Abstract
The generation of adventitious roots (ARs) is the key to the success of cuttings. The appropriate environment for AR differentiation in tea plants is acidic. However, the mechanism is unclear. In this study, pH 4.5 was suitable condition for the differentiation of AR in tea plants. At the base of cuttings, the root primordia differentiated ARs more rapidly at pH 4.5 than pH 7.0, and nine AR differentiation-related genes were found to be differentially expressed in 30 days, the result was also validated by qRT-PCR. The promoter regions of these genes contained auxin and brassinosteroid response elements. The expression levels of several genes which were involved in auxin and brassinosteroid synthesis as well as signaling at pH 4.5 compared to pH 7.0 occurred differential expression. Brassinolide (BL) and indole-3-acetic acid (IAA) could affect the differentiation of ARs under pH 4.5 and pH 7.0. By qRT-PCR analysis of genes during ARs generation, BL and IAA inhibited and promoted the expression of CsIAA14 gene, respectively, to regulate auxin signal transduction. Meanwhile, the expression levels of CsKNAT4, CsNAC2, CsNAC100, CsWRKY30 and CsLBD18 genes were up-regulated upon auxin treatment and were positively correlated with ARs differentiation.This study showed that pH 4.5 was the most suitable environment for the root primordia differentiation of AR in tea plant. Proper acidic pH conditions promoted auxin synthesis and signal transduction. The auxin initiated the expression of AR differentiation-related genes, and promoted its differentiated. BL was involved in ARs formation and elongation by regulating auxin signal transduction.
Key message
Acidic environments promote auxin synthesis and regulate root primordia differentiation to form ARs by initiating AR differentiation-related genes expression in Camellia sinensis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Camellia sinensis (L).O.Ktze is a plant belonging to the genus Camellia within the family Camelliaceae and is known as one the world's top three beverages, along with coffee and cocoa. The popularity of tea is not only due to its rich taste, but is also associated with many beneficial health effects, such as cancer prevention and weight loss (Katiyar and Mukhtar 1996; Wolfram et al. 2006). With the development of tea industry, the area planted with tea trees has been increasing. According to statistics, the area of tea plantations worldwide has now reached more than 70 million mu (one mu equals to 0.067 ha). Therefore, a large number of high-quality tea seedlings are needed for the construction of new tea plantations and the renovation of old tea plantations. Tea trees are mainly propagated by short-spike cuttings to maintain the excellent genetic characteristics of cultivars as well as to accelerate propagation and promotion (Duclercq et al. 2011; Sharma et al. 2018; Shang et al. 2019; Wang et al. 2016). Further, this method depends on the formation of adventitious roots (ARs) (Bellini et al. 2014; Luo et al. 2021).
The formation of cuttings AR reflected the plant response to the site of injury and was controlled by a complex regulation of various signaling pathways (Druege et al. 2016). The formation of plant AR was a complicated physiological, biochemical and morphogenetic process. It was influenced by various environmental and endogenous factors including pH, temperature, moisture, light, carbohydrate, injury and plant hormones (Mauriat et al. 2014; Ribeiro et al. 2016; Wang et al. 2019). Plant hormones played a crucial role in the generation of AR. Both endogenous and exogenous hormones could regulate the growth and development of AR (Cao et al. 2018; Han et al. 2014; Zhang et al. 2015; Jia et al. 2019). Brassinosteroids (BRs), as polyhydroxylated steroid hormones, were important plant root growth regulators. BR deficient or BR insensitive mutants exhibited a wide range of growth defects, including short buds and short roots (Close and Sasse 1998). Auxin (IAA) is another key hormone in plants, which has been shown to rapidly promote AR formation in the 1930s (Cooper. 1936). In addition, exogenous application of IAA could also promote AR formation during the cutting propagation in many plants (Damiano et al. 2008; Ragonezi et al. 2010; Wu 2021). Some research indicated that IAA and BR had synergistic or antagonistic effects during root development. High levels of BR promoted cell elongation in the elongation zone of the root. At the same time, low levels of BR and high levels of IAA could maintain the size of stem cells, meristem in the resting central cell and root apical meristem. The above were essential to maintain root development (Mouchel et al. 2006; Maharjan et al. 2011; Nakamura et al. 2006; Yu et al. 2023; Pacurar et al. 2014; Kim et al. 2006).
The “acid growth theory” has long held that rising concentrations of IAA could trigger extracellular acidification, which activates cell wall–loosening enzyme to degrade the cell wall and allow smooth cell proliferation (Du et al. 2020; Elke et al. 2017; Lin et al. 2021; Matgorzata and Waldemar. 2021; Liu et al. 2022). Plant cells are embedded in the cell wall, providing structural integrity, but also limiting the space available to the cell. Therefore, they must be modified for allowing cell expansion. Previous studies showed that changes in external pH will significantly affect the auxin content in plants (Ida et al. 2010; Ales et al. 2015; Radic et al. 2016). The suitable environment for AR in tea plants was acidic, with pH4.5–6.5 being the most favorable for tea plants rooting (Niu et al. 2013; Tang et al. 2019). In acidic or alkaline environments, the differentiation of AR was inhibited to varying degrees, mainly in terms of its slow or non-differentiation, which in turn led to plant death. However, its molecular mechanism has not been fully elucidated. In this study, the widely planted tea cultivar “Fuding Dabai” was used as the research object. The optimal pH conditions for AR differentiation were explored, transcriptome under appropriate pH conditions were analyzed, and the effect of external pH environment on gene expression at the base of tea cuttings was investigated. The synthesis and signal transduction of BR and IAA under suitable pH conditions as well as the molecular mechanism that promote the differentiation of AR of tea plants were also analyzed.
Materials and methods
Plant materials
Semi-woody branches of the current year from “Fuding Dabai” with a general uniform growth trend were selected and cut into short 2–3 cm spikes of one bud and one leaf. One-third of the mature leaves were cut to reduce water evaporation and then washed five times with distilled water (Wei et al. 2014). All branches were collected from the Meitan Tea Germplasm Resource Nursery of Tea Institute of Guizhou Academy of Agricultural Sciences.
pH treatment conditions
Modified Hoagland nutrient solution was used for hydroponics (Kaiser 2005), that is, KNO3 6.0 × 10–3, NH4NO3 0.5 × 10–3, MgSO4 2.0 × 10–3, C10H12N2NaFe 0.153 × 10−3, KI 0.5 × 10−5, H3BO3 0.1 × 10−3, MnSO4 0.148 × 10−3, ZnSO4 0.534 × 10−4, NaMoO4 0.103 × 10−5, CuSO4 0.157 × 10−6, COCl2 0.253 × 10−6 and Ca(NO3)2 5.759 × 10−3 (w/v). The following nine pH treatment groups were set as pH 3.5, pH 4.0, pH 4.5, pH 5.0, pH 5.5, pH 6.0, pH 6.5, pH 7.0 and pH 7.5. The nutrient solution was adjusted to the appropriate pH with 0.1 M NaOH or 0.1 M HCl. They were then added to black hydroponic pots, each containing 3 L of nutrient solution. Tea cuttings were sequentially inserted into 0.5 cm thick black foam floats plate and then incubated in pots with 60 plants per pot, and three replications. Oxygen was applied daily in the morning/evening with an oxygen pump, every 12 h. The pH was calibrated once a day, while the nutrient solution was replaced once a week. On day 45 of culture, the rooting rate, mortality, number of new roots and root length were counted for each treatment (An et al. 2020).
Morphological observation of the tissue at the base of cuttings
The samples were fixed in FAA for 3 days, dehydrated in a graded ethyl alcohol series for 60 min each (10, 30, 50, 70, 85, and 95%) and then left in 100% ethyl alcohol overnight. The samples were embedded in paraffin and then cut into thin sections of 7 μm by rotary microtome. The thin sections were double-stained with safranin and fast green. Observations were made with an optical microscope (Koufan et al. 2020).
Transcriptome sequencing
The ARs freshly protruding from the root primordia under the pH 4.5 treatment and the root primordia only under the pH 7.0 treatment were collected in a mixture of three biological replicates (30 d) for every five plants. Sampling locations are shown in Fig. S1 A. Samples were immediately stored in liquid nitrogen until transcriptome sequencing. At the same time, sufficient samples were taken from each treatment and stored in the – 80 °C refrigerator after quick freezing in liquid nitrogen for hormone content determination. RNA extraction and transcriptome sequencing of samples were completed by Shanghai Majorbio Co. Ltd.
RNA data processing and differential gene screening
The whole genome of “Shuchazao” cultivar registered on NCBI by Anhui Agricultural University was used as reference genome (version No.:GCF_004153795.1), Sequencing data were filtered using SOAPnuke (v1.5.2) (Li et al. 2008) and clear reads were mapped to the reference genome using HISAT2 (v2.0.4) (Kim et al. 2015). Bowtie2 (v2.2.5) (Langmead et al. 2012) was then used to compare clear reads to the internal coding gene set and RSEM (v1.2.12) (Li and Dewey. 2011) was used to determine the expression level of each gene. DES Eq. 2 (v1.4.5) (Love et al. 2014) was used to analyze differentially expressed genes. The screening threshold was: |log2FC|≥ 1 and adjusted p-value < 0.05. GO enrichment analysis of genes/transcripts in the gene set was performed using the software Goatools (https://github.com/tanghaibao/GOatools) Generally, p-values were corrected by default. The GO function was considered significantly enriched when the corrected p-value (FDR) was less than 0.05 (Tang et al. 2015). KEGG pathway enrichment analysis was conducted on genes/transcripts in the gene set using R software. The calculation principle was the same as for GO function enrichment analysis. By default, the KEGG pathway was considered significantly enriched when the p-value (uncorrected) was less than 0.05.
qRT-PCR analysis
The accuracy of the RNA-seq results was verified by a qRT-PCR system with 2 × RealStar Fast SYBR qPCR Mix (Genestar, Beijing, China). Primers were designed across introns using Primer Premier 6.0 (Premier Biosof International, Quebec, Canada) and then tested for primer specificity at NCBI. The RNA returned by the company after transcriptome sequencing was reversed to cDNA (TAKARA, Dalian, China),diluted and used. The glyceraldehyde-3-phosphate dehydrogenase (CsGAPDH) gene of tea plant was used as an internal reference gene. The qRT-PCR was amplified as follows: 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. Amplifications were run on an qTOWER3G fluorescence quantitative PCR instrument (Analytikjena, Germany) of Company. All verifications were performed in three biological replicates and three technical replicates. Standard deviations (SD) of the means were calculated using standard statistical methods. Gene expression was analyzed using the ∆∆Ct data analysis method and relative gene expression was calculated using the 2−∆∆Ct method (Schmittgen and Livak. 2008).
Determination of auxin content in tea plant base
Standard curve construction
The auxin IAA standard was purchased from TMstandard Company, and contained 98.9% auxin. Accurately weighed 1.81 mg auxin standard and added to a constant volume of 25 ml of methanol, the IAA content in the mother liquor was 0.0724 mg/mL. The mother liquor was diluted to the following concentrations, 0.96, 1.92, 2.89, 5.79 and 14.48 μg/mL. Detection was performed on a Shimadzu LC-2040C ultrahigh performance liquid chromatograph The method was as follows, C18 column (100 mm × 4.6 mm, 5 μm), PDA diode array detector, detection wavelength of 260 nm, flow rate at 0.5 mL/min, column temperature at 35 °C with automatic injection for injection volume of 10 μL. Mobile phase, methanol (A) and 0.1% acetic acid (B). Gradient elution of phase B was as follows, 75% 0 min, 75% 4 min, 70% 4.1 min, 70% 12 min, 60% 12.1 min, 60% 30 min, 30% 30.1 min, 30% 40 min, 60% 40.1 min, 60% 45 min, 75% 45.1 min and 75% 60 min (Fu et al. 2009). The standard curve was as follows, y = 37,642x—2208.9, R2 = 0.9999, where y represents the peak area and x represents the concentration (Fig. S1B).
Determination of auxin content
A frozen sample was taken and placed in a mortar and pestle for rapid grinding. Quickly and accurately weighed 0.2000 g, placed in a test tube, added 3 ml of methanol and sealed. The sample was extracted by ultrasound at 15 °C for 100 min in a constant temperature water bath and filtered through a 0.45 μM filter membrane to obtain the sample to be tested. The operation was the same as in 1.7.1. The resulting peak areas were taken into the standard curve to obtain the auxin concentrations.
Analysis of homeopathic elements in promoter region
The 2500 bp sequence upstream of the ATG of the AR development-related genes in the tea plant genome was intercepted(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). After analyzing the homeopathic elements,it was visualised and mapped using TBtools software.
Statistical analysis
Parametric analyses were performed using ANOVA and means were compared using Tukey's test(p < 0.05), which was conducted using SPSS 21.0 software. All data were expressed as the mean of at least three replicates and one SD. All figures were plotted in the GraphPad Prim 9.0.
Result analysis
Effects of environmental pH on the differentiation of AR in tea cuttings
The number and length of ARs at the base of tea plants cuttings showed a trend of increasing and then decreasing with rising pH. The highest number of ARs was found at pH 4.5 and pH 5.0 treatments. In particular, the longest average root length, the highest rooting rate, the highest number of new roots and the lowest mortality rate were observed at pH 4.5, with significant difference compared to other treatment groups (Fig. 1A, B). These indicators were second at pH 5.0 only to pH 4.5. The results showed that an environment of pH 4.5–5.0 was favorable to the differentiation and development of tea plants roots. Further, pH 4.5 was the most suitable acidic environment for the differentiation of tea plants roots under the experimental conditions.
Effects of different pH treatments for 45 d on the AR of cuttings. A The base of the cuttings for 45 d. Bar = 5 mm. B Effects of different pH treatments on the phenotypic indicators of AR in tea cuttings. The x-axis represents the pH value of the treatment and the y-axis represents the value of the indicator. The error bar represents ± SEs, n ≥ 3. Different letters showed significant differences (P < 0.05), while the same letters showed no significant differences (P > 0.05)
By comparing the morphological changes of cells at the base of the cuttings during AR generation and development under pH 4.5 and pH 7.0, it was found that root primordia was generated at day 22 in both pH conditions. Furthermore, the root primordia were formed by the proliferation and differentiation of parenchyma cells from the central sheath to the phloem. However, root primordia began to form root tips and break through root primordium epidermis at day 30 under pH 4.5, compared to 38 d in pH 7.0. This suggested that pH 4.5 environment promoted the differentiation of root primordia to form AR (Fig. 2).
Histological observation of ARs formation process of tea cuttings at pH 4.5 and pH 7.0. Paraffin section observation of founder cells at 0 d, 22 d, 30 d and 38 d during AR development. ARs appeared at pH 4.5 for 30 days, and pH 7.0 for 38 days. Bar = 1 mm. Arrows: purple, pericycle (Pe); dark,phloem (Ph); red, root primordia; blue, ARs formed by differentiation from root primordia. Xylem (Xy); marrow (Ma)
Analysis of gene expression levels in AR differentiation
Differences in gene expression between pH 4.5 and pH 7.0 at the base of the cuttings were compared by transcriptome sequencing. At this time, the root primordia were just protruding from the AR under the pH 4.5 environment, while at the root primordia stage under pH 7.0. The results showed that there were 5741 DEGs, including 1844 up-regulation genes and 3897 down-regulation genes (Fig. S2). The results of GO analysis and KEGG analysis of DEGs are shown in Fig. S3 and Fig. S4, respectively.
After reviewing the reported AR differentiation-related genes from other plants, the genes with the highest similarity and functional domains in the genomes of tea plants were also compared and considered as homologous sequences of tea plants. In the transcriptome data, the following nine genes were significantly differentially expressed, with six positively [CsKNAT4 (LOC114273157), CsLBD18 (LOC114307927), CsLBD33 (LOC114275146), CsNAC2 (LOC114287191), CsNAC100 (LOC114260242) and CsWRKY30 (LOC114314481)] and one negatively regulated gene [CsWRKY75 (LOC114293712)] for AR differentiation being up-regulated. The expression of a negatively regulated gene [CsIAA14 (LOC114321959)] and an auxin transport vector [CsAUX/LAX3 (LOC114282558)] were down-regulated. In order to clarify the expression levels of these genes, validation was performed by qRT-PCR and the results were consistent with the transcriptomic data (Fig. 3). Gene names and primer sequences are shown in Table. S1.
qRT-PCR was used to validate differentially expressed genes in the transcriptome of AR differentiation-related. The relative expression was calculated as 2.−ΔΔCt. The left y-axis is the result of qRT-PCR and the right y-axis is the result of RNA-seq. The error bar represents ± SEs, n ≥ 3.Different letters showed significant differences (P < 0.05), while the same letters showed no significant differences (P > 0.05)
Analysis of homeopathic elements in the promoter region
The homeopathic elements in the 2500 bp sequence upstream of the start codon of the above nine genes were analyzed. The results showed that all genes contained brassinosteroid response elements. CsKNAT4, CsLBD18, CsLBD33, CsNAC2, CsWRKY30 and CsIAA14 contained auxin response elements (Fig. 4). It suggested that the differential expression of the above genes might be related to the levels of IAA, BR as well as signal transduction.
The pH significantly affects the synthesis and signal transduction related genes expression of BR and IAA
The interaction between phytohormones BR and IAA regulated the generation and development of AR during root differentiation. Analysis of the KEGG pathway showed significant differences in the expression levels of several genes involved in BR and IAA synthesis as well as signaling at pH 4.5 compared to pH 7.0. In particular, the expression levels of key negatively regulated gene (CsIAA14) in the IAA signaling pathway and key positively regulated gene (CsBZR1) in the BR signaling pathway were significantly down-regulated. It demonstrated that the signaling pathways of IAA and BR were promoted and inhibited at pH 4.5, respectively, which was consistent with previous report of antagonistic regulation of root differentiation by IAA and BR (Fig. S5).The high performance liquid chromatography (HPLC) was used to determine the auxin content at the base of the cuttings under both treatments, showing that the auxin content was 65.4% higher at pH 4.5 than at pH 7.0 (Fig. S6). 11 DEGs in the IAA, BR synthesis and signal transduction pathways were randomly selected for qRT-PCR. The gene name and primer sequence were shown in Table. S1.The results showed that the expression levels of the 10 genes were consistent with the transcriptome, except for gene LOC114300015 (CsYUCCA6-like) (Fig. S7). This result also indicated that pH 4.5 could promote the synthesis of IAA. The above results suggested that the increase in auxin content and signal transduction at pH 4.5 might be responsible for promoting AR differentiation. In contrast, BR played an opposite role to IAA in regulating AR differentiation.
BR and IAA affect gene expression and regulate AR
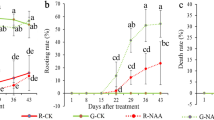
Tea plant cuttings were treated with 20 nM BL and 10 μM IAA at pH 4.5 and pH 7.0, respectively. After 45 d, the rooting rate, mortality rate, number of new roots and average root length were calculated. The results showed that the IAA treatment significantly reduced the mortality rate and increased the rooting rate of cuttings under both pH conditions. The BL treatment, on the other hand, had no significant effect, indicating that the differences in mortality rate and rooting rate between pH 4.5 and pH 7.0 were caused by IAA. Number and length are important indicators to evaluate the condition of the root system. In this study, the IAA treatment significantly increased the number and length of roots at pH 7.0 and also slightly increased the two indicators at pH 4.5. In contrast, BL treatment could inhibit the number of roots at pH 4.5 to the level at pH 7.0, but promoted root length at pH 7.0, with less effect than IAA. In conclusion, IAA and BR acted antagonistically in regulating AR production in tea plants. During the AR elongation stage, both BR and IAA served to promote root growth (Fig. 5).
Phenotypic indicators of tea plant cuttings after 45 d. The x-axis represents the pH value and hormone treatment and the y-axis represents the value of the indicator. The error bar represents ± SEs, n ≥ 3. Different letters showed significant differences (P < 0.05), while the same letters showed no significant differences (P > 0.05)
In order to investigate whether IAA and BR regulate root primordia differentiation by inducing the expression of AR differentiation-related genes, the expression levels of these genes(Sect. “Analysis of gene expression levels in AR differentiation”) at AR production stage were examined by qRT-PCR (Fig. 6). The results showed that there were significant differences in gene expression levels as influenced by BL and IAA. CsKNAT4, CsNAC2, CsNAC100, CsWRKY30 and CsLBD18 genes were significantly up-regulated by IAA induction at pH 7.0 but were not affected by BL. The expression of these genes was significantly promoted by IAA at pH 4.5, but was down-regulated to the expression level at pH 7.0 under BL treatment. This indicated that they were mainly involved in regulating root mass generation at pH 4.5 and were induced by IAA, and BL could inhibit this process. There was no significant difference in the expression level of CsLBD33 gene between BL and IAA treatments. The expression level of CsWRKY75 gene was down-regulated by IAA, which might be related to its negative regulation of root hair generation. The expression of CsAUX/LAX3 was up-regulated under BL and IAA treatment but down-regulated under pH 4.5, which was associated with the ability of exogenous IAA and BL to promote the aggregation of IAA in plants. The expression of CsIAA14 gene was significantly up-regulated by BL and down-regulated by IAA, showing opposite effects of inhibiting or promoting auxin signal transduction. In conclusion, up-regulation of auxin content and signal transduction induced expression of CsKNAT4, CsNAC2, CsNAC100, CsWRKY30 and CsLBD18 genes might be responsible for promoting AR production. BR might inhibit auxin signaling by promoting the expression of CsIAA14 gene, thereby reducing AR.
Gene expression at the stage of AR differentiated in the base of tea cuttings. The relative expression was calculated as 2.−ΔΔCt. The left y-axis is the result of qRT-PCR. The error bar represents ± SEs, n ≥ 3. Different letters showed significant differences (P < 0.05), while the same letters showed no significant differences (P > 0.05)
Discussion
Rooting rate, length and the number of AR were the key indicators for evaluating the maturity of cutting propagation (An et al. 2020). These three indicators under pH 4.5 were much higher than other treatment groups, and the mortality rate was also the lowest. The cell morphology at the base of cuttings differed between the stages of root primordium differentiation to form AR at pH 4.5 and pH 7.0, similar to the results of exogenous auxin treatment to promote AR in plants. (Quan et al. 2017). Therefore, it could be hypothesized that in the early stages, IAA was transported from the upper end to the lower end of the cutting morphology, promoting the proliferation and differentiation of the thin-walled cells from the central sheath to the phloem to form the root primordia. However, the synthesis of IAA or signal transduction was affected at the cutting base due to pH, further leading to differences in AR production in the root primordia. At pH 4.5, the content of auxin increased, inducing loosening and degradation of the cell wall at the base of the cuttings. At the same time, cell proliferation and differentiation favored the production of AR. After GO enrichment analysis of DEGs screened for transcriptome data, the highest enrichment was found for the plant secondary cell wall generation, plant cell wall generation, xylan biosynthesis and xylan metabolic process (Fig. S4 A, B). The results indicated that the cell wall was extensively disrupted and stretched, creating the conditions for AR to grow from the root primordia, which was consistent with the results of the histological sections.
The differentiation of plant roots were largely hormonally mediated (Zhang et al. 2018; Zhao et al. 2009). A growing number of studies showed that the regulation of endogenous hormone signaling was closely linked to the formation of AR. The balance of high levels of IAA and low levels of BR was crucial for the maintenance of apical resting center cells and meristem. IAA could increases the expression levels of several genes encoding BR catabolic enzymes, including BAS1, SOB7, BAT1 and BEN1. Conversely, BR could inhibit its signal transduction by regulating the expression of the IAA, an inhibitor of the IAA signaling pathway. This might form a feedback loop that helped maintain the balance between the two hormone (Bao et al. 2004; Juthamas and Wang. 2015). In the transcriptome, BR synthesis and signal transduction were inhibited and IAA synthesis and signal transduction were promoted. This balance of high levels of IAA and low levels of BR promoted the formation and development of AR. Studies of AR formation in purple grass, sugarcane and mulberry cuttings showed significant differences in the expression of several genes involved in BR and IAA signaling pathways, similar to the results of the present study (Devi et al. 2021; Li et al. 2020; Cao et al. 2018).
CsKNAT4 gene could regulate the root angulation and was expressed during the formation of root apical meristem as well as in the associated regions of cambium. The homologous gene AtKNAT6 in Arabidopsis was highly expressed when the root primordia were differentiated into AR (Qi and Zheng. 2013). The transcription factors CsLBD18 and CsLBD33 were related to callus formation and played roles in the initiation and boundary establishment of plant lateral organs. Their up-regulated expression activated cell wall loosening factors that degraded cell wall and promoted root differentiation (Lee et al. 2017). Homologous of CsNAC2 and CsNAC100 genes were demonstrated to have positive regulatory effects on AR formation in soybean and rice, respectively. Their up-regulated expression could increase rooting number and shorten rooting time (Redillas et al. 2012a, b). CsWRKY30 gene was involved in wound repair and stress response in cuttings, and its up-regulation in expression promoted plant root regeneration (Ding et al. 2015). Up-regulation of CsWRKY75 gene expression inhibited the generation of root hairs (Rishmawi et al. 2014). Down-regulation of CsIAA14 gene enhanced IAA signal transduction (Fukaki et al. 2002). Down-regulation of CsAUX/LAX3 gene, the gene encoding the IAA input vector, indicated that that IAA transport was diminished. It was hypothesized that IAA may not be synthesized and transported to the base of the cuttings, but synthesized and accumulated in the cells at the base of the cuttings. Exogenous BL and IAA could significantly affect root differentiation at pH 4.5 and pH 7.0. The expression levels of CsKNAT4, CsNAC2, CsNAC100, CsWRKY30 and CsLBD18 genes were always up-regulated when AR was promoted and down-regulated when AR was inhibited, positively correlating with AR production. Notably, the expression of CsIAA14 gene was always significantly up-regulated when BL was induced, which hindered auxin signaling. In contrast, IAA could alleviate this inhibition to a certain extent, which in turn promoted AR differentiation. This might be because IAA promoted the expression level of BR catabolism-related enzymes coding gene, reducing BR content and mitigating the induction of CsIAA14 gene. The significant increase in IAA content at the base of the cuttings at pH 4.5 compared to pH 7.0 also indicated a significant down-regulation of CsIAA14 gene expression by IAA content. This study provided valuable insights into the response of AR differentiation-related genes to phytohormones.
This study showed that pH 4.5 had the most significant promoting effect on the differentiation of tea plant roots and was the most suitable acidic environment for the differentiation of tea plant roots. The suitable acidic pH conditions promoted auxin synthesis and signal transduction. Auxin initiated the expression of genes related to AR differentiation and promoted it. BR was involved in the regulation of AR formation and elongation through the regulation of auxin signaling. In addition, the DEGs identified in this study were potential candidates for future functional analysis. It could provide a theoretical basis for further revealing the molecular mechanism that tea plants can only differentiation normally in acid environment.
Data Availability
The dataset used and/or analyzed in this study can be obtained from the author according to reasonable requirements.
References
An H, Zhang J, Xu F, Jiang S, Zhang X (2020) Transcriptomic profling and discovery of key genes involved in adventitious root formation from green cuttings of highbush blueberry (Vaccinium corymbosum L.). BMC Plant Biology 201:1–14. https://doi.org/10.1186/s12870-020-02398-0
Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z (2004) Brassinosteroids interact with auxin to promote lateral root developmentin Arabidopsis. Plant Physiol 134:1624–1631
Barbezab Elke, Dünserb Kai, Gaidoraa Angelika, Lendla Thomas, Buschac Wolfgang (2017) Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc Natl Acad Sci. 5:E4884–E4893. https://doi.org/10.1073/pnas.1613499114
Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similaritiesand diferences. Annu Rev Plant Biol. 65:639–666. https://doi.org/10.1146/annurev-arplant-050213-035645
Cao X, Du W, Shang C, Shen Q, Liu L, Cheng J (2018) Comparative transcriptome reveals circadian and hormonal control of adventitious rooting in mulberry hardwood cuttings. Acta Physiol Plant 4011:1–16. https://doi.org/10.1007/s11738-018-2772-y
Chaiwanon Juthamas, Zhi-Yong Wang (2015) Spatiotemporal Brassinosteroid Signalingand Antagonism with Auxin PatternStem Cell Dynamics in Arabidopsis Root. Curr Biol 25:1031–1042. https://doi.org/10.1016/j.cub.2015.02.046
Clouse SD, Sasse JM (1998) BRASSINOSTEROIDS:essentialregulators of plant growth and development. Annu Rev Neurosci 49:427–451
Cooper WC (1936) Transport of root-forming hormone in woody cuttings. Plant Physiol 114:779. https://doi.org/10.1104/pp.11.4.779
Damiano C, Padro MDA, Frattarelli A (2008) Propagation and establishment in vitro of myrtle (Myrtus communis L.), pomegranate (Punica granatum L.) and mulberry (Morus alba L.). Propag Ornam Plants 81:3–8
Devi J, Kaur E, Swarnkar MK, Acharya V, Bhushan S (2021) De novo transcriptome analysis provides insights into formation of in vitro adventitious root from leaf explants of Arnebia euchroma. BMC Plant Biol 211:1–16. https://doi.org/10.1186/s12870-021-03172-6
Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ (2015) Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regula-tion of ABA signaling and auxin homeostasis. Plant J 84:56–69. https://doi.org/10.1111/tpj.12958
Druege U, Franken P, Hajirezaei MR (2016) Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front Plant Sci 7:381. https://doi.org/10.3389/fpls.2016.00381
Du M, Spalding Edgar P, Gray William M (2020) Rapid auxin-mediated cell expansion. Annu Rev Plant Biol 715:1–24. https://doi.org/10.1146/annurev-arplant-073019-025907
Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011) De novo shoot organogenesis:from art to science. Trends Plant Sci 1611:597–606. https://doi.org/10.1016/j.tplants.2011.08.004
Fu JH, Zhu JF, Wang JD (2009) Determination of auxin in Arabidopsis thalianan by solid phase extraction and high-performance liquid chromatography with fluorescencedetection. Chin J Anal Chem 379:1324–1327
Fukaki Hidehiro, Tameda Satoshi, Masuda Haruka, Tasaka Masao (2002) Lateral root for-mation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 292:153–168. https://doi.org/10.1046/j.0960-7412.2001.01201.x
Han H, Sun X, Xie Y, Feng J, Zhang S (2014) Transcriptome and proteome profling of adventitious root development in hybrid larch (Larix kaempferi×Larix olgensis). BMC Plant Biol 141:1–13. https://doi.org/10.1186/s12870-014-0305-4
Hongwei An, Qinfei Song, Suzhen Niu (2021) Effect of pH value on growth and physiology of two wild tea trees. Mol Plant Breed 07:09–51
Jia K-P, Dickinson AJ, Mi J, Cui G, Xiao TT, Kharbatia NM, Guo X, Sugiono E, Aranda M, Blilou I (2019) Anchorene is a carotenoidderived regulatory metabolite required for anchor root formation in Arabidopsis. Sci Adv 511:6787. https://doi.org/10.1126/sciadv.aaw6787
Kaiser WME (2005) Mineral nutrition of plants: principles and perspectives, E. Epstein, A. Bloom, second ed., Sinauer Associates, Inc. Publishers, Sunderland, MA (2004), (400pp., US$ 79.95), ISBN: 0-87893-172-4. J Lan Hyol.16212:1380−1381. https://doi.org/10.1016/j.jplph.2005.06.002.
Katiyar S, Mukhtar H (1996) Tea in chemoprevention of cancer. Int J Oncol 8:221–238. https://doi.org/10.3892/ijo.8.2.221
Kim H, Park PJ, Hwang HJ, Lee SY, Oh MH, Kim SG (2006) Brassinosteroid signals control expression of the AXR3/IAA17 gene in the cross-talk point with auxin in root development. Biosci Biotechnol Biochem 70:768–773
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 124:357–360
Koufan M, Mazri MA, Essatte A, Moussafr S, Belkoura I, El Rhafari L, Toufk I (2020) A novel regeneration system through micrografting for Argania spinosa (L.) skeels, and confirmation of successful rootstock-scion union by histological analysis. Plant Cell Tissue Organ Culture 1422:369–378. https://doi.org/10.1007/s11240-020-01868-5
Lager Ida, Andréasson Ola, Tiffany L, Dunbar Erik Andreasson, Escobar Matthew A, Rasmusson AG (2010) Changes in external pH rapidly alter plant gene expressionand modulate auxin and elicitor responses. Plant, Cell Environ 33:1513–1528. https://doi.org/10.1111/j.1365-3040.2010.02161.x
Langmead B, Salzberg SL, Langmead B (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lee HW, Kang NY, Pandey S (2017) Dimerization in LBD16 and LBD18 transcription factors is critical for lateral root formation. Plant Physiol 174:301–311. https://doi.org/10.1080/15592324.2017.1411450
Li B, Dewey CN (2011) RSEM:accurate transcript quantification from RNA-Seq datawith or without a reference genome. BMC Bioinformatics 121:323
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714. https://doi.org/10.1093/bioinformatics/btn025
Li A, Lakshmanan P, He W, Tan H, Liu L, Liu H, Liu J, Huang D, Chen Z (2020) Transcriptome profling provides molecular insights into auxin-induced adventitious root formation in sugarcane (Saccharum spp. interspecific hybrids) microshoots. Plants 98:931. https://doi.org/10.3390/plants9080931
Li Liu, Wen Song, Shijia Huang, Kai Jiang, Moriwaki Yoshitaka, Yichuan Wang, Yongfan Men, Dan Zhang, Xing Wen, Zhifu Han, Jijie Chai, Hongwei Guo (2022) Extracellular pH sensing by plant cell-surface peptide-receptor complexes. Cell 185:1–15. https://doi.org/10.1016/j.cell.2022.07.012e
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 1512:550
Luo J, Nvsvrot T, Wang N (2021) Comparative transcriptomic analysis uncovers conserved pathways involved in adventitious root formation in poplar. Physiol Mol Biol Plants 279:1903–1918. https://doi.org/10.1007/s12298-021-01054-7
Maharjan PM, Schulz B, Choe S (2011) BIN2/DWF12 antagonis-tically transduces brassinosteroid and auxin signals in the roots ofArabidopsis. J Plant Biol 54:126–134. https://doi.org/10.1007/s12374-010-9138-3
Mauriat M, Petterle A, Bellini C, Moritz T (2014) Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by afecting auxin transport. Plant J 783:372–384. https://doi.org/10.1111/tpj.12478
Mouchel CF, Osmont KS, Hardtke CS (2006) BRX mediatesfeedback between brassinosteroid levels and auxin signalling in rootgrowth. Nature 443:458–461
Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, Nozaki H, Asami T, Yoshida S, Fujioka S (2006) Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in amanner dependent on organ type. Plant J 45:193–205
Niu SZ, Song QF, Fan WG (2013) Analysis on site soil nutrients of ancient tea trees in the southwest state of Guizhou Province. Zhejiang Nongye Xuebao (Acta Agriculturae Zhejiangensis) 256:1348–1353
Pacurar DI, Pacurar ML, Bussell JD, Schwambach J, Pop TI, Kowalczyk M, Gutierrez L, Cavel E, Chaabouni S, Ljung K (2014) Identification of new adventitious rooting mutants amongst suppressors of the Arabidopsis thaliana superroot 2 mutation. J Exp Bot 656:1605–1618. https://doi.org/10.1093/jxb/eru026
Pěnčík A, Turečková V, Paulišić S, Rolčík J, Strnad M, Mihaljević S (2015) Ammonium regulates embryogenic potential in Cucurbita pepothrough pH-mediated changes in endogenous auxin and abscisicacid.Plant Cell. Tissue Organ Culture 122:89–100. https://doi.org/10.1007/s11240-015-0752-0
Polak Małgorzata, Karcz Waldemar (2021) Some new methodological and conceptual aspects of the“acid growth theory”for the auxin action in maize (Zea mays L.)coleoptile segments: do acid-and auxin-induced rapid growth differ in their mechanisms? Int J Mol Sci 2223:17. https://doi.org/10.3390/ijms22052317
Qi B, Zheng H (2013) Modulation of root-skewing responses by KNAT1 in Arabidopsis thaliana. Plant J 76:380–392
Quan J, Meng S, Guo E, Zhang S, Zhao Z, Yang X (2017) De novo sequencing and comparative transcriptome analysis of adventitious root development induced by exogenous indole-3-butyric acid in cuttings of tetraploid black locust. BMC Genomics 181:1–14. https://doi.org/10.1186/s12864-017-3554-4
Radić S, Vujčić V, Glogoški M, Radić-Stojković M (2016) Influenceof pH and plant growth regulators on Stevia rebaudiana. Period Biol 1181:9–19. https://doi.org/10.18054/pb.2016.118.1.3420
Ragonezi C, Klimaszewska K, Castro MR, Lima M, Oliveira P, Zavattieri MA (2010) Adventitious rooting of conifers: infuence of physical and chemical factors. Trees 246:975–992. https://doi.org/10.1007/s00468-010-0488-8
Redillas Mark C.F.R., Jeong Jin S, Kim Youn S, Jung Harin, Bang Seung W, Choi Yang D, Ha Sun-Hwa, Reuzeau Christophe, Kim Ju-Kon (2012) The overexpres-sion of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805. https://doi.org/10.1111/j.1467-7652.2012.00697.x
Redillas MC, Jeong JS, Kim YS, Jung Harin, Bang Seung W, Choi Yang D, Ha Sun-Hwa, Reuzeau Christophe, Kim Ju-Kon (2012) The overexpres-sion of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805. https://doi.org/10.1111/j.1467-7652.2012.00697.x
Ribeiro CL, Silva CM, Drost DR, Novaes E, Novaes CR, Dervinis C, Kirst M (2016) Integration of genetic, genomic and transcriptomic information identifes putative regulators of adventitious root formation in Populus. BMC Plant Biol 161:1–11. https://doi.org/10.1186/s12870-016-0753-0
Rishmawi L, Pesch M, Juengst C, Schauss Astrid C, Schrader Andrea, Hülskamp Martin (2014) Non-cell-au-tonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol 165:186–195
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 36:1101–1108. https://doi.org/10.1038/nprot.2008.73
Shang C, Yang H, Ma S, Shen Q, Liu L, Hou C, Cao X, Cheng J (2019) Physiological and transcriptomic changes during the early phases of adventitious root formation in mulberry stem hardwood cuttings. Int J Mol Sci 2015:3707. https://doi.org/10.3390/ijms20153707
Sharma U, Kataria V, Shekhawat N (2018) Aeroponics for adventitious rhizogenesis in evergreen haloxeric tree Tamarix aphylla (L.) karst: infuence of exogenous auxins and cutting type. Physiol Mol Plant Pathol 241:167–174. https://doi.org/10.1007/s12298-017-0493-0
Tang DD, Liu MY, Zhang QF, Shi YZ, Ma LF, Ruan JY (2019) Effects of different nitrogen forms and pH on element absorption and organic acid content in tea plant Chaye Kexue. Tea Sci 392:49–60
Tang H,Klopfenstein D,Pedersen B(2015) GOATOOLS:tools for gene ontology.Zenodo.https://escholarship.org/uc/item/5gs1t35f
Wang P, Ma L, Li Y, Sa W, Li L, Yang R, Ma Y, Wang Q (2016) Transcriptome profiling of indole-3-butyric acid-induced adventitious root formation in softwood cuttings of the Catalpa bungei variety‘YU-1’at different developmental stages. Genes Genomics 382:145–162. https://doi.org/10.1007/s13258-015-0352-8
Wang Z, Hua J, Yin Y, Gu C, Yu C, Shi Q, Guo J, Xuan L, Yu F (2019) An integrated transcriptome and proteome analysis reveals putative regulators of adventitious root formation in Taxodium ‘Zhongshanshan.’ Int J Mol Sci 205:12–25. https://doi.org/10.3390/ijms20051225
Wei K, Wang LY, Wu LY, Zhang CC, Li HL (2014) Transcriptome analysis of indole-3-butyric acid-induced adventitious root formation in nodal cuttings of Camellia sinensis (L.). PloS one 99:e107201. https://doi.org/10.1371/journal.pone.0107201
Wenwei Lin, Xiang Zhou, Wenxin Tang, Takahashi Koji, Xue Pan, Jiawei Dai, Hong Ren, Xiaoyue Zhu, Songqin Pan, Haiyan Zheng, Gray William M, Tongda Xu, Kinoshita Toshinori, Zhenbiao Yang (2021) TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature 11:27. https://doi.org/10.1038/s41586-021-03976-4
Wolfram S, Wang Y, Thielecke F (2006) Anti-obesity effects of green tea: from bedside to bench. Mol Nutr Food Res 50:176–187. https://doi.org/10.1016/j.jnutbio.2010.06.006
Wu J (2021) Study on cutting propagation technology of Dracontomelon duperreanum Pierre. J Mt Agri Biol 401:74–78. https://doi.org/10.15958/j.cnki.sdnyswxb.2021.01.013
Zhang S, Zhao Z, Zhang L, Zhou Q (2015) Comparative proteomic analysis of tetraploid black locust (Robinia pseudoacacia L.) cuttings in different phases of adventitious root development. Trees 292:367–384. https://doi.org/10.1007/s00468-014-1116-9
Zhang Y, Nasser V, Pisanty O, Omary M, Wulf N, Di Donato M, Tal I, Hauser F, Hao P, Roth O (2018) A transportome-scale amiRNA-based screen identifes redundant roles of Arabidopsis ABCB6 and ABCB20 in auxin transport. Nat Commun 91:1–12. https://doi.org/10.1038/s41467-018-06410-y
Zhao Y, Hu Y, Dai M, Huang L, Zhou D-X (2009) The WUSCHEL related homeobox gene WOX11 is required to activate shootborne crown root development in rice. Plant Cell 213:736–748. https://doi.org/10.1105/tpc.108.061655
Zipeng Yu, Ma Jinxin, Zhang Mengyue, Li Xiaoxuan, Sun Yi, Zhang Mengxin, Ding Zhaojun (2023) Auxin promotes hypocotyl elongation by enhancing BZR1 nuclear accumulation in Arabidopsis. Sci Adv 91:1–15. https://doi.org/10.1126/sciadv.ade2493
Acknowledgements
We thank the equipment support provided by Institute of Agro-bioengineering and the College of Life Sciences of Guizhou University.
Funding
This work was supported by National Natural Science Foundation of China, "Functional analysis of Brassinosteroid transcription factor CsBZRs in Camellia sinensis under cold stress" (Grant numbers: 32160077). Talent program of the Guizhou Academy of Agricultural Sciences, "Research on functional genomics and high-value utilization technology of important economic plants such as Eucommia ulmoides"[(2022) No.02]. Guizhou Provincial Talent Office "Guizhou Provincial Characteristic Plant Germplasm Resources Utilization and Innovation Talent Base" (RCJD2018-14). Guizhou Provincial High-level Innovative Talent Training Project [QKH Talent (2016) No. 4003].
Author information
Authors and Affiliations
Contributions
The corresponding authors DGZ and YZ provided design ideas and experimental guidance for the paper, and revised the paper. KL, the author of this article, designed and operated the experiment according to the ideas of the corresponding author, and wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests. The authors have no relevant fnancial or non-fnancial interests to disclose.
Ethical approval and consent to participate
We does not contain any studies with human or animal subjects.
Consent for publication
My manuscript doesn’t contain any individual person’s data in any form (including any individual details, images or videos).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11103_2023_1383_MOESM1_ESM.tif
Supplementary file1 (TIF 11661 KB)—Transcriptome sequencing sample location map and auxin standard curve. A The red circle is the sampling site for transcriptome sequencing. B Preparation of auxin standard. The X-axis represents auxin concentration, the Y-axis represents peak area.
11103_2023_1383_MOESM2_ESM.png
Supplementary file2 (PNG 16 KB)—Number of DEGs. Red represents the DEGs number that is up−regulated, and blue represents the DEGs number that is down−regulated. The x−axis represents the number of corresponding differential genes (DEGs), and the y−axis represents the difference comparison scheme for group.
11103_2023_1383_MOESM3_ESM.tif
Supplementary file3 (TIF 13702 KB)—GO classification of the DEGs. A GO annotation for DEGs. The x−axis is the number of genes in the annotation, and the y−axis represents GO categories of gene function. B Enrichment of the GO annotation of the DEGs.The x−axis represents the percentage of DEGs belonging to the corresponding pathway, and the y−axis representsthe top 20 pathways. The sizes of bubbles represent the number of DEGs in the corresponding pathway, and the colors of the bubbles represent the enrichment P−adjust of the corresponding pathway.
11103_2023_1383_MOESM4_ESM.tif
Supplementary file4 (TIF 18831 KB)—KEGG classification of the DEGs. A and B KEGG annotation for DEGs.The x−axis is represents KEGG categories of gene function, and the y−axis is the number of genes in the annotation. C Enrichment of the KEGG annotation of the DEGs.The x−axis represents the percentage of DEGs belonging to the corresponding pathway, and the y−axis representsthe top 20 pathways. The sizes of bubbles represent the number of DEGs in the corresponding pathway, and the colors of the bubbles represent the enrichment P−adjust of the corresponding pathway.
11103_2023_1383_MOESM5_ESM.tif
Supplementary file5 (TIF 15383 KB)—Enrichment map of KEGG pathway differentially expressed genes in BR and IAA metabolism as well as signal transduction. A KEGG signaling pathway of phytohormone (map04075). B KEGG metabolic pathway of IAA (map00380). C KEGG metabolic pathway of BR (map00905). Red boxes indicate up-regulation of gene expression and blue boxes for down-regulation.
11103_2023_1383_MOESM6_ESM.tif
Supplementary file6 (TIF 1236 KB)—Auxin content at the base of cuttings in the differentiation stage of AR (30 d) under pH 7.0 and pH 4.5. Standard curve: y=37642x -2208.9, R2=0.9999. y: Peak area; x: Concentration. The x-axis represents the pH value and the y-axis represents auxin content. The error bar represents ± SEs, n ≥3. Significant differences at p<0.05 by t-test and is labeled with lower case letters.Different letters showed significant differences (P<0.05), while the same letters showed no significant differences (P>0.05).
11103_2023_1383_MOESM7_ESM.tif
Supplementary file7 (TIF 22343 KB)—The expression trend of DEGs in transcriptome was verified by qRT-PCR. The relative expression amount is calculated as 2−ΔΔCt. The left y-axis is the result of qRT-PCR, and the right y-axis is the result of RNA-seq. The error bar represents ±SEs, n≥3. Different letters showed significant differences (P<0.05), while the same letters showed no significant differences (P>0.05).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, K., Zhao, Y. & Zhao, DG. Transcriptome analysis reveals the effect of acidic environment on adventitious root differentiation in Camellia sinensis. Plant Mol Biol 113, 205–217 (2023). https://doi.org/10.1007/s11103-023-01383-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-023-01383-z