Abstract
Sesamum indicum is widely cultivated in India for its superior agronomic characters like adaptation to the warm climate of India and resistance to drought and pathogen attack. However, the yield of sesame per unit area of land is quite low. In this context, the genes involved with reproductive tissue development is very important since they are directly involved with crop productivity. Therefore, identification and isolation of anther-specific genes through PCR-Select cDNA subtraction between cDNAs of anthers and leaves was carried out. Sequencing of ~ 700 clones from the cDNA subtraction library followed by BLASTX analysis using sesame database resulted in the identification of 163 annotated unique transcripts, of which 18 transcripts showed homology with the genes of exclusive anther-/pollen-specific functions. Furthermore, anther-specific expression of some of the sesame transcripts as well as already identified anther-specific genes of Arabidopsis and rice have been monitored during development of sesame anthers. Subsequently, full-length cDNA sequence and the upstream sequence of an anther-specific β-1 3-glucanase gene was isolated. Transgenic tobacco plants were developed with GUS gene under the control of a 437-bp upstream sequence along with the first exon and first intron of the gene. Monitoring of GUS gene expression pattern indicated exclusive expression of the promoter in the tapetal cell layer before tetrad formation in the anther. Interestingly, upon treatment with ABA as well as drought stress, GUS was found to express in the vegetative parts of the plant such as leaves and roots as well.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sesame (Sesamum indicum L., Pedaliaceae) is a potential oilseed crop of India (Biabani and Pakniyat 2008). It is one of the oldest cultivated plants in the world (Ali et al. 2007). The superior oil quality and important agronomic characters have made the plant an obvious choice for human consumption. The potential of this crop from the nutritional as well as medicinal point of view has not yet been exploited to the fullest extent (Abou-Gharbia et al. 1997). The seeds of the plant are a robust source of polyunsaturated fatty acids as compared to that of other oil crops like mustard, canola, olive, etc. (Samman et al. 2008). The seeds comprise about 50% of oil which is a voluminous reservoir of ω-6 (C18:2) and ω-3 (C18:3) fatty acids. Moreover, sesame is a rich source of value-added antioxidant substances like sesamin and sesamolin which act as naturally occurring preservatives and are maneuvered extensively in the nutraceutical and pharmaceutical industries (Morris 2002).
Besides this, sesame has elite agronomic features like drought tolerance (Kadkhodaie et al. 2014); it suits very well the Indian context as a summer plant (Garai and Datta 1999) as well as ensures maximum land utility as multiple cropping is possible (Huang et al. 2008). That apart, it shows resistance against a large number of pathogens that ascertains a great benefit to the environment for lesser use of pesticides. However, the yield of sesame per unit area of land is quite low (Roy et al. 2009). In this context, the study of the genes expressed in the reproductive organs is very important since tissue-specific expression of these genes have direct involvement with crop productivity. Unfortunately, very little information about the sesame reproductive organ-specific genes and expression pattern is available for this important crop. It is a known fact that proper development of pollen is vital for fertilization of the ovule to turn it into a viable embryo. Pollen grains and their healthy nature dictate not only the degree of their survival after pollination but also their quality to produce fertile seeds. The number and quality of the pollen depend on various developmental stages beginning from the formation and division of the pollen mother cells.
It is known that meiosis starts in the microspore mother cells in the pre-tetrad stage, and the tapetum plays a major role in supplying nutrients to the developing tetrads. At the end of meiosis, free tetrads start developing within each locule. In the post-tetrad stage, dissolution of tetrads occurs due to degeneration of the callose wall surrounding the tetrads, which results in the release of individual microspores. After maturation of pollen, tapetum gets degenerated. Therefore, the single-cell layered tapetum within the anther is extremely important and plays a critical role in sporogenesis. The tapetal layer comprises a layer of cells surrounding the pollen sac, which arise from the same progenitor cells as the developing male gametophyte (Goldberg 1993; Scott et al. 1991). The integrity of the tapetal cell layer is directly proportional with the formation of fertile pollen, and any impairment in gene expression in this tissue has been shown to result in complete male sterility (Bino 1985; Chaudhury 1993; Grant et al. 1986; Horner and Rogers 1974; Izhar and Frankel 1971; Mariani et al. 1990; Shi et al. 2009). Recently, Cui et al. (2012) had already published a Plant Male Reproduction Database (PMRD) for easy access to various anther-related genes of Arabidopsis and rice. Information on some of the genes expressed in tapetum were found to be associated with the production and secretion of various kinds of proteins that enhance the processes of the pollen tube growth and germination on the stigma (Dickinson and Lewis 1973; Heslop-Harrison 1968; Kandasamy et al. 1994).
Thus, a cDNA subtraction has been carried out using the cDNAs of anther as a tester and that of the leaf as a driver for the identification of anther-specific genes from sesame. Some genes that express in the anthers along with one β-1,3-glucanase have been identified in this process. It is known that the tapetum secretes the β-1,3-glucanase enzymes also known as callase, which enables the breakdown of the callose wall around tetrads formed after the reductional division of the pollen mother cells (Frankel et al. 1969; Izhar and Frankel 1971; Mepham and Lane 1970; Stieglitz 1977). It is reported that silencing of the β-1,3-glucanase in rice resulted in male sterility due to disruption of callose degradation in the anther locules at the early microspore development (Wang et al. 2009). Some of the β-1,3-glucanase genes belonging to the groups H and K are known to have flower-/reproductive-specific expression. Two homologues of this from Arabidopsis thaliana belonging to group K are reported to have anther-specific expression At4g14080 and At3g23770. These anther-specific genes play a role in the dissolution of the callose deposition in the tetrads after each round of reductional division and also in nourishing the male gametophytes by the breakdown of complex sugars (Bucciaglia et al. 2003).
This family of β-1,3-glucanase gene is reported to have several other multifaceted functions in the plant developmental and defense-related processes. For example, homologues of this class of the β-1,3-glucanase gene are known to function as a cryoprotectant in the skin of Vitis vinifera cv. Cardinal (Romero et al. 2008); on the other hand, they also help in acclimatization to freezing conditions in winter rye (Secale cereale L.) (Yaish et al. 2006). Moreover, these endoglucanases also play a significant role in secondary cell wall formation by contributing cellulose crystallization process (Glass et al. 2015).
It is speculated that the gene has significant roles in seed development, germination, and ripening (Doxey et al. 2007). The role of the β-1,3-glucanase gene in response to stress is also known to be extremely significant and important. In fungal cell walls, the degradation of β-1,3-glucan is thought to contribute toward fungal cell wall destabilization. It also causes the release of cell wall-associated immune elicitors that further stimulate defense responses (Leubner-Metzger and Meins 2000). Thus, this class of genes is not only involved in the plants’ physiological and developmental processes but also with both biotic and abiotic stresses.
In this paper, we have emphasized the validation of anther-specific expression of the β-1,3-glucanase gene and further isolated the full-length gene and the upstream sequence from sesame. The upstream sequence fused to the GUS reporter gene and cloned into a plant expression vector. In vivo functional assay of the upstream element was monitored by a transgenic approach using the model plant Nicotiana tabecum. Interestingly, we also found that the promoter was activated in other parts of the plant upon treatment with hormones like ABA and also during drought stress. Therefore, in this paper, we are reporting the identification and isolation of some anther-specific genes of sesame, as well as a novel β-1,3-glucanase gene, and its promoter sequence that shows anther-specific expression as well as a unique expression pattern in other parts of the plant during drought stress. This is a unique observation of resource utilization by a single gene during tissue development and stress management.
Results
Identification of Sesame Anthers with Intact Tapetum Tissue for cDNA Subtraction
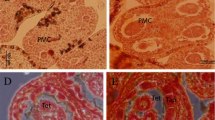
Anthers were collected from buds of different sizes ranging from 1- to 8-day-old buds of Sesamum indicum. Transverse sections were made from the anthers to determine the integrity of the sporogenous tissue. The cross sections of anthers from the 3–4-day-old buds appeared to have an intact sporogenous tissue with an integral tapetal cell layer (Fig. 1). Extraction of total RNAs from the selected anthers as well as from leaves of sesame followed by purification of polyA mRNA was carried out for identification of anther-specific genes by cDNA subtraction. The mRNAs isolated from the anthers with intact tapetum (3–4 days) and leaves were used as tester and driver, respectively, using the PCR-Select cDNA Subtraction Kit from Clontech. Two successive hybridizations were performed to equalize and enrich differentially expressed sequences (Supplementary Fig. S1 a). A secondary PCR amplification was then performed using nested primers (APP and 5PP) designed from the adapter sequence to reduce non-specific PCR products and enrich the differentially expressed sequences (Supplementary Fig. S1 b). Enrichment of the library with anther-specific cDNA sequences was checked by the amount of amplification of a sesame-specific GAPDH gene (obtained from NCBI database) before and after cDNA subtraction. The abundance of this housekeeping gene decreased significantly in the subtracted library, whereas it remained almost unchanged in the non-subtracted library. This result indicated that effective and efficient subtraction had taken place (Supplementary Fig. S1 c).
Different stages of anther during flower development of sesame: a 1–16-day-old buds; b anthers in different stages arranged according to days of maturation (from left to right). The flower buds of Sesamum indicum were arranged in a row and classified as SI (very early stage of sporogenous tissue development), SII (well-defined sporogenous tissue with intact tapetum), and SIII (anther with matured pollen grain). c The 4-day-old anther was found to have intact sporogenous tissue and thus the tapetal layer and used for cDNA subtraction
The subtracted and enriched transcripts were cloned into the pGEM-T plasmid vector for subsequent sequencing of ~ 700 clones from the cDNA subtraction library followed by homology-based annotation using NCBI, BLASTX, resulting in the identification of 163 unique transcripts without repetition. Out of these, 153 (Supplementary Table S1 ) sequences showed > 60% homology with the sesame expressed sequence tags (ESTs) deposited in the NCBI database. Out of which, 18 transcripts showed homology with anther-/pollen-specific genes (Table 1), 117 transcripts showed homology with known genes, 18 transcripts showed homology with uncharacterized ESTs, and 9 transcripts did not show any homology when sesame was used as reference organism. These EST sequences of sesame were submitted to the NCBI database under the accession numbers HO710134 to HO710204 and JK755160–JK755244 (Supplementary Table S1 ). The cDNA subtraction library was validated by RT-PCR using a few randomly selected clones and normalized using sesame-specific GAPDH (Fig. 2). One of these sequences was that of β-1,3-glucanase (BISI27f) which distinctly showed expression only in the anther tissue and no expression in the leaf tissue.
Validation of the cDNA subtraction library enriched with anther-specific transcripts through RT-PCR: Primers were designed from the sequences of randomly selected transcripts of the clones of cDNA subtraction library using the NCBI Primer-BLAST software. The RT-PCR was performed from both anther and leaf cDNA with those primers at various concentrations of template cDNA ranging from 1:10, 1:30, to 1:50. Among the six ESTs, BISI27f (β-1,3-glucanase), BISI325f, BISI328f, and BISI480f were showed anther specificity compared the BISI21f and BISI371f which showed expression in both anther and leaf tissues. Expression of GAPDH in both leaves and anthers indicate normalization of cDNA for RT-PCR
Validation of Anther-Specific Transcripts from Sesame
Among these EST sequences of S. indicum, one EST sequence (SiBG, NCBI accession no. HO710138) shows high homology with anther-specific β-1,3-glucanase gene (accession no. emb CAA82271.1) of Nicotiana tabacum apart from several other β-1,3-glucanase genes from different plants. To validate the anther-specific expression of the gene, RT-PCR was done using SiBG-specific (BISI27F) forward and reverse primers (Supplementary Table S2 ) in different tissues like, leaf, stem, root, petals, anthers, gynoecium, and the fruits of sesame by using the cDNAs from different tissues. It was found that this particular homologue of the β-1,3-glucanase gene expressed specifically in the anthers and almost undetectable in other tissues in S. indicum (Fig. 3). Apart from that, expression of the 17 ESTs, which showed homology with anther/pollen-specific genes were further validated in all the different tissues like the leaf, stem, root, petals, anthers, gynoecium, and the fruits of sesame, through RT-PCR (Supplementary Fig. S2 ). Out of the 17 transcripts, 12 transcripts could be detected in RT-PCR. No amplification was observed in 5 transcripts, namely, accession numbers HO710166, HO710158, HO710149, JK755225, and JK755241.
Expression of the β-1,3-glucanase gene in different tissues of the sesame plant: cDNAs from seven different tissues of sesame were used for monitoring the expression of the β-1,3-glucanase gene through RT-PCR. The plant parts include root, leaf, stem, petals, pistils, fruit, and anthers. The upper panel of the figure showed that the β-1,3-glucanase gene was exclusively expressed in anther but not in other tissues. The sesame-specific GAPDH was taken as endogenous internal control of all the tissues
Isolation of Upstream Sequence of β-1,3-glucanase
5′ RACE was carried out to identify the 5′ nucleotide sequence of the β-1,3-glucanase gene (SiBG), using a gene-specific reverse primer (BG5RCR1) designed from 466–661 bp of SiBG sequence and the forward primer (RAP1) with 3′ poly G and adapter sequence (supplied with the kit; Supplementary Table S2). Sequence analysis of the 5′ RACE-amplified 475-bp 5′ sequence of SiBG gene (Fig. 4) showed the highest homology with the 5′-coding region of (Nicotiana tabacum β-1,3-glucanase) NiBG gene and a probable ATG along with a − 10-bp sequence from 5′ UTR were identified on BLASTX analysis. Thereafter, the rest of the 5′ UTR region and upstream sequence of the SiBG gene was PCR amplified with the adapter-specific forward and gene-specific reverse primer at + 171 bp (GS Primer 3) in the coding region, using the EcoRV restricted and adapter ligated genomic DNA of S. indicum following the protocol of “Genome Walker” kit of Clontech. Sequence analysis of the PCR-amplified 710-bp sequence obtained by genome walking through BLASTN and BLASTX showed a 437-bp sequence before ATG and an 85-bp sequence, which is exactly matching with the 5′ RACE sequence. The next 104 bp of the genome-walking sequence did not show any match with the 5′ RACE-amplified sequence, but after that, an 84-bp sequence showed complete match between the sequences (Supplementary Fig. S3). The sequence of 104 bp, which did not show any homology, was found to start with GT and end with AG, indicating the possible existence of the first intron. The 437-bp nucleotide of the genome-walking clone considered as upstream sequences since it did not show any significant homology with any plant-specific β-1,3-glucanase gene. The 86-bp sequence downstream of ATG, as well as an 85-bp sequence on both sides of the probable intron of 104 bp, showed almost 99% homology (except the intron) with a software-generated full-length cDNA sequence of glucan endo-1,3-beta-glucosidase, of S. indicum, (XM_011088785.1) deposited by a separate laboratory. This deposited sequence has not only helped us to establish the co-linearity between the unknown upstream sequence with the coding sequence of the gene but also to identify the translational start codon ATG (Supplementary Fig. S4). Motif scan analysis using PLACE software of this 710-bp sequence indicated the presence of three POLLENILEAT52 motifs each in upstream and the first intron whereas six GTGANTG10 motifs in upstream and two in the first intron (Fig. 4). Both these motifs are known for its anther-/pollen-specific expression in plants and repetitions of these anther-specific cis-acting elements within 437-bp upstream and 104-bp first intron sequences ensure a probable anther-specific expression of the gene. Also, some ABA-responsive/ABA-related motifs like WRKY71OS, DPBFCOREDCDC3, DRE2COREZMRAB17, EBOXBNNAPA, LTRECOREATCOR15, and MYCCONSENSUSAT were also present in the sequence of this upstream element. The overall results indicate that the upstream sequence is very promising for directing gene expression in an anther-specific manner.
In silico comparison of the upstream sequences between TA29, A9, and SiBGproplus: A comparison of upstream sequences of sesame β-1,3-glucanase (SiBGproplus), TA29 of Nicotiana tabacum, and A9 of Brassica oleracea based upon their location of anther-/pollen-specific motifs like GTGANTG10 and POLLENILEAT52 through a schematic representation
Cloning into Binary Vector and Expression in Plant
The 632-bp sequence of SiBGproplus (containing 437-bp upstream sequence before ATG, 85-bp sequence of the first exon, 104 bp of first intron and an additional 5 bp from the second exon) was amplified by genomic PCR, using sequence-specific forward and reverse primers containing HindIII and BamHI restriction sites, respectively. The amplified fragment was cloned in the same HindIII and BamHI sites of the pBI221 vector by replacing the CaMV 35S promoter with the SiBG (proplus) sequence and fused it with the GUS reporter gene. The entire cassette containing the GUS reporter gene under the control of SiBG (proplus) sequence along with the NOS terminator was excised from the clone of pBI221 by digestion with HindIII and EcoRI, and the fragment was then cloned into the binary vector, pPZPY112 (Supplementary Fig. S5 ). The transgenic Nicotiana tabacum plants were raised by transforming the leaf discs of N. tabacum (SR1) using the LBA4404 Agrobacterium strain containing chimeric pPZPY112 with the GUS reporter gene under the control of the SiBG upstream sequence to monitor the anther-/tapetum-specific expression pattern of the upstream sequence.
The positive tobacco transformants were screened by PCR using SiBGproplus-specific primers. The PCR-positive plants were grown in soil, and on the appearance of floral buds (4–5 days old), the anther was stained in X-Gluc solution for histochemical GUS assay. The cross section of the anthers showed GUS expression in the tapetal layer, just below the inner anther wall (Fig. 5f). However, no expression was observed in other floral parts like sepals or petals and neither in any vegetative parts of the plant like a leaf, stem, or root (Fig. 5a–e).
Expression pattern of SiBGproplus::GUS in different parts of the transgenic Nicotiana tabacum: A. Graphical representation of the SiBGproplus::GUS construct B. Histochemical assay of the GUS expression pattern in different parts of the SiBGproplus::GUS transgenic Nicotiana tabacum. No GUS expression was observed in the (a) leaves, (b) sepals, (c) petals, (d) root, or (e) stem. The transverse section of anthers (f) showed GUS expression exclusively in the tapetal layer, located within anther
Expression Pattern of Various Anther Developmental Genes of Arabidopsis and Rice in Sesame
Some anther developmental genes like MS1, MS2, MYB26, TIP2, MADS3, CYP704B2, and GAMYB were taken to compare the expression pattern of such genes in different stages of sesame anther in respect to leaf tissue of sesame. MS1, MS2, and MYB26 were downregulated in all the stages in anther tissue compared to the leaf tissue in sesame (Fig. 6). On the other hand, the TIP2 gene was upregulated in stage I but downregulated in stages II and III whereas the genes like MADS3, CYP704B2, and GAMYB were downregulated at stage I and upregulated at later stages (II and III) of sesame anther compared to the leaf tissue indicating these genes are developmentally regulated.
Expression analysis of some orthologues genes of Arabidopsis involved in anther development at different stages of anthers in Sesame: The expression pattern analysis was performed in three different stages of anther like SI (meiotic stages of pollen mother cell), SII (tetrad stages of pollen), and SIII (filled with pollen grains). The differential expression pattern of the genes was monitored through qRT-PCR in different stages of anther of sesame compared to expression in leaves. GAPDH was taken to normalize the relative expression of the genes
Anther-Specific Expression of the Sesame β-1,3-glucanase Gene Is Stage Specific
One of the fascinating observations regarding the induction pattern of SiBGpro is a stage-specific expression during anther development. We divided the anther developmental stages of the transgenic tobacco lines into three different stages based on days of maturity and the expression pattern of GUS gene. For synchronization of the GUS gene expression with pollen development, iron-acetocarmine staining of the pollen mother cells of the three stages of anthers was carried out simultaneously. We have considered the anthers from days 1 to 3 as stage I, from days 4 to 6 as stage II, and days 7 to 8 as stage III (Fig. 7a). It was quite interesting to note that no visible GUS expression on histochemical of staining was observed in the stage I (1–3 days old) anthers where the pollen mother cells were at the meiotic stages ranging from prophase I through telophase II (Fig. 7b). However, tapetal cells of stage II (4 to 6 days old) anthers having pollen mother cells in the tetrad stage of meiosis (young microspore stage) showed very high GUS expression (Fig. 7c). In stage III, where most of the pollen mother cells were in mature bipartite and tripartite pollen (matured pollen stage) along with very few in the tetrad stage, the GUS expression in tapetal cells at this stage showed a significant reduction in histochemical staining (Fig. 7c). Thus, it was clear that the highest promoter activity of SiBG(proplus) is correlated with the stage of tetrad dissolution, which is required for the maturation of the pollen grains. Both the specificity of expression on the tissue and the stage of development were a captivating observation.
Histochemical monitoring of the GUS gene expression under the SiBGproplus in different stages of anther in Nicotiana tabacum: a Sizes of floral buds representing the stages 1, 2, and 3, which were marked for studying the expression pattern of SiBGproplus of sesame in tobacco. b Anthers of the respective stages depicting their size. c Histochemical GUS staining of anthers followed by transverse sections to show the stage-specific GUS expression in tapetum tissue. GUS expression in stage 2 is found to be most prominent and stage 3 exhibits very less GUS expression. d Meiotic stages of pollen mother cells (PMC) of the three stages of anthers. Stage 1 shows late meiosis and Stage 2 shows the tetrad stage of the PMC. Stage 3 shows matured pollen grains. All images have been taken under ×40 magnification of the compound light microscope
Drought Stress and ABA Treatment Can Induce Anther-Specific SiBG Promoter in Other Parts of the Plants
As already stated, we have found certain ABA-responsive motifs within the sequence of the identified SiBGPro sequence. Thus, to study the changes in the expression pattern of the β-1,3-glucanase gene upon external hormone treatment, the full-grown T1 plants of the transgenic tobacco lines were treated with varying concentrations of ABA. We observed expression of the SiBGPro in leaves and roots of the plants upon treatment with 20 and 30 μM of ABA, as evident from the GUS expression pattern (Fig. 8). This indicates the function of the gene during stress as well. Therefore, to study the function of the promoter on drought/dehydration stress, we kept our transgenic tobacco lines under drought stress for 7 days. It was observed that dehydration response resulted in induction of GUS expression in the leaves and roots of the transgenic tobacco plants (Fig. 9). In sesame also, we have tested the expression of β-1,3-glucanase (SiBG) on external application of ABA at a concentration of 20 μM as well as exposing sesame plants under drought. It turned out that, in the sesame, the transcript levels of β-1,3-glucanase (SiBG) were distinctly detected in leaves on drought and ABA treatment whereas no transcript was detected by RT-PCR in the control leaves (Supplementary Fig. S6 ). In both cases, RT-PCR analysis of the ABA and drought-treated sesame cDNA using the primers NBG-f (+ 1 bp to + 21 bp) and NBG-r (+ 132 bp to + 150 bp) designed from 5′ end of the gene showed induction of the gene. Furthermore, sequence analysis (data not shown) of the 150-bp amplified sequence showed exact splicing of the first intron of the gene (Supplementary Fig. S7 ).
Induction of the promoter of the sesame β-1,3-glucanase (SiBGproplus) on ABA treatment: Leaves and roots of the transgenic lines of Nicotiana showed GUS expression upon treatment with varying concentrations of ABA. Comparably less GUS staining was found in the leaves a upon 10 μM ABA treatment; however, a significant increase in GUS expression is seen upon treatment with 20 and 30 μM ABA in leaves (b, c), respectively. Similar treatment was also done in the case of the roots. The roots also showed GUS expression upon d 10, e 20, and f 30 μM ABA treatment
Histochemical GUS staining of SiBGPro::GUS transgenic line of Nicotiana tabacum: The transgenic SiBGPro::GUS lines of Nicotiana were treated in a drought condition for 7 days. When the wilting was just initiated, the histochemical GUS staining was performed from the wilted leaves and the root of the plant as well. The a leaves and b root both exhibited strong GUS activity
Discussion
Yield potential of sesame per unit area of land is less compared to that of most other oil-producing crops in India. However, a higher content of PUFAs (Samman et al. 2008), oleic and linoleic acid (about 43% each), palmitic acid (9%), and stearic acid (4%) (Rao and Rao 1981) in sesame oil compared to that of most of the other commercially potential oilseeds demands increase in yield potential of this crop. Moreover, a number of beneficiary attributes and eco-friendly traits of this crop are needed to be exploited for minimization and increase in edible oil production. In this connection, one of the most important areas is to have a wholesome idea about the kind of genes expressed in the reproductive organs concerning the male gamete of this plant since they determine the proper development of the pollen grains, which eventually determine the rate of fertilization. Therefore, identification of the spatiotemporal expression pattern of the anther-specific genes and the mechanism of controlling tissue specificity of the genes are an obligatory requirement.
Subtraction Library Revealed Several Genes that Express in the Anthers
We constructed a subtraction library using early stages (3–4 days old) of sesame anther cDNA as the tester and leaf as the driver. BLASTX analysis of the nucleotide sequences of the individual clones of the library showed homology of 18 transcripts with various genes which are already reported to have a substantial role in the process of microsporogenesis, anther differentiation, and eventually pollen development (Table 1). Out of these 18 genes, 13 were detected in the sesame anther through RT-PCR, and all of them showed anther specificity. The rest of the 5 genes were found to be associated with the function of pollen in mature stages of development. Homology of the 18 gene sequences with already functionally characterized genes in the development of the male gametophyte is an indicator of the fact that the subtraction has delivered the desired anther-specific genes as compared to the leaves of sesame.
Tapetum-Specific Induction of SiBG Upstream Sequence in N. tabacum
The SiBG of sesame was found to have a high degree of homology with the anther-specific endo-type β-1,3-glucanase gene of Sesamum indicum and the Arabidopsis thaliana homologues At4g14080 and At3g23770 which exhibit anther-specific expression. These classes of genes are also known as callase and carry out a very important pollen development function of dissolution of tetrad wall which is surrounded by thick deposition of a callose (β-1,3-glucan) wall (Leubner-Metzger and Meins 2000). These callase enzymes are part of an enzyme complex that is secreted from the tapetal cell layer at the late stage of meiosis. The callase complex of lily consists of a 32-kDa endo-type β-glucanase and a 62-kDa exo-type β-1,3-glucanase. The endo-type enzyme seems to be most important for the degradation of the callose walls, while the exotype β-1,3-glucanase is involved in further hydrolysis of released oligosaccharides. Temporal alterations of β-glucanase expression, or failure to express β-glucanase, lead to abnormality in dissolution of the tetrad callose walls, which has been shown to be a primary cause of male sterility in cytoplasmic male-sterile lines of petunia (Frankel et al. 1969), sorghum (Warmke and Overman 1972), and soybean (Jin et al. 1997). To pinpoint the expression pattern of the sesame endo-type β-1,3-glucanase with the development of anther, the 437-bp upstream, 85-bp first exon, and 104-bp first intron along with 5 bp of second exon sequences (SiBGpro) were cloned (accession no. KT246471) and fused with GUS gene for monitoring in vivo expression patterns. Motif scan analysis showed the presence of a number of anther-/tapetum-specific motifs like GTGANTG10 and POLLENILEAT52 in the upstream as well as in the first intron of this gene. In plants, several reports have established that intron plays a vital role on enhancing expression and function of a number of genes like Arabidopsis phosphoribosyl anthranilate transferase 1 (PAT1) (Rose 2002), the first intron of maize shrunken-1 (Sh1) (Clancy and Hannah 2002), and petunia actin-depolymerizing factor 1 (PhADF1) (Mun et al. 2002). Apart from that, introns are found to be involved with the spatial or temporal expression patterns of flowering genes such as agamous (AG) (Deyholos and Sieburth 2000) and flowering locus C (FLC) (Sheldon et al. 2002). Transgenic tobacco plants with the chimeric GUS gene under the control of SiBGpro showed blue staining exclusively in the tapetal layer of the anthers and no other part of the transgenic tobacco plants, which indicated not only anther specificity but also proper splicing of the intron. Furthermore, the GUS expression was only found in the later stage of meiosis where microspores were in the tetrad stage indicting possible involvement of the gene in the dissolution of the callose deposition. Cytology with the anthers of the transgenic plants at different stages showed induction of the promoter confined in the tapetum of the anther between the telophase II to tetrad stage of gametogenesis. This phenomenon of breaking down the complex sugars for nourishing the male gametophytes after each round of reductional division has been already reported (Bucciaglia et al. 2003). The other promoters, TA29 and A9, which are known for its exclusive anther-specific expression also showed lots of similarity with the SiBGpro regarding repeated presence of the GTGANTG10 and POLLENILEAT52 motifs. TA29 in Nicotiana tabacum is known to express from the time of tetrad formation up to the stage of free microspore formation (Koltunow et al. 1990; Schrauwen et al. 1996). Similarly, the A9 promoter from Brassica napus expresses in the tapetum layer only after meiosis until tetrad release (Schrauwen et al. 1996). This information also supports the expression pattern SiBG in the tobacco anther.
Tapetum-Specific Expression of SiBG Upstream Sequence Altered During Stress
Interestingly, the SiBGPro sequence was found to be also adorned with a number of various ABA-responsive motifs apart from anther-specific motifs, indicating the possibility of ABA-induced expression. Induction of SIBG gene in sesame upon treatment with PEG and on external application of ABA indicated the stress-induced expression of the gene. Furthermore, induction of this promoter on external ABA treatment as well as during PEG mediated induction of drought stress in the vegetative parts of the plants like leaves, root, and stem apart from anther in transgenic Nicotiana tabecum pointed toward a very intriguing possibility of the multifarious function of a developmental gene under various stress conditions. Expression of the GUS gene only at a higher concentration (> 20 μM) of ABA in the leaves (Supplementary Fig. S7 ) signified that a relatively strong ABA-dependent stress response can direct the expression of the gene in other parts of the plant as well. Besides that, it also signifies that hormone concentration also controls the tissue specificity of the genes during the diverse developmental processes and plant resistance to biotic and abiotic stresses (Creelman and Mullet 1997; Kessler and Baldwin 2001). Enhanced expression of the SiBGpro on dehydration stress upon PEG treatment for 3 days in the transgenic tobacco lines as well as sesame also explain the biological significance of the gene since upregulated expression of the β-1,3-glucanase gene is also known upon dehydration stress (Bray 2004). It has also been reported that the Arabidopsis β-1,3-glucanase (AtBG1) protein is responsible for the release of ABA from ABA-GE and that ABA deconjugation plays a significant role in providing an ABA pool for plants that allows them to adjust to changing physiological and environmental conditions (Lee et al. 2006). Hence, the direct relationship between this gene and ABA is important in the activation of this under various kinds of signals. This suggests that the plant can divert a tissue-specific gene to other parts of the plant under stresses in a concentration-dependent ABA response above a certain threshold level to make an economical use of its resources. Therefore, the tissue specificity and stress-inducible nature of this promoter could grant wide applicability in plant biotechnology.
Methods
Plant Growth Conditions, Selection of Appropriate Anther Stage, and RNA Isolation
Sesamum indicum variety Rama obtained from ICAR, New Delhi, were grown under field condition. The plants took 2 months to reach flowering conditions. On appearance of floral buds, they were collected at 1-day intervals and at different sizes and stages ranging from 2-day-old bud to 8-day-old buds. Cross sections were made from the anthers of these bud and observed under light microscope to get an indication of the integrity of the sporogenous tissue and the tapetum. Other floral parts like the sepals, petals, and gynoecium were also taken from 4-day-old buds. Leaves, roots, and stems were collected from 14-day-old plants.
Total RNA was isolated from the selected anthers and leaf tissues by using the NucleoSpin RNA Plant kit (Macherey–Nagel, Germany) according to the manufacturer’s protocol. Then, the total RNA was treated with RNase-free DNaseI provided in the kit for 37 °C for 45 min to remove any contaminating genomic DNA. After phenol:chloroform extraction, the total RNA was alcohol precipitated and dissolved in RNase-free water. After checking, the integrity of the total RNA in 1% agarose followed by spectrophotometric quantitation (1 O.D. is equal to 40 μg/ml of RNA) was dissolved in RNase-free water. For preparing cDNA subtraction library, mRNA was purified from the total RNA by passing it through oligo (dT) cellulose column according to Sambrook et al.
Preparation of cDNA Subtraction Library and Identification of Anther-Specific Genes
The first-strand cDNA synthesis was carried out using 2 μg of mRNA using Superscript III Reverse Transcriptase (Invitrogen) in a reaction volume of 20 μl with the oligo d(T) primer according to the manufacturer’s protocol. A cDNA subtraction library was constructed using the cDNA prepared from mRNA of 3–4-day-old anthers with intact tapetum as tester and the cDNA synthesized from leaf tissue as driver, using PCR-Select cDNA Subtraction Kit from Clontech (according to the manufacturer’s instructions). The obtained cDNA was digested with RsaI in a 12 μl reaction. This was followed by the ligation of two adapters to the digested fragments. These fragments were then hybridized in a 200 μl reaction volume followed by the PCR reactions as elucidated in the manufacturer’s protocol. Adapter-/anther-specific sequences were then enriched by a secondary PCR. Efficiency of subtraction procedure was monitored by using the sesame-specific GADPH gene as the internal control.
The anther-specific sequences obtained after PCR-Select subtraction were cloned in pGEM-T vector (Promega) and transformed in Escherichia coli (DH5α) for subsequent analysis. The clones were randomly picked up and sequenced individually. The sequencing was done using an automated DNA sequencer (Big Dye Terminator Automated Sequencer, Applied Biosystems) and primers (M13F and M13R).
Primers were designed using Primer BLAST program from NCBI. Primers were checked through the RT-PCR-mediated amplification of a single band of appropriate size.
Isolation of Full-Length SiBG Gene and Their Upstream Elements
The full-length cDNA sequences of the SiBG gene were isolated by 5′ RACE according to the protocol of Thermo Fisher Scientific Kit. The forward primer was supplied with the kit and the reverse primer was designed from the 661-bp sequence of the cDNA sequence generated from the cDNA subtraction library. The PCR-amplified fragment was gel eluted using gel extraction kit (QIAGEN) and ligated with pGEMT vector followed by TA cloning and transformed into DH5α strain of E. coli. The ampicillin-resistant white colonies appearing in X-gal/IPTG plates were checked through colony PCR and then sequenced. The obtained sequence was then used to design primers to carry out isolation of the upstream element of SiBG gene by using 5′ walking procedure using sesame genomic DNA following the protocol of Chatterjee et al. (2013).
Construction of Chimeric SiBGproplus-GUS Construct for Plant Transformation
The PCR-amplified SiBG upstream element along with the first exon, first intron, and 5 bp of the second exon fused with BamHI and HindIII sites was cloned in plasmid vector pBI221 at the BamHI and HindIII sites and fused with GUS gene by replacing the CAMV35S promoter. The newly modified vector was named pBI221 (− 35S, + SiBGproplus). The cassette containing the SiBGproplus fused with GUS was cut out from the pBI221 (− 35S/+ SiBGproplus) by digestion with HindIII and EcoRI and ligated at the same site of the MCS of pPZPY112 binary vector. The chimeric pPZPY112 vector with the GUS reporter gene under SiBGproplus was then finally introduced within Agrobacterium strain LBA4404 for raising transgenic lines.
Development of Transgenic Nicotiana tabacum Lines
The protocol for the stable transformation of N. tabacum was performed as described by Horsch (1985) with slight modifications. The Nicotiana plants were maintained in sterile tissue culture conditions by micropropagation in Murashige-Skoog (MS) media with 0.8% agarose. The leaf discs cut out from 12-day-old plants were used as explants for the purpose of transformation. Explants were transferred to MS medium supplemented with 3 mg/l of BAP (RM) and incubated for 48 h prior to infection. The Agrobacterium strain LBA4404 harboring the chimeric pPZPY112 with SiBGproplus-GUS was grown for 48 h in LB medium containing 50 mg/l rifampicin and 25 mg/l of chloramphenicol at 28 °C. This was followed by fresh inoculation with previous culture in LB medium and grown until the O.D. reaches 0.8. The cells were centrifuged and resuspended in 25 ml of liquid MS medium. Then, the 48 h pre-incubated leaf discs (explants) of Nicotiana tabacum were infected by dipping them in Agrobacterium suspension for 30 min. The explants were then replaced back into the same plate for 48 h for co-cultivation. This was done in a culture room under scattered light. After the period of co-cultivation, explants were rinsed twice for 5 min each with 25 ml of sterile water and were plated on MS medium supplemented with 3 mg/l of BAP, 50 mg/l of kanamycin, and 300 mg/l of cefotaxim with 0.8% agar for 10 days. Following this, the explants were transferred to fresh medium with the same constituents but the concentration of selection antibiotic kanamycin was increased to 100 mg/l. Dark green plantlets that regenerated/differentiated on this medium from the cut edge of leaf discs were transferred to the MS basal medium suplemented with 100 mg/l kanamycin selection and subsequent subculture was followed to make a full-grown plant.
Selection of Positive Transformants and Their Hardening
The positive integration of the gene cassette within the regenerated plantlets was checked by direct PCR method with the kanamycin and promoter-specific primer (KanF/KanR and SiBGproF/SiBGproR, respectively) using the KAPA3G Direct PCR kit (Kapa Biosystems), using the manufacturer’s protocol. The lines which showed positive and correct amplification with all combinations of primers including SiBGpro and Kan primers were hardened and grown till floral buds appeared. GUS histochemical assay of different parts of the plant was performed as stated below. Thus, the positive transgenics were confirmed through this assay.
Aceto-Carmine Staining of Anthers
One percent of the aceto-carmine staining solution was prepared by adding carmine powder to boiling 45% glacial acetic acid, cooled rapidly, and then filtered into a dark glass. Five milliliters per gram of 10% ferric chloride (FeCl2.6H2O) was added to the solution to intensify the stain. The anthers of proper stage from the transgenic lines were taken and stained after making very fine sections to release the pollen mother cells from the anther lobes. These were then observed under the compound microscope under both ×10, ×40, and ×100 magnifications.
Treatment with ABA
The transgenic and vector control plants were treated with three different working concentrations of ABA. Half MSO solution of ABA was prepared from 1 M methanolic stock solution which was in turn prepared from the standard hormone provided by HiMedia. Of ABA, 10, 20, and 30 μM were sprayed onto the transgenic whole plants and kept at 24 °C for 6 h as described in plant growth conditions. After the treatment duration, the leaves from the respective plants were transferred to GUS solution for further histochemical assay.
Dehydration Stress Treatment
Full-grown transgenic lines of Nicotiana tabacum were subjected to dehydration stress by withholding water for 10 days after transfer to soil conditions. Leaves of these plants were detached from the whole plants on completion of the dehydration stress and then subjected to GUS histochemical assay.
Abbreviations
- ABA :
-
Abscisic acid
- bp:
-
Base pair
- PMC:
-
Pollen mother cells
- CDS`:
-
Coding DNA sequence
- cDNA:
-
Complementary deoxyribonucleic acid
- kb:
-
Kilobase
- MS:
-
Murashige and Skoog
- PDB:
-
Protein data bank
- PCR:
-
Polymerase chain reaction
- RNase A:
-
Ribonuclease A
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- T1:
-
First-generation transgenic line
References
Abou-Gharbia HA, Shahidi F, Adel A, Shehata Y, Youssef MM (1997) Effects of processing on oxidative stability of sesame oil extracted from intact and dehulled seeds. J Am Oil Chem Soc 74(3):215–221. https://doi.org/10.1007/s11746-997-0126-9
Ali GM, Yasumoto S, Katsuta M (2007) Assessment of genetic diversity in sesame (Sesamum indicum L.) detected by amplified fragment length polymorphism (AFLP) markers. Electron J Biotechnol 10(1):0–0. https://doi.org/10.2225/vol10-issue1-fulltext-16
Biabani AR, Pakniyat H (2008) Evaluation of seed yield-related characters in sesame (Sesamum indicum L.) using factor and path analysis. Pak J Biol Sci 11(8):1157–1160. https://doi.org/10.3923/pjbs.2008.1157.1160
Bino RJ (1985) Histological aspects of microsporogenesis in fertile, cytoplasmic male sterile and restored fertilePetunia hybrida. Theoret Appl. Genetics 69(4):423–428. https://doi.org/10.1007/BF00570912
Bray EA (2004) Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J Exp Bot 55(407):2331–2341
Bucciaglia PA, Zimmermann E, Smith AG (2003) Functional analysis of a β-1,3-glucanase gene (Tag 1) with anther-specific RNA and protein accumulation using antisense RNA inhibition. J Plant Physiol 160(11):1367–1373. https://doi.org/10.1078/0176-1617-01207
Chatterjee M, Mazumder M, Basu D (2013) Functional analysis of the promoter of a glycosyl hydrolase gene induced in resistant Sinapis alba by Alternaria brassicicola. Phytopathology 103(8):841–850. https://doi.org/10.1094/PHYTO-11-12-0303-R
Chaudhury A (1993) Nuclear genes controlling male fertility. Plant Cell 5(10):1277–1283. https://doi.org/10.1105/tpc.5.10.1277
Clancy M, Hannah LC (2002) Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol 130(2):918–929. https://doi.org/10.1104/pp.008235
Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48(1):355–381. https://doi.org/10.1146/annurev.arplant.48.1.355
Cui X, Wang Q, Yin W, Xu H, Wilson ZA, Wei C, Pan S, Zhang D (2012) PMRD: a curated database for genes and mutants involved in plant male reproduction. BMC Plant Biol 12(1):215. https://doi.org/10.1186/1471-2229-12-215
Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12(10):1799–1810. https://doi.org/10.1105/tpc.12.10.1799
Dickinson HG, Lewis D (1973) The formation of the tryphine coating the pollen grains of Raphanus, and its properties relating to the self-incompatibility system. Proc R Soc London Ser B Biol Sci 184(1075):149–165. https://doi.org/10.1098/rspb.1973.0040
Doxey AC, Yaish MWF, Moffatt BA, Griffith M, McConkey BJ (2007) Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol Biol Evol 24(4):1045–1055. https://doi.org/10.1093/molbev/msm024
Frankel R, Izhar S, Nitsan J (1969) Timing of callase activity and cytoplasmic male sterility in petunia. Biochem Genet 3(5):451–455. https://doi.org/10.1007/BF00485605
Garai AK, Datta JK (1999) Influence of plant growth regulators on growth, morpho-physiological characters and yield of summer sesame (Sesamum indicum L. cv. Rama) under moisture stress. Acta Physiol Plant 21(3):277–281. https://doi.org/10.1007/s11738-999-0043-7
Glass M, Barkwill S, Unda F, Mansfield S (2015) Endo-β-1,4-glucanases impact plant cell wall development by influencing cellulose crystallization. J Integr Plant Biol 57(4):396–410. https://doi.org/10.1111/jipb.12353
Goldberg RB (1993) Anther development: basic principles and practical applications. Plant Cell 5(10):1217–1229. https://doi.org/10.1105/tpc.5.10.1217
Grant I, Beversdorf WD, Peterson RL (1986) A comparative light and electron microscopic study of microspore and tapetal development in male fertile and cytoplasmic male sterile oilseed rape (Brassica napus). Can J Bot 64(5):1055–1068. https://doi.org/10.1139/b86-144
Heslop-Harrison J (1968) Pollen wall development. Science 161(3838):230–237. https://doi.org/10.1126/science.161.3838.230
Horner HT Jr, Rogers MA (1974) A comparative light and electron microscopic study of microsporogenesis in male-fertile and cytoplasmic male-sterile pepper (Capsicum annuum). Can J Bot 52(3):435–441. https://doi.org/10.1139/b74-056
Horsch R (1985) A simple and general method for transferring genes into plants. Science 227(4691):1229–1231. https://doi.org/10.1126/science.227.4691.1229
Huang G, Liu X, Liu L, Ye F, Zhang M, Shu Y (2008) Comprehensive evaluation of multiple cropping systems on upland red soil. Front Biol China 3(3):344–350. https://doi.org/10.1007/s11515-008-0047-5
Izhar S, Frankel R (1971) Mechanism of male sterility in petunia: the relationship between pH, callase activity in the anthers, and the breakdown of the microsporogenesis. Theoret Appl Genetics 41(3):104–108. https://doi.org/10.1007/BF00277751
Jin W, Horner HT, Palmer RG (1997) Genetics and cytology of a new genic male-sterile soybean [Glycine max (L.) Merr.] Sex Plant Reprod 10(1):13–21. https://doi.org/10.1007/s004970050062
Kadkhodaie A, Razmjoo J, Zahedi M, Pessarakli M (2014) Selecting sesame genotypes for drought tolerance based on some physiochemical traits. Agron J 106(1):111. https://doi.org/10.2134/agronj2013.0260
Kandasamy MK, Nasrallah JB, Nasrallah ME (1994) Pollen-pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development 120:3405–3418
Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291(5511):2141–2144. https://doi.org/10.1126/science.291.5511.2141
Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2(12):1201–1224. https://doi.org/10.1105/tpc.2.12.1201
Lee KH, Piao HL, Kim H-Y, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee I-J, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126(6):1109–1120. https://doi.org/10.1016/j.cell.2006.07.034
Leubner-Metzger G, Meins F (2000) Sense transformation reveals a novel role for class I β-1,3-glucanase in tobacco seed germination. Plant J 23(2):215–221. https://doi.org/10.1046/j.1365-313x.2000.00773.x
Mariani C, Beuckeleer MD, Truettner J, Leemans J, Goldberg RB (1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347(6295):737–741. https://doi.org/10.1038/347737a0
Mepham RH, Lane GR (1970) Observations on the fine structure of developing microspores ofTradescantia bracteata. Protoplasma 70(1):1–20. https://doi.org/10.1007/BF01276839
Morris, J.B., 2002. Food, industrial, nutraceutical, and pharmaceutical uses of sesame genetic resources. Trends in new crops and new uses 153–156
Mun J-H, Lee S-Y, Yu H-J, Jeong Y-M, Shin M-Y, Kim H, Lee I, Kim S-G (2002) Petunia actin-depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene 292(1-2):233–243. https://doi.org/10.1016/S0378-1119(02)00646-7
Rao VP, Rao SP (1981) Chemical composition and fatty acid profile of high yielding varieties of oilseeds. Indian J Agric Sci 51:703–707
Romero I, Fernandez-Caballero C, Goñi O, Escribano MI, Merodio C, Sanchez-Ballesta MT (2008) Functionality of a class I beta-1,3-glucanase from skin of table grapes berries. Plant Sci 174(6):641–648. https://doi.org/10.1016/j.plantsci.2008.03.019
Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8(11):1444–1453. https://doi.org/10.1017/S1355838202020551
Roy N, Abdullah S, Jahan MS (2009) Yield performance of sesame (Sesamum indicum L.) varieties at varying levels of row spacing. Res J Agric Biol Sci 5:823–827
Samman S, Chow JWY, Foster MJ, Ahmad ZI, Phuyal JL, Petocz P (2008) Fatty acid composition of edible oils derived from certified organic and conventional agricultural methods. Food Chem 109(3):670–674. https://doi.org/10.1016/j.foodchem.2007.12.067
Schrauwen J a M, Mettenmeyer T, Croes AF, Wullems GJ (1996) Tapetum-specific genes: what role do they play in male gametophyte development? Acta Botanica Neerlandica 45(1):1–15. https://doi.org/10.1111/j.1438-8677.1996.tb00491.x
Scott R, Hodge R, Paul W, Draper J (1991) The molecular biology of anther differentiation. Plant Sci 80(1-2):167–191. https://doi.org/10.1016/0168-9452(91)90281-C
Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of flowering locus C and for the epigenetic maintenance of repression. Plant Cell 14(10):2527–2537. https://doi.org/10.1105/tpc.004564
Shi Y, Zhao S, Yao J (2009) Premature tapetum degeneration: a major cause of abortive pollen development in photoperiod sensitive genic male sterility in rice. J Integr Plant Biol 51(8):774–781. https://doi.org/10.1111/j.1744-7909.2009.00849.x
Stieglitz H (1977) Role of beta-1,3-glucanase in postmeiotic microspore release. Dev Biol 57(1):87–97. https://doi.org/10.1016/0012-1606(77)90356-6
Wang W, Bianchi L, Scali M, Liu L, Bini L, Cresti M (2009) Proteomic analysis of β-1,3-glucanase in grape berry tissues. Acta Physiol Plant 31(3):597–604. https://doi.org/10.1007/s11738-008-0269-9
Warmke HE, Overman MA (1972) Cytoplasmic male sterility in sorghum. I. Callose behavior in fertile and sterile anthers. J Hered 63(3):103–108. https://doi.org/10.1093/oxfordjournals.jhered.a108244
Yaish MWF, Doxey AC, McConkey BJ, Moffatt BA, Griffith M (2006) Cold-active winter rye glucanases with ice-binding capacity. Plant Physiol 141(4):1459–1472. https://doi.org/10.1104/pp.106.081935
Acknowledgements
The authors are indebted to the Director, Bose Institute, Kolkata, for providing the necessary facilities for carrying out our research. We are grateful to University Grants Commission for providing fellowship to the author. We are also indebted to Department of Biotechnology, Government of India, for funding in the form of project.
Author information
Authors and Affiliations
Contributions
The study was conceived, planned, and executed by SP, MM, and DB. All experimental works were carried out at different stages of the study by SP, SM, AB, BM, MM, US, AM, and AD. Time to time technical guidance, whenever necessary, was offered by DB, MM, and SP. The manuscript was edited and prepared by SP, MM, and DB. All authors read and approved the final manuscript prepared for submission by SP, MM, and DB.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
Supplementary Figure S1
Agarose gel profile of the subtracted cDNA population and its validation: (A) The cDNA obtained from the anthers of sesame buds was subtracted from that of the leaves following the protocol provided with the PCR-Select cDNA Subtraction Kit from Clontech. Lane 1 shows the 500 bp ladder, Lane 2 shows the test sample (subtracted cDNAs, lane 3 shows the unsubtracted control cDNA, and lane 4 shows the 100 bp ladder. (B) The subtracted cDNA population was then purification using QIAGEN purification kit. Further, the purified cDNA was alcohol precipitated and then run on a 1% Agarose gel to check the quality. Lane 1 shows the 100 bp ladder; Lane 2 shows the purified subtracted cDNA population. (C) The subtracted and unsubtracted libraries were checked for the expression of the GAPDH gene. Reduction in the expression of GAPDH in the subtracted population is an evidence of successful subtraction. Lanes 1&5 depict 18 cycles of PCR for the subtracted and unsubtracted libraries respectively. Similarly, lanes 2&6, 3&7 and 4&8 depict 23, 28 and 33 cycles of either libraries as shown. For the unsubtracted cDNA library, the expression of GADPH is strong between 18 and 33 PCR cycles. However, the expression of the same is markedly low in the subtracted cDNA population even after 33 cycles, whereas no expression is seen in from 18 to 28 PCR cycles. Thus, it is apparent that proper and efficient subtraction has taken place. C.No. Stands for a number of cycles. (PDF 629 kb)
Supplementary Figure S2
Expression pattern of the 17 selected transcripts in the different tissues of sesame: RT-PCR data showing the amplification of 12 out of 17 transcripts in anther tissue, whereas no comparable amplification of the same was detected in other tissues of the plant in the gel. GAPDH is taken as internal control. (JPEG 1249 kb)
Supplementary Figure S3
Isolation of full-length β-1,3-glucanase (BISI27f) gene by 5′ RACE and its upstream sequence: (A) Full-length β-1,3-glucanase (SiBG) gene was isolated through 5’RACE. An 813 bp long EST sequence was obtained after the cDNA subtraction which had significant homology with the Nicotiana tabecum β-1,3-glucanase gene. (B) The isolated fragment was incorporated into a TA clone and sequenced. Primers were designed from it’s 5′ region, and then 5’RACE genome walking procedure was performed to isolate the upstream sequence of the SiBG gene. A 512 nt sequence of the upstream element was isolated from the genome walking. (C) Further 5′ walking was performed in a similar way which resulted in a total of 712 nt long upstream sequences. (PDF 458 kb)
Supplementary Figure S4
(a) Schematic representation of the isolation of upstream and 1st intron nucleotide sequence of β-1,3-glucanase (BISI27f) gene through cDNA subtraction followed by 5′ RACE and PCR mediated genome walking. (b) Schematic representation of the construct containing the 437 bp upstream sequence, first exon, first intron and the fusion sequence of the sesame β 1,3-glucanase gene with the GUS reporter gene. (PPTX 51 kb)
Supplementary Figure S5
Restriction analysis of the SiBGpro-GUS cloned in the plant expression vector called pPZPY112. A. Lanes 1 & 3 show digestion of clones with the enzymes HindIII and EcoRI which gives a pop-out at the size of around 2.8 kb which corresponded with the size of the desired cassette. Lanes 2 & 4 show digestion of clones with the enzymes HindIII and BamHI which gives a pop-out of around 670 kb which corresponds to the size of the SiBGpro which was cloned between these given sites. Lane 5 is the 1 kb plus ladder. The clones were also checked for the presence of the SiBGpro sequence and that of the kanamycin resistance gene by using the specific primers in a PCR reaction. B. Lanes 1–4 show the amplicon of the promoter sequence (437 bp) in the clones and lanes 5–8 show the presence of the kanamycin resistance gene (~500 bp). (PDF 340 kb)
Supplementary Figure S6
Induction pattern of SiBG gene upon ABA and drought treatment in sesame leaves: 4 weeks old sesame plants were treated with 20 μM ABA for 6 h and 20% PEG for 48 h each. The control plants were treated only with water. In control samples (Lane 1), there was no amplification of the β-1,3-glucanase (SiBG), whereas the ABA treated (Lane 2) and the PEG treated (Lane 3) plants showed a distinct induction of the same. GAPDH was taken as the internal control in both the cases. (JPEG 148 kb)
Supplementary Figure S7
Splicing of the first intron of the gene in anthers and upon ABA and drought treatment of leaves. (a) Amplification of the SiBG gene sequence along with the first intron through RT-PCR using GUS gene specific reverse primer and SiBG gene specific 5’end forward primer (NBG-f) from the total cDNA of transgenic Nicotiana tabecum plants, treated with ABA/dehydration stress. Lane 1 shows amplification using genomic DNA which results in a band of a larger size (100 bp larger). Lanes 2 and 3 show amplification in anther cDNAs of stages II and III. Lanes 4, 5 and 6 show amplification from the cDNAs of leaves treated with ABA at 10 μM, 20 μM and 30 μM concentrations. Lane 7 showed control without stress. Lanes 8, 9 and 10 show amplification from cDNAs of leaves exposed to drought stress (days 3, 5 and 7 respectively). (b) In sesame also we have tested the expression of β-1,3-glucanase (SiBG) on external application of ABA at a concentration of 10 μM, 20 μM and 30 μM as well as exposing sesame plants under drought for 7 days. Lane 1 shows amplification using genomic DNA which results in a band of a larger size (100 bp larger). Lanes 2 and 3 show amplification in anther cDNAs of stages II and III. Lanes 4, 5 and 6 show amplification from the cDNAs of leaves treated with ABA at 10 μM, 20 μM and 30 μM concentrations. Lane 7 showed control without stress. Lanes 8, 9 and 10 show amplification from cDNAs of leaves exposed to drought stress (days 3, 5 and 7 respectively). In both the cases, RT-PCR analysis of the ABA/drought treated sesame cDNA using the primers NBG-f (+1 bp to +21 bp) and NBG-r (+132 bp to +150 bp) designed from 5’region of the gene showed induction of the gene. NBG-f – AGCTGTCGACAAGCTTCGACGGCCCGGGCTGGTAAC. NBG-r- AGCTGGATCCAGCACCTGCCAGTGAAAACC. GUS r- CAATTGCCCGGCTTTCTTGTAAC (PDF 629 kb) (PNG 81 kb)
Supplementary Table S1
(DOCX 49 kb)
Supplementary Table S2
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Parveen, S., Mazumder, M., Bhattacharya, A. et al. Identification of Anther-Specific Genes from Sesame and Functional Assessment of the Upstream Region of a Tapetum-Specific β-1,3-glucanase Gene. Plant Mol Biol Rep 36, 149–161 (2018). https://doi.org/10.1007/s11105-017-1054-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-017-1054-y