress, leading to adverse effects such as osmotic imbalance, nutrient deficiency, and oxidative stress (Munns and Tester 2008; Thabet and Alqudah 2023). In this study, all agronomical and yield traits showed increment for SL, SS, GS, WGS, and TKW, respectively, under Se NPs treatment compared to salt stress (control). These findings showed agreement with Thabet and Alqudah (2023), highlighting the essential role of selenium nanoparticles in regulating the resilience of plant responses to salinity stress. In addition, it could be a valid selection criterion for assessing salinity stress tolerance in barley breeding programs.
Salinity stress affects a range of morphological and physiological traits in barley, and understanding the genetic basis of these traits is crucial for breeding programs aimed at enhancing salinity tolerance. Here are key QTLs associated with morphological and physiological traits in barley under salt stress. This study showed 86 SNPs (− log10 p-value ≥ 3) for all agronomic traits that were significantly detected on all chromosomes. Interestingly, highly significant associations (− log10 (p-value) > 11) were discovered on chromosomes that were found to be highly associated with GS_Se on chr 5H. Our findings also showed agreement with Thabet and Alqudah (2023), suggesting that these QTLs are fundamental for marker-assisted selection (MAS) and for the design of strategies aimed at improving the salinity tolerance of barley.
Candidate Genes
Based on GWAS analysis, a gene encoding Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein (HORVU.MOREX.r3.1HG0003270) on chromosome 1H was found at 6,697,338 bp of the top SNP (A:C) associated with enzymatic and nonenzymatic antioxidants_ related traits under both treatments. The Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase (C2GnT) family of enzymes is involved in complex glycan biosynthesis. These enzymes modify the structure of glycoproteins by catalyzing the transfer of N-acetylglucosamine (GlcNAc) in a beta-1,6 linkage to a mannose core, a process integral to the formation of complex or hybrid N-glycans (Bierhuizen et al. 1995). In plants, this process is vital for protein folding, stability, and function, which are crucial under stress conditions such as salinity (Strasser 2016). Glycosylation, particularly N-glycosylation, is a significant posttranslational modification that affects protein conformation, distribution, stability, and function. Under salinity stress, the demand for proper protein folding and stabilization is heightened due to the increased production of stress-related proteins (Solá and Griebenow 2009). The role of C2GnT enzymes in this context is to ensure the structural integrity and functionality of these proteins, particularly those involved in stress response pathways (Nagae et al. 2020). For example, glycosylated proteins in the plasma membrane can affect ion transport mechanisms which are essential for maintaining ion homeostasis in a saline environment (Kang et al. 2008). Research has suggested that complex N-glycans may have roles in signaling processes related to plant stress responses. The modification of glycoproteins by C2GnT enzymes could influence the interactions between hormones and their receptors or other key signaling molecules, thereby affecting gene expression patterns necessary for salinity tolerance (Saint-Jore-Dupas et al. 2006). Moreover, properly glycosylated proteins, facilitated by the action of C2GnT enzymes, are crucial for the stability and function of antioxidant enzymes involved in ROS scavenging. This helps in mitigating oxidative damage during salinity stress (Apel and Hirt 2004; Hasanuzzaman et al. 2021).
Similarly, the reliable region with high significant SNP (A:G) at position 8,959,918 bp was mapped on chromosome 1H. Within this region, HORVU.MOREX.r3.1HG0004230 gene was annotated as elongation factor was identified. Elongation factors play crucial roles in protein synthesis within cells, and their functions can be particularly important for plants under environmental stress, such as salinity (Shin et al. 2009). Salinity stress is known to affect plant growth and productivity significantly, especially in barley, one of the more salt-sensitive cereals. Understanding the role of elongation factors in response to salinity stress in barley involves exploring molecular and physiological pathways (Thabet et al. 2021a, b). Elongation factors like eEF (eukaryotic elongation factor) are integral to the process of translation, or protein synthesis, in the cell. Under salinity stress, certain proteins that protect the plant or repair damage are in high demand, and the role of elongation factors becomes critical (Gao et al. 2019). These factors may ensure the synthesis of stress-related proteins, including osmoprotectants, antioxidants, and ion transport regulators that help barley cope with high salinity. For instance, Gao et al. (2019) reported that overexpression of the elongation factor GmEF4 can improve tolerance to salinity and drought in soybean. This indicates that manipulating the components of the translation machinery, such as elongation factors, could be a viable strategy for enhancing salinity stress tolerance in barley (Athar et al. 2022). Understanding the role of elongation factors in salinity stress response opens avenues for genetic improvements in barley. Through traditional breeding or genetic engineering, varieties of barley could be developed that overexpress specific elongation factors, potentially enhancing their salinity tolerance (Munns and Tester 2008). In conclusion, elongation factors are central to the protein synthesis machinery of the cell and play a crucial role in the response of barley to salinity stress. Their involvement in the regulation of stress-related proteins, gene expression, osmotic stress adaptation, and interaction with other stress response pathways highlights their importance. Further research in this area could help improve plant resilience in saline environments, enhancing food security in affected areas.
Moreover, high-LD genetic loci were mapped on chromosome 1H with the (T:C) SNP that was located at position (58,238,555–58,239,948 bp). Within this region, a gene mapped as polyadenylate-binding protein 1-B-binding protein, HORVU.MOREX.r3.1HG0018170, was detected for GS_Se, WGS_C, and WGS_Se. Polyadenylate-binding protein 1 (PABP1) is a critical component of cellular machinery in eukaryotes, involved in the regulation of mRNA stability and translation. PABP1-binding protein (PABP1-BP) serves as a modulator of PABP1 activities (Brook et al. 2012). PABP1-BP interacts with PABP1 and can modulate its function. Under normal conditions, PABP1 binds to the poly(A) tail of mRNA molecules, influencing their stability and the efficiency of translation. During salinity stress, plants need to rapidly alter their protein synthesis patterns, favoring the production of stress-responsive proteins (Urquidi Camacho et al. 2020). PABP1-BP could potentially regulate these processes by influencing PABP1’s interaction with mRNAs, thereby affecting the synthesis of proteins crucial for salinity stress response (Addo-Quaye et al. 2008). Salinity stress typically activates various signaling pathways in plants, including the mitogen-activated protein kinase (MAPK) cascade. While the direct connection between PABP1-BP and MAPK pathways in barley under salinity stress has not been explicitly established, studies in other organisms suggest that PABP1-BP could potentially interact with components of signal transduction pathways, thereby affecting the cellular response to salinity stress (Jerome Jeyakumar et al. 2020). Further research is needed to elucidate the precise roles of PABP1-BP in barley’s response to salinity stress, including its potential interaction with PABP1, involvement in stress granule dynamics, regulation of signal transduction pathways, and role in PCD. Such studies would enhance our understanding of stress response resilience mechanisms in barley and could offer new strategies for improving salinity tolerance in this important crop.
As well, receptor-like protein kinase is involved in salt tolerance, and HORVU.MOREX.r3.6HG0596150 was found at position (374,041,230–374,043,818 bp) of the (T:C) SNP on chromosome 6H for antioxidants in both conditions. Receptor-like kinases (RLKs) are a group of cell-surface receptors that play vital roles in perceiving extracellular signals and transducing these into intracellular responses in plants (Jose et al. 2020). They are crucial for various physiological processes, including growth, development, and stress responses. RLKs are integral to the plant’s ability to perceive, respond, and adapt to the increased salinity in its environment (Xiao and Zhou 2023). RLKs are known to perceive stress signals and initiate stress response pathways. Under salinity stress, specific RLKs in barley may recognize the increased salt concentration or the resultant cellular effects as a stress signal, which triggers a cascade of downstream signaling processes (Xiao and Zhou 2023). Upon perceiving the stress signal, RLKs activate various intracellular signaling pathways. This includes the mitogen-activated protein kinase (MAPK) signaling cascade, which is a critical component of the stress response in plants. Through phosphorylation events, the signal is transduced within the cell, eventually leading to the activation of specific transcription factors and gene expression (Ahanger et al. 2017). One of the challenges plants face during salinity stress is the excessive accumulation of sodium ions (Na+). Some RLKs are known to regulate the activity of ion transporters. In barley, these RLKs might influence the expression or activity of sodium transporters, helping to maintain ion homeostasis by controlling uptake, compartmentalization, or exclusion of Na+ (Zhu 2003). Furthermore, certain RLKs interact with ABA signaling pathways, potentially affecting stomatal closure, seed germination, and other processes that found to be involved in salinity stress (Berens et al. 2017). Therefore, understanding the specific RLKs expressed in barley, their signal transduction pathways, and their interactions with other proteins will be crucial for developing salinity-tolerant barley varieties through breeding and genetic engineering.
Ultimately, significant SNP (G:A) was detected on chromosome 7H at position (521,369,195- 521,370,360 bp) that found to be associated with our trait of interest. Within this region, a gene mapped as peroxidase, HORVU.MOREX.r3.7HG0718950, was detected for SS_Se, SL_C, and SL_Se. Peroxidases are a group of enzymes that play crucial roles in various biological processes, including lignification, suberization, cross-linking of cell wall proteins, defense against pathogens, and responses to abiotic stresses (Tognolli et al. 2002). Under salt stress, peroxidases participate in reactive oxygen species (ROS) scavenging, modulation of signaling pathways, and enhancement of antioxidant defense and contribute to the lignification process which reinforces cell walls (Kidwai et al. 2020). Under salinity stress, excessive accumulation of salts can lead to increased production of ROS, which is harmful to cellular structures and can lead to oxidative damage. Peroxidases, particularly class III peroxidases, are vital for the scavenging of ROS, thereby protecting cells from oxidative stress. They catalyze the reduction of H2O2 (a primary ROS) using various phenolic compounds as substrates (Hasanuzzaman et al. 2021). Under salinity stress, the balance between the production and scavenging of ROS is disrupted. Peroxidases work in conjunction with other antioxidants like SOD, CAT, and APX to mitigate oxidative damage, thereby contributing to stress tolerance (Kesawat et al. 2023). Furthermore, salinity stress can affect the integrity and permeability of plant cell walls. Peroxidases are involved in the lignification process, which reinforces cell walls and provides an enhanced barrier against the excessive uptake of Na+ and Cl− ions. This process is crucial for maintaining ion homeostasis in plant cells under saline conditions (Liu et al. 2021). Peroxidases can influence various signaling pathways associated with plant stress responses. For example, they are thought to participate in ABA-mediated signaling pathways during salinity stress, contributing to stomatal closure to prevent water loss and regulate seed germination under stressful conditions (Balasubramaniam et al. 2023; Zhao et al. 2021). Certain peroxidases are involved in the detoxification processes that contribute to salt tolerance. They participate in the metabolism of toxic compounds produced under stress conditions, including certain phenolic compounds that are generated in response to salt stress (Naliwajski and Skłodowska 2021). Therefore, understanding the specific peroxidases expressed during salinity stress and their precise functions requires detailed molecular, biochemical, and physiological studies. Advances in genomic tools and techniques, including transcriptomic and proteomic analyses, are facilitating this research, potentially enabling the development of barley cultivars with enhanced salinity tolerance through genetic engineering or selective breeding.
Conclusion
In conclusion, the comprehensive study on genetic mapping in barley has illuminated the significant role that specific genomic loci and quantitative trait nucleotides (QTNs) play in enhancing stress resilience, particularly in the context of salt tolerance. This research underscores the critical interplay between these genetic regions and the plant’s inherent antioxidant defenses, a relationship that emerges as a cornerstone in the plant’s ability to thrive in saline environments. Based on GWAS, significant SNP (G:A) on chromosome 7H at position (521,369,195- 521,370,360 bp) was detected to be associated with our trait of interest under selenium nanoparticles application. Within this region, a gene mapped as peroxidase, HORVU.MOREX.r3.7HG0718950, was detected for SS_Se, SL_C, and SL_Se, suggesting the essential role of this gene in regulating the resilience of plant responses to salinity stress after application of selenium nanoparticles. By identifying and understanding the function of these pivotal genomic positions, we unlock new varieties for the advancement of breeding programs. This knowledge paves the way for the development of barley cultivars with improved salt tolerance, a breakthrough with substantial implications for agricultural sustainability and food security. These findings are especially relevant in the face of global challenges such as climate change and soil salinization, which pose increasing threats to crop productivity and agricultural landscapes worldwide. Furthermore, the insights gained from this study extend beyond barley. They provide a valuable genetic blueprint that could potentially be extrapolated to other important cereal crops, thereby broadening the scope of resilient agriculture. Ultimately, this research represents a significant stride forward in our scientific understanding and offers tangible pathways for the practical application of genetic findings to secure and enhance global food production under stress conditions.
Data Availability
No datasets were generated or analysed during the current study.
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762
Agbolade JO, David O, Ajiboye A, Kioko J, Jolayemi O, Olawuni I, Ojo M, Akomolafe G, Adekoya M, Komolafe R (2019) Morpho-physiological effect of selenium on salinity-stressed wheat (Triticum aestivum L.). Journal of Biological Research - Bollettino Della Società Italiana Di Biologia Sperimentale, 92(1). https://doi.org/10.4081/jbr.2019.7650
Ahanger MA, Akram NA, Ashraf M, Alyemeni MN, Wijaya L (2017) Ahmad PJBp. Signal Transduction and Biotechnology in Response to Environmental Stresses 61:401–416
Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T (2014) Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS ONE 9:e113120
Alqudah AM, Sallam A, Stephen Baenziger P, Börner A (2020) GWAS: Fast-forwarding gene identification and characterization in temperate cereals: Lessons from Barley - a review. J Adv Res 22:119–135
Al-Tamimi N, Brien C, Oakey H, Berger B, Saade S, Ho YS, Schmöckel SM, Tester M, Negrão S (2016) Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat Commun 7:13342
Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
H-u-R Athar, Zulfiqar F, Moosa A, Ashraf M, Zafar ZU, Zhang L, Ahmed N, Kalaji HM, Nafees M, Hossain MA, Islam MS, El Sabagh A, Siddique KHM (2022) Salt stress proteins in plants: An overview. Front Plant Sci 13:999058. https://doi.org/10.3389/fpls.2022.999058
Atta K, Mondal S, Gorai S, Singh AP, Kumari A, Ghosh T, Roy A, Hembram S, Gaikwad DJ, Mondal S, Bhattacharya S, Jha UC, Jespersen D (2023) Impacts of salinity stress on crop plants: Improving salt tolerance through genetic and molecular dissection. Front Plant Sci 14:1241736. https://doi.org/10.3389/fpls.2023.1241736
Balasubramaniam T, Shen G, Esmaeili N (2023) Zhang H (2023) Plants’ response mechanisms to salinity stress. Plants 12:2253. https://doi.org/10.3390/plants12122253
Bano I, Skalickova S, Sajjad H, Skladanka J, Horky P (2021) Uses of Selenium Nanoparticles in the Plant Production 11:2229
Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K (2017) Evolution of Hormone Signaling Networks in Plant Defense 55:401–425
Bierhuizen MF, Maemura K, Kudo S, Fukuda M (1995) Genomic organization of core 2 and I branching beta-1,6-N-acetylglucosaminyltransferases. Implication for evolution of the beta-1,6-N-acetylglucosaminyltransferase gene family. Glycobiology 5:417–425
Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK (2012) The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J 441:803–812
Dahnke W (1988) Whitney DJRcstpftNCR. Measurement of Soil Salinity 499:32–34
De Kok L, Maas F, Godeke J, Haaksma A (1986) Kuiper P (1986) Glutathione, a tripeptide which may function as a temporary storage compound of excessive reduced sulphur in H2S fumigated spinach plants. Plant Soil 91:349–352. https://doi.org/10.1007/BF02198121
Devi MS, Srinivasan S, Muthuvel A (2023) Selenium nanomaterial is a promising nanotechnology for biomedical and environmental remediation: A detailed review. Biocatal Agric Biotechnol 51:102766
Feng X, Lai Z, Lin Y, Lai G, Lian CJBg (2015) Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA Group) 16:1–16
Gao Y, Ma J, Zheng JC, Chen J, Chen M, Zhou YB, Fu JD, Xu ZS, Ma YZ (2019) The elongation factor GmEF4 is involved in the response to drought and salt tolerance in soybean. Int J Mol Sci 20(12):3001. https://doi.org/10.3390/ijms20123001. PMID: 31248195; PMCID: PMC6627591
Garratt LC, Janagoudar BS, Lowe KC, Anthony P, Power JB, Davey MR (2002) Salinity tolerance and antioxidant status in cotton cultures. Free Radical Biol Med 33:502–511
Gudkov SV, Shafeev GA, Glinushkin AP, Shkirin AV, Barmina EV, Rakov II, Simakin AV, Kislov AV, Astashev ME, Vodeneev VA, Kalinitchenko VP (2020) Production and use of selenium nanoparticles as fertilizers. ACS Omega 5:17767–17774
Hasanuzzaman M, Raihan MRH, Masud AAC, Rahman K, Nowroz F, Rahman M, Nahar K, Fujita M (2021) Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int J Mol Sci 22(17):9326. https://doi.org/10.3390/ijms22179326. PMID: 34502233; PMCID: PMC8430727
Jackson MJEC, NJ (1958) Soil chemical analysis prentice Hall Inc 498:183–204
Jerome Jeyakumar JM, Ali A, Wang W-M, Thiruvengadam M (2020) Characterizing the role of the miR156-SPL network in plant development and stress response. Plants (Basel) 9(9):1206. https://doi.org/10.3390/plants9091206. PMID: 32942558; PMCID: PMC7570127
Jose J, Ghantasala S, Roy Choudhury S (2020) Arabidopsis transmembrane receptor-like kinases (RLKs): A bridge between extracellular signal and intracellular regulatory machinery. Int J Mol Sci 21(11):4000. https://doi.org/10.3390/ijms21114000. PMID: 32503273; PMCID: PMC7313013
Kamran M, Parveen A, Ahmar S, Malik Z, Hussain S, Chattha MS, Saleem MH, Adil M, Heidari P, Chen J-TJIjoms (2020) An overview of hazardous impacts of soil salinity in crops, tolerance mechanisms, and amelioration through selenium supplementation 21:148
Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, Lee SY, von Schaewen A, Koiwa H (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci U S A 105:5933–5938
Kesawat MS, Satheesh N, Kherawat BS, Kumar A, Kim H-U, Chung SM, Kumar M (2023) Regulation of reactive oxygen species during salt stress in plants and their crosstalk with other signaling molecules mdash. Current Perspectives and Future Directions 12:864
Kidwai M, Ahmad IZ, Chakrabarty D (2020) Class III peroxidase: An indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep 39:1381–1393
Liu J, Zhang W, Long S, Zhao C (2021) Maintenance of cell wall integrity under high salinity. Int J Mol Sci 22(6):3260. https://doi.org/10.3390/ijms22063260. PMID: 33806816; PMCID: PMC8004791
Maehly AC, Chance B (1954) The assay of catalases and peroxidases. Methods Biochem Anal 1:357–424
Meints B, Hayes PMJPBR (2019) Breeding naked barley for food, feed, and malt 43:95–119
Milner SG, Jost M, Taketa S, Mazon ER, Himmelbach A, Oppermann M, Weise S, Knupffer H, Basterrechea M, Konig P, Schuler D, Sharma R, Pasam RK, Rutten T, Guo G, Xu D, Zhang J, Herren G, Muller T, Krattinger SG, Keller B, Jiang Y, Gonzalez MY, Zhao Y, Habekuss A, Farber S, Ordon F, Lange M, Borner A, Graner A, Reif JC, Scholz U, Mascher M, Stein N (2019) GeneBank genomics highlights the diversity of a global barley collection. Nat Genet 51:319–326
Monat C, Padmarasu S, Lux T, Wicker T, Gundlach H, Himmelbach A, Ens J, Li C, Muehlbauer GJ, Schulman AH, Waugh R, Braumann I, Pozniak C, Scholz U, Mayer KFX, Spannagl M, Stein N, Mascher M (2019) TRITEX: Chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol 20:284. https://doi.org/10.1186/s13059-019-1899-5
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, Bose J, Byrt CS, Chen ZH (2020) Foster KJJNP. Energy Costs of Salt Tolerance in Crop Plants 225:1072–1090
Nagae M, Yamaguchi Y, Taniguchi N, Kizuka Y (2020) 3D structure and function of glycosyltransferases involved in N-glycan maturation. Int J Mol Sci 21(2):437. https://doi.org/10.3390/ijms21020437. PMID: 31936666; PMCID: PMC7014118
Naliwajski M, Skłodowska M (2021) The relationship between the antioxidant system and proline metabolism in the leaves of cucumber plants acclimated to salt stress. Cells 10(3):609. https://doi.org/10.3390/cells10030609. PMID: 33801884; PMCID: PMC7998282
Nikolić N, Ghirardelli A, Schiavon M, Masin R (2023) Effects of the salinity-temperature interaction on seed germination and early seedling development: A comparative study of crop and weed species. BMC Plant Biol 23:446
Noreen S, Sultan M, Akhter MS, Shah KH, Ummara U, Manzoor H, Ulfat M, Alyemeni MN, Ahmad P (2021) Foliar fertigation of ascorbic acid and zinc improves growth, antioxidant enzyme activity and harvest index in barley (Hordeum vulgare L.) grown under salt stress. Plant Physiol Biochem 158:244–254
Pyrzynska K, Sentkowska A (2022) Biosynthesis of selenium nanoparticles using plant extracts. J Nanostructure Chem 12:467–480
Rao MB, Tanksale AM, Ghatge MS, Deshpande VV (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Rasouli F, Yun P, Kiani-Pouya A, Movahedi A, Rasouli M, Salehi M, Shabala S (2024) One size does not fit all: Different strategies employed by triticale and barley plants to deal with soil salinity. Environ Exp Bot 218:105585
Saint-Jore-Dupas C, Nebenführ A, Al B, Follet-Gueye M-L, Plasson C, Hawes C, Driouich A, Lc F, Vr G (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18:3182–3200
Sairam RK, Deshmukh PS, Shukla DS (1997) Tolerance of drought and temperature stress in relation to increased antioxidant enzyme activity in wheat 178:171–178
Samynathan R, Venkidasamy B, Ramya K, Muthuramalingam P, Shin H, Kumari PS, Thangavel S, Sivanesan I (2023) A recent update on the impact of nano-selenium on plant growth, metabolism, and stress tolerance. Plants 12:853. https://doi.org/10.3390/plants12040853
Shin D, Moon S-J, Park SR, Kim B-G, Byun M-O (2009) Elongation factor 1α from A. thaliana functions as molecular chaperone and confers resistance to salt stress in yeast and plants. Plant Sci 177:156–160
Solá RJ, Griebenow K (2009) Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci 98:1223–1245
Song J, Yu S, Yang R, Xiao J, Liu J (2023) Opportunities for the use of selenium nanoparticles in agriculture. NanoImpact 31:100478
Strasser R (2016) Plant protein glycosylation. Glycobiology 26:926–939
Tabur S, Avci ZD, Özmen S (2021) Exogenous salicylic acid application against mitodepressive and clastogenic effects induced by salt stress in barley apical meristems. Biologia 76:341–350
Thabet SG, Alqudah AM (2023) New genetic insights into improving barley cope with salt stress via regulating mineral accumulation, cellular ion homeostasis, and membrane trafficking. Environ Exp Bot 208:105252
Thabet SG, Moursi YS, Karam MA, Graner A, Alqudah AM (2018) Genetic basis of drought tolerance during seed germination in barley. PLoS ONE 13:e0206682
Thabet SG, Moursi YS, Karam MA, Börner A, Alqudah AM (2020) Natural variation uncovers candidate genes for barley spikelet number and grain yield under drought stress 11:533. https://doi.org/10.3390/genes11050533
Thabet SG, Alomari DZ, Alqudah AM (2021a) Exploring natural diversity reveals alleles to enhance antioxidant system in barley under salt stress. Plant Physiol Biochem 166:789–798
Thabet SG, Moursi YS, Sallam A, Karam MA, Alqudah AM (2021b) Genetic associations uncover candidate SNP markers and genes associated with salt tolerance during seedling developmental phase in barley. Environ Exp Bot 188:104499
Thabet SG, Alomari DZ, Börner A, Brinch-Pedersen H, Alqudah AM (2022b) Elucidating the genetic architecture controlling antioxidant status and ionic balance in barley under salt stress. Plant Mol Biol 110:287–300
Thabet SG, Alomari DZ, Börner A, Brinch-Pedersen H, Alqudah AM (2022a) Elucidating the genetic architecture controlling antioxidant status and ionic balance in barley under salt stress. Plant Mol Biol 110(3):287–300. https://doi.org/10.1007/s11103-022-01302-8. Epub 2022 Aug 2. PMID: 35918559
Thabet SG, Alqahtani MD, Jabbour AA, Alqudah AM (2023) Genetic associations underpinning the metabolite-mediated salt stress tolerance in barley. Plant Mol Biol Rep. https://doi.org/10.1007/s11105-023-01408-3
Tobe K, Zhang L, Omasa KJSSR (2003) Alleviatory effects of calcium on the toxicity of sodium, potassium and magnesium chlorides to seed germination in three non-halophytes 13:47–54
Tognolli M, Penel C, Greppin H, Simon P (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288:129–138
Urquidi Camacho RA, Lokdarshi A, von Arnim AG (2020) Translational gene regulation in plants: A green new deal. Wiley Interdisciplinary Reviews RNA 11:e1597
Wu H, Zhu M, Shabala L, Zhou M, Shabala SJJoipb (2015) K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: A case study for barley 57:171–185
Xiao F, Zhou H (2023) Plant salt response: Perception, signaling, and tolerance. Front Plant Sci 13:1053699. https://doi.org/10.3389/fpls.2022.1053699
Zhang X, Zhang L, Chen Y, Wang S, Fang Y, Zhang X, Wu Y, Xue D (2021) Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul 94:49–60
Zhao S, Zhang Q, Liu M, Zhou H, Ma C, Wang P (2021) Regulation of plant responses to salt stress. Int J Mol Sci 22:4609. https://doi.org/10.3390/ijms22094609
Zhu J-K (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Zohra E, Ikram M, Omar AA, Hussain M, Satti SH, Raja NI, Mashwani Z-U-R, Ehsan M (2021) Potential applications of biogenic selenium nanoparticles in alleviating biotic and abiotic stresses in plants: A comprehensive insight on the mechanistic approach and future perspectives 10:456–475
Acknowledgements
The authors would like to thank the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) (grant number IMSIU- RG23025)
Funding
This work was supported and funded by the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) (grant number IMSIU- RG23025).
Author information
Authors and Affiliations
Contributions
AMA, AEBSA, MSA, SMH, and SGT designed the experiment and analyzed the data. SGT and AMA wrote the manuscript. SGT conceived the idea and participated in the interpretation of the results.
Corresponding author
Ethics declarations
Competing Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alqudah, A.M., Elkelish, A., Alammari, B.S. et al. Genetic Mapping Determining the Key Genomic Loci/QTNs for Stress Resilience via Controlling Antioxidant Defenses in Barley Under Salt Stress. Plant Mol Biol Rep (2024). https://doi.org/10.1007/s11105-024-01488-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11105-024-01488-9