Abstract
Grape berries are considered recalcitrant materials in proteomic analysis, because berry tissues contain large amounts of secondary metabolites, especially phenolic compounds, which severely interfere with protein extraction and electrophoresis separation. We report hereby a PVPP/TCA-based protein extraction protocol for grape berries. Phenolic compounds in berry extracts were removed with repeated PVPP cleanups, and proteins were recovered with TCA precipitation. Protein resolution in 2-D gels was gradually improved with the increase of PVPP cleanup steps. By the protocol, about 760 protein spots of berry tissues were clearly resolved in 2-D gels with CBB staining. This protocol was also used to analyze β-1,3-glucanase (EC 3.2.1.39) in berry tissues. An anti-synthetic peptide antibody was prepared against 15 amino acid sequence residing on the surface of β-1,3-glucanase molecule. It detected two major spots in 2-D blots of berry extracts. The spots were identified by MALDI-TOF analysis as β-1,3-glucanase. The present study validates that β-1,3-glucanase is present in higher abundance in berry skins than in pulps, and in red berries than in white berries. Therefore, β-1,3-glucanase displays a tissue-specific expression. The preferential accumulation of β-1,3-glucanase in skins may be relevant to berry ripening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grapevine (Vitis vinifera L.) and related species attract intensive genomic and proteomic studies because of their economic importance worldwide for both fruit and beverage. Particularly, the knowledge of proteins present in grape berries and juices is important to winemakers and juice processors (Vivier and Pretorius 2002).
Grape berries consist of three major parts (skin, pulp and seed), all of which contribute to the aroma, color, and flavor characters of wine (Grimplet et al. 2007). Skin and pulp contain high amounts of phenolic compounds, e.g., tannins, anthocyanins, terpenes, etc. (Saravanan and Rose 2004), which severely interfere with protein extraction and electrophoresis separation. In ripe berries, more than 99% of the pulp cell volume is occupied by large vacuoles; sugar content can be as high as 26%, while protein content only represents 0.05% of the pulp fresh weight (Mathar et al. 2002; Sarry et al. 2004). Therefore, it is difficult to extract protein from grape tissues for proteomics analysis. Recently, protein extraction for grape berry clusters was optimized, and the phenol-based protocols were found to be superior to the TCA/acetone methods (Vincent et al. 2006). Likewise, the phenol-based method was used in proteome analysis of grape skins (Deytieux et al. 2007; Negri et al. 2008). However, the phenol-based methods are complicated and time-consuming. It would be interesting to develop a simple, non-phenol based protein extraction protocol for grape berries.
β-1,3-Gucanase is normally classified as a pathogenesis-related (PR) protein, induced upon pathogen attack (Menu-Bouaouiche et al. 2003). In grapevine, the expression of β-1,3-glucanase was reported in leaves (Renault et al. 1996), cell cultures (Deloire et al. 1997), and in berries exposed to pathogens and their elicitors (Jacobs et al. 1990; Renault et al. 1996), and wounding treatment (Derckel et al. 1998; Kraeva et al. 1998), but β-1,3-glucanase activity was low or undetectable in preveraison berries (Robinson et al. 1977; Jacobs et al. 1990). Recently, many proteins relevant to grape berry ripening were identified (Sarry et al. 2004; da Silva et al. 2005, Deytieux et al. 2007; Giribaldi et al. 2007; Negri et al. 2008), of which β-1,3-glucanase was found to be differentially expressed during berry ripening both at the proteome and at the transcriptome levels. Thus, there are still inconsistent results concerning the presence and activity of β-1,3-glucanases in berries.
In the present paper, a simple protocol based on PVPP/TCA was developed for protein extraction of grape berries and the expression of β-1,3-glucanase in berry tissues was evaluated with immunoblotting and MALDI-TOF analysis.
Materials and methods
Grape berries
Two high-yielding grapevine (Vitis vinifera) cv. Sangiovese and Trebbiano were used in the study. Sangiovese is the top red cultivar in Italy, and serves as the basis of such prestigious wines as Brunello and Chianti (Poni et al. 2006). Trebbiano is Italy’s most commonly planted white grape cultivar, utilized in the composition of the Chianti wine. The average fresh weight was 1.6 and 3.0 g, and the skin-to-pulp ratio (w/w) was 21.5 and 13.1%, respectively, for Sangiovese and Trebbiano berries. Ripe, healthy berries were collected from grapevines grown in the botanical garden in Siena (Italy). Samples were frozen in liquid N2 and stored at −80°C until use.
Antibody production
A 15 amino acid sequence (NH2-GLFLPNKQPKYTINF-COOH), corresponding to the conserved region (108–122 amino acid residues) of grapevine β-1,3-glucanase, was chosen for antibody production. A 3-D structure analysis showed the sequence resides on the surface of the β-1,3-glucanase molecule (http://cl.sdsc.edu/ce.html). The 15 amino acid peptide was chemically synthesized and purified by high performance liquid chromatography, and its sequence was checked by MS. About 20 μg of the synthetic peptide in 0.5 ml of phosphate buffer (pH 7.4) was mixed with 1.0 ml of Freund’s complete adjuvant. The mixture was injected into a 4-month-old New Zealand white female rabbit to produce polyclonal antibodies. This operation was repeated twice at 15-day intervals. After the final booster, the rabbit was bled and the anti-serum was separated by keeping the blood at 4°C overnight. The antiserum containing polyclonal antibodies was used in immunoblotting.
Berry protein extraction
PVPP powder (Sigma) was acidified as described previously (Charmont et al. 2005), so as to eliminate contaminants and increase polymerization. Briefly, PVPP was boiled in 10% HCl (0.1 g/mL) for 10 min and then allowed to settle down for 2 h. The resulting aqueous phase was discarded by means of aspiration. The slurry was repeatedly rinsed using distilled water until neutral pH. Finally, pure water was added to the slurry until 10% (w/v) PVPP. The PVPP suspension was stored at 4°C before use.
For berry protein extraction, skin and pulp were separated by peeling the frozen berries and removing seeds. Skin (1 g) and pulp (5 g) were separately homogenized in 5 ml extraction medium (50 mM Tris–HCl, pH 7.5, 2% SDS, 50 mM DTT, 2 mM PMSF) in a mortar (4°C). In parallel, the crude extract was adjusted to pH 8.0 with concentrated NaOH, which made the extract turn from red to blue, due to the action of anthocyanins mainly present in he skin. The homogenates were clarified by centrifugation at 16,000g for 10 min (4°C). The supernatants were transferred into new tubes and supplemented with PVPP (final 2% w/v; alternatively, the clarified extract can be added into the tube with appropriate amount of PVPP pellets.) The mixture was shaken for 10 min and then centrifuged as above. PVPP cleanup was typically conducted three times until the extract became white or yellowish. Afterwards, protein was precipitated with 10% (w/v) TCA for 1 h on ice and collected by centrifugation (15 min, 16,000g, 4°C). Protein pellets were washed with cold 10% TCA once and 80% acetone twice. Air-dried proteins were dissolved in SDS sample buffer of 2-DE rehydration solution for electrophoresis analysis. Protein was quantified by Bradford method using a Bio-Rad protein-dye reagent.
Electrophoresis and immunoblotting
IEF was performed in a Multiphor II system (GE Healthcare, USA). Proteins (100 μg) dissolved in rehydration buffer (6 M urea, 2 M thiourea, 20 mM DTT, 2% CHAPS, 1% pH 4–7 IPG buffer) were loaded via overnight rehydration into 7 cm IPG dryStrip (pH 4–7 NL). IEF was programmed as follows: 1 h at 300 V, 1 h at 3,500 V, and finally 3 h at 3,500 V (15°C). Focused strips were equilibrated in two steps with an equilibration solution (75 mM Tris–HCl, 6 M urea, 1% SDS, 20% glycerol, 0.001% bromophenol blue, plus 1% DTT in the first step and 2.5% iodoacetamide in the second). SDS-PAGE was carried out in 12.5% gel using Bio-Rad Mini-Protean II apparatus.
After electrophoresis, proteins were visualized with colloidal CBB G, or electroblotted onto polyvinylidene difluoride membrane (Hybond-P, GE healthcare). The 1-D and 2-D images were processed using Quantity One and PDQUEST software (Bio-Rad), respectively (Wang et al. 2003).
For immunoblotting, protein blots were blocked with 3% BSA overnight (4°C) and then incubated with the antiserum (diluted 1:5,000) for 1 h. After washes, the blots were incubated with peroxidase-labeled anti-rabbit IgG (GE Healthcare) at a dilution of 1:8,000 for 1 h. The detection system was ECL plus Western blotting reagents (GE Healthcare). The chemiluminescent signal was detected on Kodak X-OMAT film (Sigma).
MALDI-TOF MS
Proteins detected by the anti-synthetic peptide antibody were excised from a parallel, stained 2-D gel and subjected to in-gel digestion with trypsin (Jensen et al. 1999). Proteins were first reduced (10 mM DTT) and then alkylated (50 mM iodoacetic acid). Following vacuum-drying, the gel pieces were incubated with modified porcine trypsin (10 ng/μl, Promega) in 50 mM ammonium bicarbonate for 16 h at 37°C. The supernatants were collected, vacuum-dried, re-dissolved in 10 μl 0.1% TFA and 0.5 μl added onto a matrix consisting of 0.5 μl of 5 mg/ml 2–5-dihydroxybenzoic acid in water:acetonitrile (2:1) with 0.1% TFA.
The digested fragments were analyzed on an Ettan MALDI-TOF Pro mass spectrometer (GE Healthcare). The ion acceleration voltage was 20 kV. Each spectrum was internally calibrated with the masses of two trypsin autolysis products. MS spectra were used to search the NCBI nonredundant database for homologous sequences with Mascot software (http://www.matrixscience.com, Matrix Science, London). Protein identification was achieved on the basis of corresponding experimental and theoretical peptide-mass-fingerprinting data using a peptide mass tolerance of 50 ppm, carbamylation modification of cysteine residues and allowing a single missed tryptic cleavage. Theoretical Mr and pI of identified proteins were predicted by sequence entry at http://www.expasy.ch/tools/pi_tools.html.
Results
Grape berry protein extraction
Ripe Sangioves berries were used for developing protein extraction protocol because they have a high skin-to-pulp ratio (21.5%) and phenolic compounds are abundant in skins. Berry proteins were extracted in aqueous medium, phenolic compounds in extracts were removed by PVPP cleanup and proteins were recovered by TCA precipitation. Rather than plus water-insoluble PVPP powder in the medium, we used the PVPP suspension, which had been treated to be free of water-soluble PVP (incompatible with TCA).
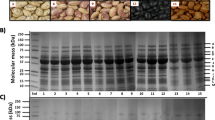
A step-by-step improvement on berry protein separation in 2-D gels was observed with the increase of PVPP cleanup steps (Fig. 1); 2-D maps with no PVPP cleanup (Fig. 1a), PVPP cleanup once (Fig. 1b) and twice (Fig. 1c) were characterized by less well-resolved spots and severe horizontal tailings. An appropriate resolution of berry proteins was obtained after PVPP cleanup thrice (Fig. 1d). Obviously, PVPP cleanup is an effective step to remove phenolic substances and to improve berry protein separation. Visually, three times PVPP cleanup can take away most of the pigment substances and result in a white or yellowish extract. Using the PVPP/TCA protocol, the average protein yield was approximately 107 μg/g for de-seeded berries (with skin and pulp), and about 760 protein spots were clearly resolved and reproducibly obtained in 2-D gels.
Effect of PVPP cleanup on grape protein separation in 2-D gels. Protein was extracted from ripe Sangiovese berries (skin and pulp), without PVPP cleanup (a), or subjected to PVPP cleanup once (b), twice (c) and thrice (d), and finally, TCA precipitation. About 100 μg of proteins were resolved by IEF on 7 cm IPG strips, followed by SDS-PAGE in 12.5% gels. Gels were visualized with colloidal CBB
Furthermore, the pH effect on PVPP cleanup was examined. The pH of unbuffered berry extracts ranges from 3.4 to 4.2, depending on cultivars. The protein profiles of pH 8.0 extract and pH 3.5 extract were almost the same, with similar spot number and gel quality (Fig. 2). Thus, the pH of berry extracts had no visible effect on PVPP cleanup. There is no need to adjust the pH of berry extracts before PVPP cleanup, though it was reported that a low pH is favorable for phenolic compounds binding to PVPP or PVP molecules (Toth and Pavia 2001).
The pH effect of berry extracts on removing phenolic compounds by PVPP cleanup. Protein was extracted from ripe Sangiovese berries (skin and pulp), with pH adjustment (a, pH 8.0) or without pH adjustment (b, pH 3.5) of crude extracts, and subjected to PVPP cleanup thrice and TCA precipitation. About 150 μg of proteins were separated by IEF with 7 cm IPG dry strips and then by SDS-PAGE (12.5% gel). Gels were visualized with colloidal CBB
Immunodetection of β-1,3-glucanase in grape berry tissues
The use of synthetic peptide as immunogen is applicable when a complete protein is not available in sufficient quantities to carry out an adequate immunization response (Humphrey et al. 1990). Theoretically, a 14 or 15-amino acid peptide is of a sufficient length to elicit an effective antibody response. In our experiment, a 15 amino acid peptide was used for producing antibody against grapevine β-1,3-glucanase.
A 3D-structure analysis showed the 15 amino acid sequence (108–122) on the surface area of the folded β-1,3-glucanase protein. The most conserved region in the 15 amino acid sequence is QPK, which is a β-strand between two irregular coil fragments in β-1,3-glucanase (Fig. 3). Sequence regions with β-strand or amphipthic helix character have been found to be antigenic (Parker and Hodges 1991). Perhaps, QPK determines the epitope of the antibody.
A 3-D structure analysis of β-1,3-glucanase. The potential epitope region (108–122 residues) resides in the surface of β-1,3-glucanase. α-Helix is shown in white, β-strand in black, irregular coil in grey. The structure was drawn by spdbv software (http://www.expasy.ch/spdbv/)
The immunoblot analysis showed that the antibody was highly specific for the immunogen peptide and displayed a strong reactivity even at the dilution 1:5,000 (Fig. 4a). The preimmune rabbit serum was not reactive (data not shown). In ripe Sangioves berry (including skin and pulp) extract, the antibody reacted with a 21 kD polypeptide (Fig. 4b, see arrowhead), which consisted of two isoforms with pIs 5.8 and 6.4, respectively (Fig. 5). The pI 6.4 spot was more abundant in either skin or pulp, as estimated by its signal intensity. It was excised from stained 2-D gel and subjected to MS analysis. Peptide mass list was used to search databases for homologous sequences. As a result, five tryptic peptide mass peaks matched to grapevine β-1,3-glucanase (Table 1). Thus, the identity of the protein recognized by the antibody was confirmed by MALDI-TOF MS. The size and pI of β-1,3-glucanase isoforms reported here were similar to those in a recent study (Deytieux et al. 2007).
Specificity of the antibody designed against grapevine β-1,3-glucanase. a CBB-stained gel (lane 1) and blot (lane 2) of the synthetic peptide NH2-GLFLPNKQPKYTINF-COOH (1 μg). The arrowhead indicates the peptide (1.8 kD). b CBB-stained gel (lane 1) and blot (lane 2) of Sangiovese berry protein (28 μg). The arrowhead indicates the immunoreactive band (21 kD)
The 2-D immunodetection of β-1,3-glucanase in grape berry tissues. Sangiovese berry protein (100 μg) was extracted by PVPP/TCA method and resolved by IEF using 7 cm pH 4–7 IPG dryStrip. Secondary SDS-PAGE was carried out on a 12.5% gel. a (skin), b (pulp) CBB G-stained gels. c (skin), d (pulp) Blots probed with the anti-synthetic peptide antibody (1;5,000 dilution). Arrows indicate the two putative isoforms of β-1,3-glucanase (21 kD). On stained gels, the major isoform (pI 6.4) is indicated with a circle
The expression of β-1,3-glucanase in different grapevine tissues and cultivars was examined by immunoblot analysis. On an equal protein basis, β-1,3-glucanase was present in abundance in skin, and in less abundance in pulp, regardless of red or white berries. Furthermore, β-1,3-glucanase was richer in counterpart tissues of red berries (Sangiovese) than in white berries (Trebbiano; Fig. 6). The order of β-1,3-glucanase abundance in various berry tissues is as follows: red berry skin, white berry skin, red berry pulp and white berry pulp. Besides, no proteins were detected by the anti-synthetic peptide antibody in other tissues, including leaves, roots and stems (data not shown). Obviously, the expression of β-1,3-glucanase on the protein level differs among tissues and cultivars.
Comparative analysis of β-1,3-glucanase abundance between red and white grape cultivars. Proteins were extracted from cv. Sangiovese (red berry) and Trebbiano (white berry) by the PVPP/TCA method. Protein (25 μg/lane) was resolved by SDS-PAGE (a), blotted, and probed with the anti-synthetic peptide antibody (b). 1 red berry skin, 2 white berry skin, 3, red berry pulp, 4 white berry pulp. The arrow indicates β-1,3-glucanase (21 kD)
Discussion
Removal of phenolic compounds from grape berry extracts using PVPP
Grape berries contain a high content of phenolic compounds, but a low content of proteins. During tissue homogenization, phenolic compounds released from berry tissues can form strong links with proteins, and the resulting complexes are more hydrophobic and susceptible to protein aggregation and precipitation. An effective removal of phenolic compounds from plant extracts is by using vinyl pyrrolidone polymers, e.g., water-soluble PVP and water-insoluble PVPP. PVP(P) can forms hydrogen bonds with phenolic compounds. The precipitating PVP(P)–phenolic complex can be eliminated from the extracts by centrifugation (Toth et al. 2001).
Nevertheless, commercial PVPP may contain a small fraction of PVP (our observation). PVP is incompatible with conventional acetone, alcohol and TCA precipitations, and co-precipitates with protein, thus interfering with subsequent electrophoresis analysis. To overcome this problem, it is necessary to process PVPP by acidification to increase polymerization and to eliminate contamination of PVP. Moreover, the use of the suspension overcomes the volumetric losses of proteins, which is another problem accompanying the use of PVPP powder.
Using the protocol reported here, an average of 760 spots from deseeded berry tissues can be visualized in mini-2-D gels with colloidal CBB staining, which represents a significant improvement compared to the number of spots previously detected in grape berry (Hsu and Heatherbell 1987; Tesnière and Robin 1992) and the quality of 2-D gel maps recently reported (Sarry et al. 2004; Vincent et al. 2006; Deytieux et al. 2007). Besides, the PVPP/TCA protocol is simpler and of lower toxicity than phenol-based protein extraction methods.
The expression of β-1,3-glucanase in grape berry is related to ripening
β-1,3-Glucanase plays various functions in cell division, seed germination, bud dormancy, flower formation and fruit ripening (Morohashi and Matsushima 2000; Buchner et al. 2002; Tian et al. 2007) and in pathogenesis-related (PR) responses (Menu-Bouaouiche et al. 2003). Previous studies reported the induced expression of β-1,3-glucanase in grapevine leaves and berries under fungal attack and stress conditions (Jacobs et al. 1990; Renault et al. 1996, 2000; Deloire et al. 1997; Kraeva et al. 1998), while β-1,3-glucanase activity was low or undetectable in grape berries (Robinson et al. 1977; Jacobs et al. 1990), and no β-1,3-glucanase activity was detected in grape at any stage of berry development (Derckel et al. 1998).
In an analysis of grape skin proteome, two isoforms of β-1,3-glucanase were identified, and their abundance increased as the berry ripened, which was suggested to correlate with the increase in the activities of both isoforms during skin ripening (Deytieux et al. 2007). Likewise, a putative β-1,3-glucanase protein (CTG1029513) was predicted to be differentially expressed during the first stage of berry development (da Silva et al. 2005). The analysis of mRNA expression profiling in grape berry tissues revealed a difference in glucanase expression, and five β-1,3-glucanase genes were expressed differentially among tissues, of which two genes (AF239617 and TC44117) were found to be highly expressed in grape skins (Grimplet et al. 2007). Recently, the grapevine genome was completely sequenced. The sequence CAB60154 (gi|6273716) in the NCBI data bank represents the complete sequence of grapevine β-1,3-glucanase (CAO23358-gi|157348466; Jaillon et al. 2007). Most recently, Negri et al. (2008) analyzed proteome changes in the skin of the grape cultivar Barbera among different stages of ripening, and two spots were identified corresponding to β-1,3-glucanase, which accumulated after veraison.
In the present study, grape berries were taken from the botanical garden with excellent disease control procedures, and regular monitoring of the vines gave no indication of disease being present. No visible signs of wounding or pathogen attack were detected in the berries tested. Reasonably speaking, the abundant presence of β-1,3-glucanase in grape skins found in the present study is unlikely to be pathogen-induced and is likely to be ripening-related, in agreement with recent studies (Sarry et al. 2004; Deytieux et al. 2007; Grimplet et al. 2007; Negri et al. 2008). Grape skin, consisting of the outermost cuticle, the epidermis and the hypodermis, forms a protective structure or a barrier against invading pathogen. β-1,3-Glucanase is well documented to function as a PR protein; therefore, its preferential accumulation in berry skins may be an adaptive defense mechanism.
Abbreviations
- 2-DE:
-
Two-dimensional electrophoresis
- CBB:
-
Coomassie brilliant blue
- DTT:
-
Dithiothreitol
- IEF:
-
Isoelectric focusing
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization time of flight
- MS:
-
Mass spectrometry
- pI:
-
Isoelectric point
- PMSF:
-
Phenylmethanesulfonyl fluoride
- PR:
-
Pathogenesis-related
- PVP:
-
Polyvinylpyrrolidone
- PVPP:
-
Polyvinylpolypyrrolidone
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCA:
-
Trichloroacetic acid
- TFA:
-
Trifluoroacetic acid
References
Buchner P, Rochat C, Wuillème S, Boutin JP (2002) Characterization of a tissue-specific and developmentally regulated β-1, 3-glucanase gene in pea (Pisum sativum). Plant Mol Biol 49:171–186. doi:10.1023/A:1014910900312
Charmont S, Jamet E, Pont-Lezica R, Canut H (2005) Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings: improved recovery following removal of phenolic compounds. Phytochem 66:453–461. doi:10.1016/j.phytochem.2004.12.013
da Silva FG, Iandolino A, Al-Kayal F, Bohlmann MC, Cushman MA, Lim H, Ergul A, Figueroa R, Kabuloglu EK, Osborne C, Rowe J, Tattersall E, Leslie A, Xu J, Baek JM, Cramer GR, Cushman JC, Cook DR (2005) Characterizing the grape transcriptome: analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol 139:574–597. doi:10.1104/pp.105.065748
Deloire A, Kraeva E, Mauro MC, Bonnet E, Bierne J (1997) Immunodetection of chitinase-like and β-1,3-glucanase-like proteins secreted in vitro by embryogenic and non-embryogenic cells of grapevines. Vitis 36:51–52
Derckel JP, Audran JC, Haye B, Lambert B, Legendre L (1998) Characterization, induction by wounding and salicylic acid and activity against Botrytis cinerea of chitinases and β-1,3-glucanases of ripening grape berries. Physiol Plant 104:56–64. doi:10.1034/j.1399-3054.1998.1040108.x
Deytieux C, Geny L, Lapaillerie D, Claveroll S (2007) Proteome analysis of grape skins during ripening. J Exp Bot 58:1851–1862. doi:10.1093/jxb/erm049
Giribaldi M, Perugini I, Sauvage FX, Shubert A (2007) Analysis of protein changes during grape berry ripening by 2-DE and MALDITOF. Proteomics 7:3154–3170. doi:10.1002/pmic.200600974
Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Wheatley K, Schlauch A, Cramer GR (2007) Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8:187. doi:10.1186/1471-2164-8-187
Hsu JC, Heatherbell DA (1987) Isolation and characterization of soluble proteins in grapes, grape juice, and wine. Am J Enol Vitic 38:6–10
Humphrey PA, Wong AJ, Vogelstein B, Zalutsky MR, Fuller GN, Archer GE, Friedman HS, Kwatra MM, Bigner SH, Bigner DD (1990) Anti-synthetic peptide antibody reacting at the fusion junction of deletion-mutant epidermal growth factor receptors in human glioblastoma. Proc Natl Acad Sci USA 87:4207–4211. doi:10.1073/pnas.87.11.4207
Jacobs AK, Dry IB, Robinson SP (1990) Induction of different pathogenesis-related cDNAs in grapevine infected with powdery mildew and treated with ethephon. Plant Pathol 48:325–336. doi:10.1046/j.1365-3059.1999.00343.x
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyere C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pe ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quetier F, Wincker P (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467. doi:10.1038/nature06148
Jensen ON, Wilm M, Shevchenko A, Mann M (1999) Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. In: Link A (ed) 2-D Proteome analysis protocols (Methods in molecular biology series), vol 112. Humana, New York, pp 513–530
Kraeva E, Tesnière C, Terrier N, Romieu C, Sauvage FX, Bierne J, Deloire A (1998) Transcription of a β-1,3-glucanase gene in grape berries in a late developmental period, or earlier after wounding treatments. Vitis 37:107–111
Mathar H, Basha SM, Lu J (2002) Variation in berry protein composition of muscadine cultivars. Am J Enol Vitic 53:87–91
Menu-Bouaouiche L, Vriet C, Peumans WJ, Barre A, Van Damme EJM, Rouge P (2003) A molecular basis for the endo-β-1,3-glucanase activity of the thaumatin-like proteins from edible fruits. Biochimie 85:123–131. doi:10.1016/S0300-9084(03)00058-0
Morohashi Y, Matsushima H (2000) Development of β-1,3-glucanase activity in germinated tomato seeds. J Exp Bot 51:1381–1387. doi:10.1093/jexbot/51.349.1381
Negri AS, Prinsi B, Rossoni M, Failla O, Scienza A, Cocucci M, Espen L (2008) Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics 9:378. doi:10.1186/1471-2164-9-378
Parker JMR, Hodges RS (1991) Prediction of surface and interior regions in proteins. Part I: Linear tripeptide sequences identify structural boundaries in proteins. Pept Res 4:347–354
Poni S, Casakini L, Bernizzoni F, Civardi S, Intrieri C (2006) Effects of early defoliation on shoot photosynthesis, yield components, and grape composition. Am J Enol Vitic 57:397–407
Renault AS, Deloire A, Bierne J (1996) Pathogenesis-related proteins in grapevines induced by salicylic acid and Botrytis cinerea. Vitis 35:49–52
Renault AS, Deloire A, Letinois I, Kraeva E, Tesniere C, Ageorges A, Redon C, Bierne J (2000) β-1,3-Glucanase gene expression in grapevine leaves as a response to infection with Botrytis cinerea. Am J Enol Vitic 51:81–87
Robinson SP, Jacobs AK, Dry IB (1977) A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol 114:771–778. doi:10.1104/pp.114.3.771
Saravanan RS, Rose JKC (2004) A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4:2522–2532. doi:10.1002/pmic.200300789
Sarry JE, Sommerer N, Sauvage FX, Bergoin A, Rossignol M, Albagnac G, Romieu C (2004) Grape berry biochemistry revisited upon proteomic analysis of the mesocarp. Proteomics 4:201–215. doi:10.1002/pmic.200300499
Tesnière C, Robin JP (1992) Two-dimensional electrophoresis of the total polypeptides in ripe red grape berries. Electrophoresis 13:93–96. doi:10.1002/elps.1150130118
Tian SP, Yao HJ, Deng X, Xu XB, Qin GZ, Chan ZL (2007) Characterization and expression of β-1,3-glucanase genes in jujube fruit induced by the microbial biocontrol agent Cryptococcus laurentii. Phytopathol 97:260–268. doi:10.1094/PHYTO-97-3-0260
Toth GB, Pavia H (2001) Removal of dissolved brown algal phlorotannins using insoluble polyvinylpolypyrrolidone (PVPP). J Chem Ecol 27:1899–1910. doi:10.1023/A:1010421128190
Vincent D, Wheatley MD, Cramer GR (2006) Optimization of protein extraction for mature grape berry clusters. Electrophoresis 27:1853–1865. doi:10.1002/elps.200500698
Vivier MA, Pretorius IS (2002) Genetically tailored grapevines for the wine industry. Trends Biotechnol 20:472–478. doi:10.1016/S0167-7799(02)02058-9
Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresti M (2003) Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24:2369–2375. doi:10.1002/elps.200305500
Acknowledgments
Funding for this study was provided by ARSIA (Regione Toscana, Italy). Our special thanks are due to Drs. Elisabetta Sensi and Rita Vignani for designing the synthetic peptide.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Stobiecki.
Rights and permissions
About this article
Cite this article
Wang, W., Bianchi, L., Scali, M. et al. Proteomic analysis of β-1,3-glucanase in grape berry tissues. Acta Physiol Plant 31, 597–604 (2009). https://doi.org/10.1007/s11738-008-0269-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-008-0269-9