Abstract
Taxodium ‘zhongshansa’, an extremely flood-tolerant tree species, has been widely planted in eastern China due to its environmental attributes. Although much progress has been made in understanding its physiological response to flooding stress, as well as its rapid propagation and variety selection, little is known regarding its molecular genetics due primarily to a lack of reliable molecular markers. In this paper, 108,692 expressed sequence tags (ESTs) with a total size of 69.3 Mb derived from T. ‘zhongshansa’ were analyzed. And 10,038 simple sequence repeat (SSR) loci were identified from 8137 SSR-containing EST sequences. The average SSR frequency in the transcriptome was one in 6.90 kb of EST sequences. The most abundant repeat type was mononucleotide (6581, 65.56 %), followed by trinucleotide (2246, 22.37 %) and dinucleotide (1080, 10.76 %). After filtering unqualified SSR loci, loci with mononucleotide repeats and compound SSRs, 1958 EST-SSR loci were selected to design SSR makers. To validate this set of SSR markers, 503 primer pairs were randomly selected to amplify across 12 genomic DNA templates from three Taxodium species. Two hundred fifty-seven (51.09 %) out of 503 primer pairs amplified the expected products, of which 81 and 176 could amplify polymorphic and monomorphic bands, respectively. The functional categorization of EST sequences containing randomly selected markers revealed that 264 (52.49 %) had homology with known proteins. This set of EST-SSR markers will provide a valuable genetic and genomic tool for further genetic research in Taxodium, such as genetic map construction, quantitative trait loci mapping, and marker assisted selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Taxodium is well known for its extreme tolerance to waterlogging, thus having great ecological and economic potential (Qi et al. 2014). Taxodium ‘zhongshansa’ are interspecies hybrid clones generated from three Taxodium species, Taxodium mucronatum, Taxodium distichum, and Taxodium ascendens (Yu et al. 2011). T. ‘zhongshansa’ are versatile trees for ornamental use in urban areas, ecological use in wetlands, and economic use for timber production (Ying and Yu 2005; Hu et al. 2012; Zhang et al. 2012; Xu et al. 2013). Until now, progress has been made on understanding the physiological response to flooding stress, as well as superior clone selection and rapid propagation of T. ‘zhongshansa’ (Ying and Yu 2005). Several hybrid clones, such as ‘zhongshansa 302’, ‘zhongshansa 118’, and ‘zhongshansa 405’, have been selected and widely planted in eastern China (Yu et al. 2011). Nevertheless, little is known about the molecular genetics due primarily to a lack of reliable molecular markers in Toxadium. This greatly hinders exploring the underlying mechanisms of economically important traits and molecular breeding. Hence, there is an urgent need to develop reliable molecular makers for Taxodium.
DNA-based molecular markers provide powerful tools for constructing genetic linkage maps, assessing the genetic diversity of germplasm, quantifying population genetic structures, relatedness, evolution, determining quantitative trait loci (QTLs), and comparative genomics (Siongng et al. 2009; Koelling et al. 2012; Varshney et al. 2005; Cavagnaro et al. 2011). Microsatellite markers or simple sequence repeats (SSRs) are short units of 1–6 nucleotide tandem repeat sequences, occurring in both coding and noncoding regions of eukaryotic genomes (Tautz and Renz 1984). Due to their high level of polymorphisms, transferability, reproducibility, codominant inheritance, and the ease of detection, SSRs are the best choice for molecular markers in genetics research (Powell et al. 1996; Gupta et al. 2003).
Currently, there are two types of SSRs based on their origin: genomic SSRs (gSSRs) and expressed sequence tag derived SSR (EST-SSRs) (Yang et al. 2013). Traditional methods of developing SSRs from genomic DNA are costly and time-consuming, involving the construction of genomic DNA libraries, probe hybridization, cloning, and sequencing. EST-SSRs, also called gene-based SSRs or genetic markers, can be developed efficiently and cost-effectively with advent of Next Generation Sequencing (NGS) systems (Zhai et al. 2014). Using the Illumina system, the short reads-based de novo assembly of contigs does not require a reference sequence and a large numbers of SSR markers can be developed using this method (Iorizzo et al. 2011). Compared with the SSR markers derived from the large-scale cloning and sequencing of DNA or insufficient public EST libraries, the development of EST-SSRs from transcriptome sequences provides adequate resources for mining large numbers of SSR markers quickly in non-model organisms (Barbará et al. 2007; Mardis 2008).
EST-SSR markers have many advantages in molecular genetics research. (1) They can detect variation in the expressed portion of the genome, allowing gene tagging to bridge between significant genetic traits and genes; (2) they can be developed at low cost and high speed from EST databases; (3) once developed, these markers, unlike genomic SSRs, may be used across a number of related species (Gupta et al. 2003); and (4) they are codominant and reproducible (Saha et al. 2004). Therefore, EST-SSR markers have been developed in many important plant, animal, and microbial species, such as wheat (Nicot et al. 2004), shrimp (Franklin et al. 2004), and Auricularia polytricha (Zhou et al. 2014).
The aim of this study was to develop EST-SSR markers as genomic tools for T. ‘zhongshansa’. The transcriptome data of ‘zhongshansa 406’ were obtained by Illumina sequencing (Qi et al. 2014). MIcroSAtellite (MISA) was used to detect SSR loci in the ESTs (Thiel et al. 2003), and Primer3.0 was used for designing primers to amplify EST-SSR molecular markers (Cardle et al. 2000). Moreover, the characteristics of EST-SSRs were analyzed, and the polymorphisms and cross-species transferability of a subset of EST-SSR primers had been estimated. To our knowledge, this study is the first to develop SSR markers in the genus of Taxodium.
Materials and Methods
Plant Materials
Altogether, 12 genotypes from three Taxodium species were taken as plant materials, of which five genotypes came from Taxodium mucronatum Tenore (TM01, TM02, TM03, TM04, and TM05), four genotypes from Taxodium distichum (L.) Rich (TD01, TD02, TD03, and TD04), and two genotypes from T. ascendens Brongn (TB01 and TB02), and one hybrid clone T. ‘zhongshansa405’ (TA 405), which is an interspecies hybrid clone generated from the cross of T. mucronatum (♀) × T. distichum (♂). The plant samples were collected from Nanjing Botanical Garden in April 2014. Young leaves, collected from each tree, were transported to the laboratory in an ice box and stored in a refrigerator at −80 °C prior to DNA extraction.
Identification SSRs in ESTs and Primer Design
MIcroSAtellite (http://pgrc.ipk-gatersleben.de/misa/misa.html) was used to identify SSR loci in the transcriptome data. The criteria were set for the detection of mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide motifs with a minimum unit size of ten, six, five, five, five, and five repeats, respectively. When the distance between the two microsatellites was less than 100 bp, they were considered as a composite microsatellite. The output data included unigene IDs, repeat motifs, number of repeats, and start and end positions. Primers were designed using Primer 3.0 software (available online at http://www.genome.wi.mit.edu/genome_software/other/primer3.html ). Region 50 bp from each end of the repeat was excluded from primer site consideration. All primers were designed to have similar annealing temperatures to allow for uniform polymerase chain reaction (PCR) cycling conditions. The range of optimal primer sizes was 17–25 bp, of PCR product sizes was 100–300 bp, and of the GC content was from 40–60 %, with 50 % as the optimal.
DNA Extraction and PCR Amplification
DNA was isolated from the fresh leaves following the CTAB method (Doyle and Doyle 1987). PCR reactions were carried out in a 10 μL reaction mix consisting of 3.75 mmol L−1 MgCl2, 0.4 mmol L−1dNTPs, 0.75 μmol L−1 of each primer, 0.5 U of TaqDNA polymerase, and 20 ng of genomic DNA. The PCR amplification protocol consisted of an initial denaturation at 94 °C for 3 min, followed by 30 cycles of 30 s at 94 °C, 45 s at the annealing temperature of 59 °C, and 45 s at 72 °C, ending with a final extension at 72 °C for 7 min. Amplification products were separated on 12 % polyacrylamide gels and stained with ethidium bromide for SSR band detection.
Polymorphism and Transferability Detection of EST-SSRs
To verify the effectiveness of this set of SSR markers, 503 primer pairs were randomly selected to amplify across 12 genomic DNA templates from three Taxodium species. The primers were considered polymorphic when a locus had at least two alleles at any frequency. The SSR marker diversity index (Na, observed number of alleles; Alleles freq, Alleles frequency) was estimated by POPGENE version 1.31 (Yeh et al. 1999). Polymorphism information content (PIC) was calculated using the formula developed by Anderson et al. (1993). Additionally, the selected EST-SSR markers were assessed across three closely related species of Taxodium. The amplification ratios and polymorphic parameters (Pa, the number of polymorphic loci; Na, observed number of alleles; and Ho, observed heterozygosity of polymorphic SSRs) across three species were summarized to analyze their transferability and polymorphic variation.
Functional Annotation of EST-SSRs
An overview of the functional categories of the 503 EST-SSRs was produced by a Gene Ontology (GO) analysis using Blast2GO v2.5 software. The GO analysis assigned the sequences into three categories, ‘molecular function (MF)’, ‘biological process (BP)’, and ‘cellular component (CC)’. The terms in each category were further analyzed.
Results
Frequency and Distribution of EST-SSRs in Taxodium ‘zhongshansa’
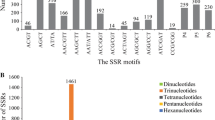
A total number of 108,692 EST sequences with a total size of 69.309123 Mb were examined in T. ‘zhongshansa’. Ten thousand thirty-eight SSR loci were identified by MISA in 8137 (7.49 %) SSR-containing sequence found in the EST database. The frequency of occurrence for SSR loci was one in 6.90 kb of EST sequences in T. ‘zhongshansa’. In 8137 SSR-containing ESTs, 6761 (83.09 %) ESTs contained one SSR locus and 1376 (16.91 %) contained two loci or more. In addition, 943 (9.39 % of 10,038 SSRs) SSR loci were considered as compound formation according to the predefined criteria (Table 1).
As shown in Table 2, the most frequent repeat motifs were mononucleotide repeats (6581, 65.56 %), followed by trinucleotides (2246, 22.37 %) and dinucleotides (1080, 10.76 %). Tetranucleotide (98, 0.98 %), pentanucleotide (17, 0.17 %), and hexanucleotide (16, 0.16 %) occurred less frequently. Table 2 also showed the distribution of repeat units. About 30.40 % (3052) of the EST-SSRs contained 10 repeat units, followed by 5 (15.03 %, 1509), 11 (14.72 %, 1478), and 6 (10.36 %, 1040). The least representative repeat units were 9 (0.88 %, 88). Moreover, there were 1324 potential EST-SSRs containing more than 13 repeat units of mononucleotide repeats.
EST-SSR Marker Development in Taxodium ‘zhongshansa’
Finally, 1958 EST-SSR markers were developed from 10038 SSR loci in T. ‘zhongshansa’ by Primer3.0 software. The SSR primers were named as TAxxxx, where TA denotes Taxodium. The remanent 8080 SSR loci were excluded from designing primers due to their inappropriate sequence structures such as short flanking sequences, SSR loci with mononucleotide repeats (easily leading to high mismatch ratio during DNA amplification), and compound SSRs. Their detailed information is shown in Additional file 1.
Polymorphism and Cross-Species Transferability Detection of EST-SSR Markers
A sample of 503 selected SSR primer pairs was applied to polymorphism detection across 12 genomic DNA using the optimized SSR-PCR system. Two hundred fifty-seven primer pairs (51.1 % of the sample) amplified the expected bands, while the remaining 246 pairs of primers amplified no product or products with unexpected size. Eighty one (Table 3) of the 257 primer pairs could amplify polymorphic (alleles of different size) and the other 176 (Addition file 2) amplified monomorphic products. Figure 1 showed the typical polymorphic bands and monomorphic band amplified by three of the 257 primer pairs. Finally, 174 alleles were found in 81 polymorphic SSR loci. The range of alleles detected on each locus was 1–4 with the average of 2.148. The allele frequency for all primers varied from 0.042 to 0.958 with the average of 0.466. The mean value of polymorphism information content (PIC) was 0.323, with a maximum of 0.66 and a minimum of 0.08. And the scope of the size about 81 polymorphic SSR loci is 12 to 36 bp; however, 80.25 % (65) of them varied from 12 to 20 bp (Table 3).
Typical polyacrylamide gel electrophoresis patterns of amplification results using three EST-SSR markers. Note: a and b show the polymorphic of Primer TA 30 (two alleles) and 40 (four alleles). c shows monomiphic of Primer TA 163. M represents DNA Maker (50 bp). Line 1 ~ 12 represent the amplification products across 12 genotypes (Line 1, 2, 3, 4, 5 stands for TM01, TM02, TM03, TM04, and TM05, respectively; Line 6,7, 8, 9 stands for TD01, TD02, TD03, and TD04, respectively; Line 10, 11 stands for TB01 and TB02, respectively; Line 12 stands for TA405)
Meanwhile, all 81 polymorphic SSR markers developed from T. ‘zhongshansa’ could amplify clear bands in the other three species, indicating a 100 % cross-species transferability ratio. The polymorphic variations of 81 primer pairs in the three species were further valued with three parameters of Pa, Na, and Ho (Table 4). The data of polymorphic parameters revealed weak interspecies differences among the three investigated species of Taxodium.
Functional Annotation for SSR-Containing EST Sequences
The original ESTs of the 503 EST-SSRs in T. ‘zhongshansa’ were annotated with gene ontology (GO), 264 (52.49 %) out of which had significant matching annotations in Nr (NCBI non-redundant protein sequences) and protein family (Pfam) on the basis of sequence similarity (Additional file 3). Studies on gene ontology focused on three categories, biological process (BP), molecular function (MF), and cellular component (CC) and the number and percentage for each category was BP (98, 37.12 %), MF (31, 11.74 %), and CC (135, 51.14 %). BP was further subdivided into eight groups represented by cellular process (60, 61.22 %), metabolic process (13, 13.27 %), localization (12, 12.24 %), signaling (5, 5.10 %), response to stimulus (3, 3.06 %), single organism process (2, 2.04 %), multicellular organismal process (2, 2.04 %), and developmental process (1, 1.02 %) (Fig. 2a). The detected matches for MF were binding (18, 58.06 %), catalytic activity (10, 32.26 %), channel regulator activity (2, 6.45 %), and enzyme regulator activity (1, 3.23 %) (Fig. 2b). Organelle (63, 46.67 %), cell (50, 37.04 %), membrane (15, 11.11 %), virion (6, 4.44 %), and extracellular region part (1, 0.74 %) constitute the groups in CC (Fig. 2c).
Discussion
Frequency, Type, and Distribution of SSRs in Taxodium ‘zhongshansa’
An EST database is an important resource for EST-SSR marker development (Vidushi et al. 2015). Fortunately, enormous quantities of EST sequences can now be generated from next-generation sequencing (NGS) technologies effectively (Ashrafi et al. 2012). But only a high quality transcriptome can develop a large number of functional SSR markers. The quality of transcriptome getting from Illumina platform could be assessed by the number of assembled transcripts, total bases of transcripts, mean length of transcripts, N50 statistic, and number of long transcripts (≥1 kb) (Wu et al. 2014; Jayasena et al. 2014; O’Nei and Emrich 2013). Here, N50 statistic is defined as the length of the shortest transcript in the set that contains the longest transcripts whose combined length reaches at least half of the sum of the lengths of all transcripts. Transcriptomes with larger number of assembled transcripts and total bases of transcripts are thought to be higher quality (Zhao et al. 2011). In the transcriptome of T. ‘zhongshansa’, 143,636 transcripts with a total of 106.3 Mb were assembled, of which 31,665 were long transcripts (≥1 kb). Besides, the mean length and N50 length of all transcripts in Taxodium transcriptome were 740 and 1324 bp, respectively, (Qi et al. 2014). Compared with the transcriptomes of pepper (Ashrafi et al. 2012) and seashore paspalum (Jia et al. 2015), the transcriptome in T. ‘zhongshansa’ contained larger number of transcripts and total bases of transcripts, indicating a higher quality of Taxodium transcriptome.
In addition to the quality of transcriptome, the frequency and density of SSRs were also affected by the search criteria set to identify SSRs, SSR search tools, datasets for the database mining, and the nature of species (Durand et al. 2010).
In comparison with former investigations, the number of SSR-containing ESTs (7.49 %, 8137) in T. ‘zhongshansa’ was higher than that in some other plants, such as citrus (5.83 %) (Chen et al. 2006), Jatropha curcas (6.8 %) (Yadav et al. 2011), Rosaceae (4 %) (Jung et al. 2005), grape (2.5 %) (Scott et al. 2000), wheat (1.72 %), rice (1.92 %), maize (0.88 %), and soybean (1.45 %) (Gao et al. 2003). These differences in the number of SSR-containing ESTs can be partly attributed to SSR search criteria and the characteristics of the EST database analyzed in (Yadav et al. 2011). The SSR search criteria can also change relative estimates of frequency of EST-SSRs (Yadav et al. 2011). The average SSR frequency is one in 6.9 kb of EST sequences in T. ‘zhongshansa’, which is lower than that in citrus (one in 5.2 kb) (Chen et al. 2006), J. curcas (one locus in 6 kb) (Yadav et al. 2011), and rosa (one in 5.5 kb) (Jung et al. 2005), but higher than that in wheat (one in 17.42 kb), rice (one in 11.81 kb), maize (one in 28.32 kb), and soybean (one in 23.80 kb) (Gao et al. 2003), showing that different microsatellite frequencies exist in diverse plants.
The highest proportion of mononucleotide repeat sequences in Taxodium ‘zhongshansa’ is in accordance with the results from J. curcas L. (Kumari et al. 2013) and wheat (Asadi and Monfared 2014), but in contrast to those from citrus (Chen et al. 2006), lily (Du et al. 2014), pineapple (Ong et al. 2012), and arabidopsis (Cardle et al. 2000), in which trinucleotides were identified as the most numerous repeats. A potential explanation for the high ratio of mononucleotide SSR loci in this study was that mononucleotide SSRs would be amplified rapidly after interspecific hybridization (Gao et al. 2011). The repeat units of 10, 5, 11, and 6 about SSR sequences account 69.73 % of the total SSRs, thus a rough trend could be discerned where the size of SSRs mainly ranged from 10 to 18 bp (Table 2). Cardle et al. (2000) demonstrated that short SSRs with size less than 20 bp have high polymorphic. The SSR motifs were also compared with the previous studies on pines (Chagné et al. 2004; Echt et al. 2011) and spruce (Rungis et al. 2004). The (A)n, (T)n, and (AT)n were also found to be the most motifs in these coniferous species. Lagercrantz et al. (1993) reported that the A/T richness of microsatellites was thought to be high in the genomic sequences of plants. And AT repeats were preferentially found at the 3′ end of the EST sequences (Rungis et al. 2004).
EST-SSR Polymorphisms and Cross-Species Transferability
This is the first report on the development and characterization of a set of functional SSR markers in Taxodium. The rate of primer amplification is 51.1 %, which is slightly lower than that in grape (Scott et al. 2000) and J. curcas L. (Yadav et al. 2011). The ratio of polymorphic primers (16.1 %) is also as low as some typical coniferous trees (Postolache et al. 2014). The coniferous trees with huge genomes have a very high content of repetitive DNA, such as transposable elements, which may lead to the low polymorphism rate (Kovach et al. 2010; Wagner et al. 2012). The PIC value, reflecting allelic diversity and frequencies among the testing individuals, can be used to evaluate the informativeness of each EST-SSR (Bostein et al. 1980). Three levels of PIC (PIC > 0.5, 0.5 > PIC > 0.25, PIC < 0.25) were generally used to judge the polymorphisms. Among 81 polymorphic primer pairs of T. ‘zhongshansa’, only eight (9.88 %) primer pairs presented high PIC value (>0.5), 43 (53.09 %) primer pairs presented moderate PIC value, ranging from 0.25 to 0.5, while the remaining 30 (37.04 %) primer pairs presented low PIC value (<0.25). The relatively lower polymorphism of EST-SSRs, compared to genomic SSRs, was confirmed in the studies of other plants (Cardle et al. 2000; Chagné et al. 2004).
The transferability of DNA markers was based on genomic similarity, thus it could reflect the genetic relationship between species (Zhang et al. 2014). Usually, EST-SSR markers have higher transferability rates between species with closer genetic relationships (Durand et al. 2010). Nevertheless, the actual transferability was also affected by other factors, such as nucleotide deletions, insertions, and substitutions at these SSR loci (Decroocq et al. 2003). In this investigation, it revealed that these EST-SSR markers developed from T. ‘zhongshansa’ were highly transferable across three closely related species. In particular, the amplification rate was 100 % for the three species belonging to Taxodium.
The polymorphic variation was not obvious between the three Taxodium species (Table 4), this might be attributed to the close relatedness and the extensive gene flow between the three species. The close relatedness was showed in the similar morphological features (Cheng 1983), the sympatric natural distributions (Florin 1963), and the high crossing compatibility (Ying and Yu 2005; Yu et al. 2011) between the three species. Indeed, Denny and Arnold (2007) treated these three species of Toxadium as one species with three botanical varieties. Besides the characteristics of high crossability, the open-pollinated mating system and wind-dispersed pollen made the extensive gene flow between the three Taxodium species possible. Undoubtedly, the long-term and frequent interspecies gene flow could greatly dilute the genetic differentiation between species. Thus, although Taxodium maintained high genetic diversity, little difference was found between the three species.
The Functional Annotation for SSR-Containing EST Sequences and its Potential Application
Gene ontology, the functional annotation schema for gene and protein sequences, has been the de facto standard in nearly all public databases (Götz et al. 2008). In this paper, the specific function of important genes was shown in more detail by GO terms. These sequences containing-SSR were mainly related with intracellular part (21), membrane (18), intracellular membrane-bounded organelle (13), intracellular organelle lumen (12), nuclear part (11), metabolism of macromolecular compound (10), and immunity (7). Further analyses of these genes should provide information about cell metabolism and may help to understand Taxodium’s resistance to waterlogging stress (Palta, et al. 2012). It is noteworthy that 81 SSRs showing high polymorphism mainly connected with cellular macromolecule metabolic process, cellular response to stress, hydrolase activity, and acting on glycosyl bonds, which may help T. ‘zhongshansa’ produce and maintain ATP to adapt to anaerobic environments (Qi et al. 2014). Thus, these SSR loci might be applied as target genes to explore the relationship between gene function and phenotype of resistance in Taxodium.
Conclusions
This paper reported the characteristics of SSRs in T. ‘zhongshansa’ EST sequences, including the frequency and distribution of SSR repeat lengths and motifs. We also identified and characterized a set of EST-SSRs and evaluated their polymorphism and cross-species transferability rates. This set of EST-SSR markers may provide a useful tool for subsequent studies in T. ‘zhongshansa’, such as construction of genetic maps, QTL mapping, and marker assisted selection. Furthermore, the functional categorization of T. ‘zhongshansa’ containing-SSR sequences revealed that these EST sequences were transcribed from many functional genes involved in cellular and molecular processes. Hence, this set of EST-SSR markers may also be used to explore the relationship between gene function and phenotype.
References
Anderson JA, Churchill GA, Autrique JE et al (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Asadi AA, Monfared SR (2014) Characterization of EST-SSR markers in durum wheat EST library and functional analysis of SSR-containing EST fragments. Mol Genet Genomics 289:625–640
Ashrafi H, Hill T, Stoffel K et al (2012) De novo assembly of the pepper transcriptome (Capsicum annuum): a benchmark for in silico discovery of SNPs, SSRs and candidate genes. BMC Genomics 13:571
Barbará T, Palma-Silva C, Paggi GM et al (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16:3759–3767
Bostein D, White RL, Sholnick M, David RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Cardle L, Ramsay L, Milbourne D et al (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 56:847–854
Cavagnaro PF, Chung SM, Manin S et al (2011) Microsatellite isolation and marker development in carrot—Genomic distribution, linkage mapping, genetic diversity analysis and marker transferability across Apiaceae. BMC Genomics 12:386
Chagné D, Chaumeil P, Ramboer A et al (2004) Cross-species transferability and mapping of genomic and cDNA SSRs in pines. Theor Appl Genet 109:1204–1214
Chen CX, Zhou P, Choi YA et al (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Cheng WC (1983) SYLVA SINICA, Volume (1). China Forestry Publishing House, Beijing, pp 318–320 (In Chinese)
Decroocq V, Favé MG, Hagen L et al (2003) Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106:912–922
Denny GC, Arnold MA (2007) Taxonomy and nomenclature of baldcypress, pondcypress, and montezuma cypress: one, two, or three species? HortTechnology 17(1):125–127
Doyle JJ, Doyle JL (1987) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Du F, Wu Y, Zhang L et al (2014) De Novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental Hybrid ‘Sorbonne’. Plant Mol Biol Report. doi:10.1007/s11105-014-0746-9
Durand J, Bodénès C, Chancerel E et al (2010) A fast and cost effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genomics 11:570
Echt CS, Saha S, Deemer DL, Nelson CD (2011) Microsatellite DNA in genomic survey sequences and unigenes of loblolly pine. Tree Genet Genomes 7:773–780
Florin R (1963) The distribution of conifer and taxad genera in time and space. Ann Geogr 20:121–132
Franklin P, Juan O, Mariuxi Z et al (2004) Development of EST-SSR markers by data mining in three species of Shrimp: Litopenaeus vannamei, Litopenaeus stylirostris, and Trachypenaeus birdy. Mar Biotechnol 7:554–569
Gao LF, Tang JF, Li HW, Jia JZ (2003) Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed 12:245–261
Gao CH, Tang ZL, Yin JM et al (2011) Characterization and comparison of gene-based simple sequence repeats across Brassica species. Mol Genet Genomics 286:161–170
Götz S, García-Gómez JM, Terol J et al (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 10:3420–3435
Gupta PK, Rustgi S, Sharma S et al (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genomics 270:315–323
Hu LJ, Yu H, Hua JF et al (2012) Comparison of photosynthetic characteristics of three clones of Taxodium ‘Zhong shanshan’ and their parents. J Plant Resour Environ 3:29–37 (In Chinese)
Iorizzo M, Senalik DA, Grzebelus D et al (2011) De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics 2011(12):389
Jayasena AS, Secco D, Levin KB et al (2014) Next generation sequencing and de novo transcriptomics to study gene evolution. Plant Methods 10:3–14
Jia X, Deng Y, Sun XB et al (2015) Characterization of the global transcriptome using Illumina sequencing and novel microsatellite marker informationin seashore paspalum. Genes Genomics 37:77–86
Jung S, Abbott A, Jesudurai C et al (2005) Frequency, type, distribution and annotation of simple sequence repeats in Rosaceae ESTs. Funct Integr Genomics 5:136–143
Koelling J, Coles MC, Matthews PD (2012) Development of new microsatellite markers (SSRs) for Humulus lupulus. Mol Breed 30:479–484
Kovach A, Wegrzyn JL, Parra G et al (2010) The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genomics 11:420
Kumari M, Grover A, Yadav PV et al (2013) Development of EST-SSR markers through data mining and their use for genetic diversity study in Indian accessions of Jatropha curcas L.: a potential energy crop. Genes Genomics 35:661–670
Lagercrantz U, Ellegren H, Anderson L (1993) The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res 21:1111–1115
Mardis ER (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24:133–141
Nicot N, Chiquet V, Gandon B et al (2004) Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs). Theor Appl Genet 109:800–805
O’Nei ST, Emrich SJ (2013) Assessing De Novo transcriptome assemblymetrics for consistency and utility. BMC Genomics 14:465
Ong WD, Voo CL, Kumar SV (2012) Development of ESTs and data mining of pineapple EST-SSRs. Mol Biol Rep 39:5889–5896
Palta MM, Doyle TW, Jackson CR et al (2012) Changes in diameter growth of Taxodium distichumin response to flow alterations in the savannah river. Wetlands 32:59–71
Postolache D, Leonarduzzi C, Piotti A et al (2014) Transcriptome versus genomic microsatellite markers: highly informative multiplexes for genotyping Abies alba Mill. and Congeneric Species. Plant Mol Biol Report 32:750–760
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Qi BY, Yang Y, Yin YL et al (2014) De novo sequencing, assembly, and analysis of the Taxodium ‘Zhongshansa’ roots and shoots transcriptome in response to short-term waterlogging. BMC Plant Biol 2014(14):201. doi:10.1186/s12870-014-0201-y
Rungis D, Be’rube’ Y, Zhang J et al (2004) Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theor Appl Genet 109:1283–1294
Saha MC, Mian R, Zwonitzer JC et al (2004) An SSR- and AFLP-based genetic linkage map of tall fescue (Festuca arundinacea Schreb.). Theor Appl Genet 110:323–336
Scott KD, Eggler P, Seaton G et al (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Siongng KK, Lee SL, Tsumura Y et al (2009) Expressed sequence tag–simple sequence repeats isolated from Shorea leprosula and their transferability to 36 species within the Dipterocarpaceae. Mol Ecol Resour 9:393–398
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12:4127–4131
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Vidushi M, Irshad AR, Praveen A et al (2015) Development of chemical and EST-SSR markers for Ocimum genus. Ind Crop Prod 63:65–70
Wagner S, Gerber S, Petit RJ (2012) Two highly informative dinucleotide SSR multiplexes for the conifer Larix decidua (European larch). Mol Ecol Resour 12:717–725
Wu HB, Gong H, Liu P et al (2014) Largescale development of EST-SSR markers in sponge gourd via transcriptome sequencing. Mol Breed 34:1903–1915
Xu XW, Wan FX, Yang D (2013) Effects of three planting patterns of ascendens mucronatum on soil fertility in Shanghai costal areas. J Nanjing For Univ 1:163–167, In Chinese
Yadav HK, Ranjan A, Asif MH et al (2011) EST-derived SSR markers in Jatropha curcas L.: development, characterization, polymorphism, and transferability across the species/genera. Tree Genet Genomes 7:207–219
Yang J, Dai PF, Zhou TH et al (2013) Genetic diversity and structure of wintersweet (Chimonanthus praecox) revealedby EST-SSR markers. Sci Hortic 150:1–10
Yeh F, Yang RC, Boyle T et al (1999) POPGENE, version 1.32: the user friendly software for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, AB, Canada
Ying YL, Yu CG (2005) Taxodium ‘zhongshans’. China Forestry Publishing House, Beijing (In Chinese)
Yu CG, Yin YL, Xu JH (2011) Four hybrid varieties of Taxodium. Sci Silvae Sin 5:181–182 (In Chinese)
Zhai LL, Xu L, Wang Y et al (2014) Novel and useful genic-SSR markers from de novo transcriptome sequencing of radish (Raphanus sativus L.). Mol Breed 33:611–624
Zhang XF, Luo ZF, Pan B, Wang X (2012) Study of process for ACQ-D preserved ‘Taxodim Zhongshansha 302’ plywood manufacture. China For Prod Ind 4:28–50 (In Chinese)
Zhang MY, Fan L, Liu QZ (2014) A novel set of EST-derived SSR markers for pear and cross-species transferability in Rosaceae. Plant Mol Biol Report 32:290–302
Zhao QY, Wang Y, Kong YM (2011) Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinforma 12(Suppl 14):S2
Zhou Y, Chen LF, Fan XZ, Bian YB (2014) De Novo assembly of Auricularia polytricha transcriptome using Illumina sequencing for gene discovery and SSR marker identification. PLoS ONE 9:1–16
Acknowledgments
We are grateful to Dr. Meng Xu for comments. The authors also thank Novogene Bioinformatics Technology (Beijing, China) for assisting with transcriptome sequencing. This research was supported by Public service platform of science and technology of Jiangsu province (BM2012058), the Key Project in the Provincial Science & Technology Pillar Program (agriculture) of Jiangsu (BE2012343), the Agricultural Science and Technology Innovation Project of Jiangsu Province (CX132046), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Data Accessibility
Illumina sequencing data from ‘zhongshansa’ roots and shoots were deposited in the NCBI SRA database under accession number SRP043177.
The primers successfully designed and additional supplemental materials: Support information.
Yanli Cheng,Ying Yang and Ziyang Wang are co-first authors
Huogen Li holds a PhD, Nanjing Forestry University
Electronic supplementary material
Below is the link to the electronic supplementary material.
Additional file 1
The detail information of 1,958 primer pairs designed from containing-SSR ESTs (XLSX 526 kb)
Additional file 2
The detail information of 176 monomorphic primer pairs (XLSX 1359 kb)
Additional file 3
The detail information for the 264 GO annotations (XLSX 513 kb)
Rights and permissions
About this article
Cite this article
Cheng, Y., Yang, Y., Wang, Z. et al. Development and Characterization of EST-SSR Markers in Taxodium ‘zhongshansa’. Plant Mol Biol Rep 33, 1804–1814 (2015). https://doi.org/10.1007/s11105-015-0875-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0875-9