Abstract
Diamine putrescine (Put) and polyamines; spermidine (Spd) and spermine (Spm) are essential component of every cell because of their involvement in the regulation of cell division, growth and development. The aim of this study is to enhance the levels of Put during fruit development and see its implications in ripening and quality of tomato fruits. Transgenic tomato plants over-expressing mouse ornithine decarboxylase gene under the control of fruit-specific promoter (2A11) were developed. Transgenic fruits exhibited enhanced levels of Put, Spd and Spm, with a concomitant reduction in ethylene levels, rate of respiration and physiological loss of water. Consequently such fruits displayed significant delay of on-vine ripening and prolonged shelf life over untransformed fruits. The activation of Put biosynthetic pathway at the onset of ripening in transgenic fruits is also consistent with the improvement of qualitative traits such as total soluble solids, titratable acids and total sugars. Such changes were associated with alteration in expression pattern of ripening specific genes. Transgenic fruits were also fortified with important nutraceuticals like lycopene, ascorbate and antioxidants. Therefore, these transgenic tomatoes would be useful for the improvement of tomato cultivars through breeding approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ripening in climacteric fruits is regarded as necessary evil wherein it imparts desirable flavor, colour and texture to the fruit (Giovannoni 2001). Desptite enhancing the fruit quality, ripening causes considerable post-harvest losses due to excessive softening and over-ripening (Payasi and Sanwal 2010). Several attempts have been made to introduce delayed ripening trait in climacteric fruits, and the most widely used strategies are the down-regulation of ripening induced genes, including ethylene metabolism, modulation of fruit softening or cell wall metabolism (Hamilton et al. 1990; Oeller et al. 1991; Lashbrook et al. 1998; Brummell et al. 1999; Wang et al. 2005; Xie et al. 2014). However, these strategies have certain limitations. For instance, the manipulation of ethylene pathway resulted in delayed ripening however, overall fruit quality was also affected (Xiong et al. 2005). Moreover, most of these strategies suffered a setback due to the functional redundancy shown by other members of such gene families (Gupta et al. 2013).
Diamine putrescine (Put) and polyamines (PAs); spermidine (Spd) and spermine (Spm) are omnipresent polycationic small molecules that are present in all living cells. They regulate a plethora of biological processes, including gene expression and cell signalling, and are absolutely essential for normal growth and development of various organisms (Alcázar et al. 2010; Igarashi and Kashiwagi 2010; Kusano et al. 2008; Schwartz et al. 2011; Takahashi and Kakehi 2010). PA biosynthesis pathway initiates with the production of diamine Put, an obligate precursor of higher PAs, Spd and Spm, directly by decarboxylation of ornithine by ornithine decarboxylase (ODC; EC 4.1.1.17). In addition to the ODC pathway, plants and bacteria have an alternate pathway for Put biogenesis, which involve decarboxylation of arginine by arginine decarboxylase (ADC; EC 4.1.1.19). Spd and Spm are synthesized from Put by the sequential addition of amino propyl groups, and these reactions are catalyzed by Spd and Spm synthases (SPDS/SPMS/; EC 2.5.1.16/EC2.5.1.22) respectively. The amino propyl groups are donated by the decarboxylated S-adenosylmethionine (SAM), which in turn is produced from SAM by SAM decarboxylase (SAMDC; EC 2.5.1.1.6) (Sauter et al. 2013).
PAs are implicated in early fruit development and ripening. They act as anti-ripening and anti-senescence agents, by antagonizing ethylene action (a ripening hormone) in fruit and vegetative tissues (Cassol and Mattoo 2002). The exogenous treatment of PAs in various fruits has been shown to retard fruit colour change, respiration rate and senescence, delay ethylene emission and fruit ripening, increase fruit firmness and cell wall stability and reduced mechanical stress and chilling symptoms (Apelbaum et al. 1981; Valero et al. 2002). PAs, along with salicylic acid also regulate ethylene biosynthesis by influencing ACC synthase transcript accumulation (Li et al. 1992; Handa and Mattoo 2010). PAs and ethylene affect plant growth and senescence antagonistically by sharing SAM as a precursor, where a plant cell can divert SAM pools either into PA biosynthesis, ethylene biosynthesis, or both (Kumar and Rajam 2004; Lasanajak et al. 2014). Tomato transgenics over-expressing yeast SAMDC and yeast Spd synthase (SPDS) showed enhanced lycopene content, fruit juice viscosity and ethylene levels along with the increased Spd and Spm production (Mehta et al. 2002; Nambeesan et al. 2010). The fruits of these transgenics accumulating higher levels of Spd and Spm also exhibited quantitative changes in transcript profile of ethylene receptor populations and component(s) of the ethylene signalling pathway. These studies suggest that PA-ethylene nexus plays a crucial role in fruit ripening. Therefore, while looking at multifunctional and regulatory aspects of PAs and ethylene, it is possible that controlled manipulation of these key regulators rather than a structural or a regulatory gene operating in single branch of the biosynthesis pathway may result in better improvement of fruit shelf life and quality traits. Hence the utilization of the metabolic interactions of pathways related to post-harvest characteristics like the competitive interaction of PAs with that of ethylene seems to be an alternative strategy for the modulation of fruit ripening and quality. Accordingly, PA biosynthetic enzymes qualify as targets to control progression of ripening in various fruits. In this regard, most of the studies have exploited the higher PAs (Spd and Spm), but little has been reported for the utilization of Put. Interestingly, the three PAs have been shown to influence fruit ripening differentially, where the effects of diamine Put generally contrast with that of Spd and Spm (Handa and Mattoo 2010). This warrants further investigation to demonstrate the role of individual PAs in fruit ripening. Therefore, in this study, we over-expressed Put biosynthesis gene, ODC in tomato under the control of fruit-specific promoter (2A11) to demonstrate its role in ripening.
Results and discussion
Development of homozygous transgenic tomato plants over-expressing ODC transgene
The selection of ODC gene source i.e., mouse was based on the availability of gene sequence from said source at the commencement of study and also because of its least homology (at the nucleotide level, ~45 %) to its tomato counterpart. Several independent tomato transformants were developed using 2A11-ODC gene construct. Initial screening of primary transformants (T0) by PCR using primers specific to NPT-II and m-ODC genes revealed the presence of the transgenes. DNA blot analysis of T0 plants detailed the copy number of transgene, which varied from one to multiple in different events. Out of seven lines analyzed, four lines (LeODC22, LeODC23, LeODC27 and LeODC32) showed single copy insertion while rest of the lines (LeODC30 and LeODC44) showed multiple copy insertion (Supplementary Fig. 2). In order to negate the possibility of differential expression due to variable copy number (Hobbs et al. 1993), single copy lines (also confirmed as homozygous lines) were utilized in the study. The results of segregation analysis conferred that LeODC22, LeODC23, LeODC27 and LeODC32 lines as homozygous for ODC transgene integration (Supplementary Fig. 2). The morphology of transgenic plants was normal. They were fertile with pollen viability ranging from 90 to 95 %, which is similar to that of untransformed plants (UT) (Supplementary Fig. 4). The onset of ripening in transgenic fruits was at par with the UT fruits (data not shown).
Stability of m-ODC gene expression across generations
Northern hybridization studies showed strong fruit-specific m-ODC transgene expression across four generations of transgenic tomato (i.e. T0–T3). m-ODC transcripts were first noticed at the mature green (MG) stage and their titers increased during breaker (BR) and pink red (PR) stage followed by a sharp decline at red ripe (RR) stage (Fig. 1). Transcript accumulation was concomitant with the activity of fruit-specific promoter (2A11). This promoter has been very well characterized from tomato plant, where 2A11 gene expresses strictly in a fruit-specific manner (Pear et al. 1989).
Stable expression of transgene across four generations (T0–T3) of LeODC transgenic tomato restricted to ripening fruits. Northern blot analysis was done using radio-labeled m-ODC gene fragment to probe transgene transcript accumulation in T3 generation fruits. Radio-labeled SlACTIN gene probe was used to ascertain ACTIN gene levels as internal control. MG mature green, BR breaker, PR pink red, RR red ripe, stages of fruit ripening
Polyamine levels during different stages of fruit ripening
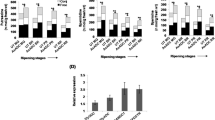
The fruits of UT tomato plants showed maximum PA titres in MG tissue, followed by a steep fall till RR stage. In contrast to the free and conjugated fractions, the bound fraction was present at relatively low level in UT fruits (Fig. 2, Supplementary File 1). Put and Spd concentrations declined during ripening with the Spm being in very low levels and negligible at final stage. As expected, transgenic fruits exhibited an overall increase in PAs (up to 2.0 fold) over UT fruits, with significant increase in Put (1.6–1.9 fold) due to the over-expression of ODC transgene. The increased Put accumulation was also accompanied by concomitant increase in higher PAs, viz., Spd (1.5-fold) and Spm (1.5–1.85 fold). In fruits of LeODC transgenics, high concentration of PAs were recorded at BR stage and thereafter, PA levels subsided although the concentrations were still significantly high levels as compared to UT fruits. The free (2.0 fold) and bound (1.5 fold) fractions contributed majorly to the enhanced PA pools as compared to conjugated fraction (Fig. 2a, Supplementary File 1). There was no significant change in mRNA levels of native tomato ODC and ADC genes, but SAMDC1 and SPDSYN transcripts increased in transgenic fruits over UT fruits (Fig. 2b, Supplementary Fig. 5a). In fact, SAMDC1 has been suggested to be predominantly responsible for SAMDC activity in ripening fruits (Gupta et al. 2013). Put upsurge in transgenic fruits is mainly because of the over-expression of m-ODC gene, while increased SAMDC1 and SPDSYN transcripts is accountable for the enhanced Spd and Spm content in transgenic fruits. These results demonstrated that the over-expression of heterologous ODC in tomato fruits has led to the up-regulation of the whole PA biosynthetic pathway and resulted in PA accumulation. Similar observations were obtained in transgenic wheat plants expressing an oat ADC (Bassie et al. 2007). In an another study, Put has been shown to enhance the conversion of precursor SAMDC into active SAMDC, leading to increased amounts of active enzyme and caused the activation of the PA biosynthesis (Kameji and Pegg 1987).
PA levels in transgenic and UT fruits at MG, BR, PR and RR stages in T3 generation. PA estimation was done thrice with tissue extract from third generation of UT and transgenic fruits, in 10 % PCA by TLC method. Three biological replicates were maintained in the experiment (n = 3 × 3 = 9). † and ‡ denote significant differences in free and bound fractions respectively from UT at 5 % level. a Put, Spd and Spm levels in transgenic and UT fruits. b Relative expression levels of PA biosynthesis genes in UT and T3 transgenic fruits. Relative quantification was performed by qRT-PCR using comparative D cycle threshold (CT) method. Expression levels of all the genes were normalized to SlyActin levels. The data represent the average of three biological replicates, each with two technical replicates and was plotted with error bars showing standard deviation. *Significant at p ≥ 0.05 over UT control. Expression analysis was done with three biological replicates (sample size = 3 × 2)
Ethylene production and expression profile of ethylene biosynthesis genes
Ethylene holds the well established status of a ripening inducer and stimulator in climacteric fruits. While it accelerates different pathways needed for completion of ripening, it also adversely affects the post-harvest storage of fruits. Our results showed that ethylene biosynthesis was significantly impaired in LeODC transgenic fruits. There was ~40 % reduction in ethylene levels in transgenic fruits over UT fruits (Fig. 3a). Further, fruit-specific homologues of 1-aminocyclopropane-1-carboxylate synthase (ACS) and 1-aminocyclopropane-1-carboxylate oxidase (ACO) genes displayed transcript reduction in transgenic fruits (Fig. 3b, Supplementary Fig. 5b) and this has resulted in low levels of ethylene in transgenic fruits over UT fruits. There are several studies, which reported the negative correlation between PA levels and ACS and ACO expression. For instance, PAs have been shown to inhibit ACS transcript accumulation (Handa and Mattoo 2010; Li et al. 1992; Ziosi et al. 2006). PAs and ethylene intersect biosynthetically at a common point for precursor, SAM (Winer and Apelbaum 1986). Thus, an inverse relationship exists between PAs and ethylene biosynthesis during fruit ripening which is modulated by changes in native SAMDC activity. Therefore, in the present study, the over-production of Put and its conversion into Spd and Spm might have resulted in limiting SAM flux towards ethylene biosynthesis. A recent report by Lasanajak et al. (2014) further substantiates our results. Further, these results are also consistent with results obtained with the use of PA inhibitors (Kushad and Dumbroff 1991; Parra-Lobato and Gomez-Jimenez 2011) and transgenic approaches employed for the fruit quality improvement (Kumar et al. 1996; Lasanajak et al. 2014; Madhulatha et al. 2014). However, Mehta et al. (2002) reported elevated ethylene levels in tomato transgenics over-expressing yeast SAMDC gene, and suggested that the SAM is not a limiting factor for polyamine and ethylene biosynthesis.
Ethylene levels in fruits of UT and transgenic tomato plants. a Ethylene evolution was measured in UT and transgenic fruits (RR) through GC, in four generations (T0–T3). Data shown is the representation of mean of nine biological replicates in each of the ten independent experiments from T3 generation (n = 9 × 10 = 90). b Relative expression levels of ethylene biosynthesis genes in UT and transgenic fruits. Relative quantification was performed by qRT-PCR using comparative D cycle threshold (CT) method. Expression levels of all the genes were normalized to SlyActin levels. The data represent the average of three biological replicates, each with two technical replicates and was plotted with error bars showing standard deviation. *Significant at p ≥ 0.05 over UT control. Expression analysis was done in T3 generation with three biological replicates (sample size = 3 × 2)
Improvement in storage attributes of transgenic fruits
Data obtained from four generations, i.e. T0–T3 of transgenic fruits consistently showed decreased rate of respiration (CO2 levels), reduced physiological loss of water (PLW), delayed on-vine ripening and extended shelf life (Fig. 4). LeODC transgenic fruits recorded 15–40 % reduction in CO2 evolution when compared to the UT ones (Fig. 4b). However, LeODC22 and LeODC32 did not show reduction in respiratory activity, which could be attributed to the position effect of transgene integration. PLW was also slowed down in transgenic fruits (Fig. 4c). There was ~7 days delay in on-vine transition from BR to RR stage as compared to UT fruits (Fig. 4a). Skin wrinkles were seen in UT fruits after 6–7 days whereas transgenic fruits started showing sign of senescence after 10–15 days of storage at room temperature (Fig. 4d). Fruits kept at 4 °C at RR stage showed approximately 20–25 days of shelf life without showing chilling symptoms, whereas UT fruits started rotting after 10–12 days. Therefore, transgenic fruits showed an extended shelf life of ~7 days at room temperature and ~15 days at 4 °C as compared to UT fruits. Transgenic fruits (RR) were also found to be firmer than UT fruits (Data not shown).
Keeping qualities of UT and transgenic tomato fruits. Data is the representation of T3 generation showing means of nine biological replicates in each of the ten independent experiments (n = 9 × 10 = 90). Each set of experiment was repeated in four generations (T0–T3) with reproducible results. a Delayed on-vine ripening. b Respiratory activity in ripening fruits from UT and transgenic plants was measured through gas analyzer. c Rate of physiological loss of water was monitored for 10 days in ripe fruits kept at room temperature. d Shelf life of ripe fruits kept at room temperature. e Relative expression levels of cell wall hydrolyzing genes in UT and LeODC fruits. Relative quantification was performed by qRT-PCR using comparative D cycle threshold (CT) method. Expression levels of all the genes were normalized to SlyActin levels. The data represent the average of three biological replicates, each with two technical replicates and was plotted with error bars showing standard deviation. *Significant at p ≥ 0.05 over UT control. Expression analysis was done in T3 generation with three biological replicates (sample size = 3 × 2)
Put treatments in different fruits have shown to reduce their respiration rate by hampering ethylene production (Khan et al. 2008; Serrano et al. 2003; Torrigiani et al. 2012). Decline in respiratory activity and PLW in transgenic fruits supports for the delayed on-vine and extended shelf life. The prolonged shelf life is also attributed to low ethylene levels in transgenic tomato fruits since the ethylene levels in harvested tomato fruit bears a negative correlation with its shelf life (Guillén et al. 2007).
The textural softening during fruit ripening results from separation and dissolution of cell wall components. Consequently cell wall stability and rigidity majorly determine the processing characteristics, firmness and quality of tomato fruits. Various cell wall depolymerizing enzymes like polysaccharide hydrolases/glycoside hydrolase, transglycosylases, lyases and expansins get induced during ripening and are responsible for ripening related firmness loss (Brummell 2006). The expansin 1 (EXP1), β-galactosidase 4 (TBG4), polygalacturonase (PG) and xyloglucan endotransglycosylase/hydrolases 5 (XTH5) genes have been shown to be specifically expressed during fruit ripening and play major role in fruit softening (Pirrello et al. 2009). Expression analysis of both EXP1 and TBG4 revealed elevated transcript levels in transgenic fruits over UT fruits (Fig. 4e, Supplementary Fig. 5c). PG showed no difference in transcript abundance but considerable reduction was observed in XTH5 expression in transgenic fruits over UT fruits (Fig. 4e, Supplementary Fig. 5c).
Although our findings on expression of cell wall loosening genes are in conflict with the data on enhanced shelf life of fruits, the increase in bound PA fractions in transgenic fruits seems to be important for the enhanced shelf life possibly by stabilizing the membranes. Put, Spd and Spm are covalently associated with pectic polysaccharides in plant cell walls (D’Orazi and Bagni 1987). The interference with PA biosynthesis has been shown to hamper cell wall formation making it amorphous, while the exogenous treatment of PAs can reverse these changes (Berta et al. 1997). Knee and Bartley (1981) have demonstrated that SAM methylation of the free carboxylic groups in pectin leads to disruption of calcium (Ca) cross linkages of adjacent polyuronides, which causes loss in cohesion of cell walls. The negatively charged carboxyl groups (COO–) of uronic acids in pectinic material are normally available for cross-linkages through multivalent cations, mainly calcium. In transgenic fruits, since SAM precursor is diverted to PA synthesis, it was not readily available for disruption of calcium bridges. The effect of enhanced PAs on maintaining fruit wall rigidity is also ascribable to their action similar to Ca2+ in forming cross-linkage to the COO– group of the pectic substances in the cell wall. This cross-linking reportedly blocks the access of degrading enzymes, such as pectin methyl esterase, pectinesterase and PG, reducing the rate of softening during storage (Mirdehghan et al. 2007). Thus, in spite of the enhanced expression of cell wall modifying genes, the increased cellular PAs might be involved in the stabilization of cell wall integrity by reducing cell wall permeability and consequently leading to the lower PLW in transgenic fruits. These results indicate that the increase in PAs during fruit development would decrease the rate of fruit ripening.
Improved nutritional quality in LeODC fruits
Total soluble solids (TSS) content is the key component in determining fruit flavour and juice viscosity and is of special significance for processing industry (Ferguson and Boyd 2002). It comprises of sugars, minerals and acids contents. In the present study, TSS was increased by 30–50 %, in juice of the transgenic fruits as compared to UT fruit juice (Table 1). Transgenic fruits also exhibited ~50 % increase in total sugars and ~2.0 fold increase in titratable acids (TAs) (Table 1). Since organic acids (e.g. citric acid) are the substrates of respiration, which is an ethylene-dependent factor, the lower rate of respiration in transgenic fruits might have resulted in the accumulation of TAs in such fruits (Defilippi et al. 2004).
Comparison of ascorbic acid (AsA) levels between transgenic and UT fruits revealed ~60 % higher levels in fruits of LeODC27 transgenic line over UT fruits (Table 1). AsA is a primary antioxidant and an important phytopharmaceutical found in tomato fruits. Ascorbate functions as a cofactor in ethylene biosynthesis (Watada et al. 1976). AsA levels decline during ripening and senescence, which is correlated with its consumption in ethylene biosynthesis pathway. In present study, the enhanced PA levels might have caused ascorbate accumulation in transgenic fruits by impeding with ethylene biosynthesis. Yahia et al. (2001) also reported the association between the increased cellular concentration of Put with higher ascorbic acid in fruits. High levels of total antioxidants (~fivefold increase) was also recorded in transgenic fruits (Table 1).
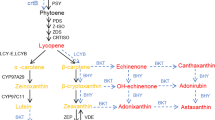
The transgenic fruits were visually more dark red in colour than UT fruits, which is linked with the lycopene build-up in such fruits. Transgenic fruits accumulated ~10–40 % more lycopene content than UT fruits (Table 1). The expression pattern of lycopene biosynthesis genes viz., 1-deoxy-d-xylulose 5-phosphate synthase (DXS1) and phytoene synthase (PSY1) revealed significant increase in their mRNA titres, while transcript profile of lycopene epsilon-cyclase (LES) which is a lycopene catabolic gene, did not show any significant difference from UT control (Fig. 5, Supplementary Fig. 5d). Therefore, the up-regulated expression of these transcripts accounts for the enhanced lycopene content in transgenic fruits over UT fruits. The enhanced PA levels might be involved in the increased levels of lycopene and expression of related genes possibly by stabilization of mRNA turnover or regulation of lycopene biosynthesis genes. It has been reported that reduced degradation of membrane phospholipids in fruits tend to reduce the turnover of phospholipids and free fatty acids. Since fatty acid biosynthesis and carotenoid biosynthesis share the common precursor (Acetyl CoenzymeA), reduced demand for acetyl CoA for fatty acid biosynthesis in transgenic fruits may tend to enhance their availability for carotene and lycopene biosynthesis (Oke et al. 2003). This could also explain the enhanced lycopene in transgenic fruits. Both these arguments are in agreement with the reports of Mehta et al. (2002), Neily et al. (2011) and Madhulatha et al. (2014) on transgenic tomato with over-expression of yeast SAMDC and SPDSYN and human SAMDC gene. Therefore, PA accumulation in transgenic fruits appears to cause an increase in the phytonutrients in fruits.
Expression pattern of lycopene biosynthesis genes in T3 fruits. Relative quantification was performed by qRT-PCR using comparative D cycle threshold (CT) method. Expression levels of all the genes were normalized to SlyActin levels. The data represent the average of three biological replicates, each with two technical replicates and was plotted with error bars showing standard deviation. *Significant at p ≥ 0.05 over UT control. Expression analysis was done with three biological replicates (sample size = 3 × 2)
In conclusion, the present study proclaims that the PA accumulation in fruits due to the over-expression of a Put biosynthesis gene, ODC can inhibit ethylene emission, reduce the perishability and improve quality of tomato fruits. These results also suggest that Put may regulate fruit ripening in tomato.
Materials and methods
Plant material
The seeds of tomato (Solanum lycopersicum Mill. cv. Pusa Ruby) were procured from National Seeds Corporation, New Delhi. Fully expanded cotyledons from 10 to 12 days old tomato seedlings grown in pots containing garden soil and vermiculite (1:1) under controlled growth conditions (26 ± 1 °C, 16 h photoperiod with irradiance of 40 μE mol m−2 s−1) were utilized for transformation experiments.
Development of LeODC transgenic tomato plants
A 4.0 kb fruit-specific 2A11 promoter fragment was excised from pGEM-T-2A11 (gift from Dr. V. S. Reddy, International Centre for Genetic Engineering and Biotechnology, New Delhi) by SmaI and EcoRI restriction digestion, and was ligated in pBinAR binary vector, replacing CaMV 35S promoter. Mouse ODC (m-ODC) gene (1.7 kb) was fished out from pBSK-ODC (gift from Prof. Tony Pegg, Hershey, USA) plasmid by restriction digestion with EcoRI and BamHI enzymes. The m-ODC gene fragment was end filled using klenow fragment and was subsequently ligated into SmaI site of pBin2A11. The pBin-T-DNA contained sequences encoding the enzyme neomycin phosphotransferase II (NPT-II) from the Tn5 transposon of E. coli (K12), under the control of NOS promoter from Agrobacterium tumefaciens (Supplementary Fig. 1). The resultant binary vector, pBin2A11ODC (Supplementary Fig. 1), was mobilized into A. tumefaciens strain LBA4404. The transformed A. tumefaciens was used for tomato transformation according to a protocol described by Madhulatha et al. (2007). The expression of NPT-II activity was used as a selectable trait to screen transformed plants. Transformed and untransformed (UT) regenerants, with well-developed roots were then transferred to plastic pots and following hardening they were grown in transgenic green-house/net-house for further studies.
Integration and segregation analysis of transgene in tomato transformants
Genomic DNA was isolated from the leaves of UT and tomato transformants using CTAB method (Doyle and Doyle 1990). Utilizing 100 ng of genomic DNA, primary transformants were screened by PCR for the presence of transgene (m-ODC) as well as NPT-II marker gene. The primer pairs used for the amplification of 750 bp fragment of NPT-II gene at 59 °C annealing, were 5′-TCAGAAGAACTCGTCAAGAA-3′ and 5′-ATGGGGATTGAACAAGATGG-3′ and for 570 bp amplicon of m-ODC gene at 59 °C annealing, were 5′-ATACCATGGTTTTGGCAGCCATAATGG-3′ and 5′-ATAGGCGCGCCGCTCCTATACCGAACTGA-3′.
Genomic DNA (10 µg), digested with XbaI enzyme was used for Southern blot hybridization using radio-labelled m-ODC probe. The probe was made using nick translation kit according to manufacturer’s guidelines (Bangalore Genei, www.bangloregenei.com/). Blots were prepared using the standard protocol (Sambrook et al. 1989), using nylon membrane (Hybond N, Pharmacia). Pre-hybridization and hybridization were carried out according to Sambrook et al. (1989).
The transgene segregation pattern was scored based on kanamycin resistant phenotype and transgene-specific PCR analysis. A schematic diagram illustrating segregation analysis across different generations has been presented in Supplementary Fig. 3. The leaves from T1, T2 and T3 seedlings (100 seedlings, raised on a 1:1 ratio of soil:vermiculite) of each independent line were surface-sterilized and placed on antibiotic (50 mg/l kanamycin) amended MS basal or shoot regeneration medium to check for the segregation of the transgene based on kanamycin-resistant phenotype. The explants were scored for kanamycin resistance after 15 days of culture. The leaves which survived on antibiotic-amended medium were considered as positive segregants and others that could not survive and died/bleached/leaf yellowing were considered negative segregants and were discarded (Supplementary Fig. 2). Genomic DNA from T1, T2 and T3 positive segregants (forty each) of each transgenic lines along with few UT plants were utilized for segregation analysis studies. Segregation pattern was confirmed by PCR in all the forty progenies from T1, T2 and T3 generations using m-ODC specific primers.
Segregation pattern of homozygous lines was also confirmed in T3 progenies by Southern blot analysis. Genomic DNA (10 µg, digested with XbaI enzyme) from three T3 progenies of few homozygous lines was hybridized with radio-labelled m-ODC probe. Preparation of blots and hybridization was carried out as described earlier.
In vitro pollen germination assay
Pollen grains were collected from freshly dehisced anthers of UT and transgenic lines. Pollen were suspended in pollen germination medium (10 % Sucrose, 1,618 µM H3BO4, 1,270 µM Ca(NO3)2, 811 µM MgSO4, 989 µM KNO3) (Sinha and Rajam 2013) and incubated for 2 h at 26 ± 2 °C in the dark. Pollen germination was recorded under a light microscope. At least 500 pollen grains were counted from each slide and percentage pollen germination was determined by repeating the experiment thrice.
Determination of different ripening stages in transgenic fruits
Flowers were tagged at anthesis and the days were noted for the fruit formation. Average days from MG (full size and firm) to reach BR and BR to RR were noted for the UT fruit based on the amount of colour each stage fruit will develop (Mehta et al. 2002). The number of average days was then applied for harvesting fruits from transgenic lines, which although now differ in their colour appearance but are at same age as that of their respective controls.
Expression analysis
Pericarp tissue (~100 mg; MG, BR, PR and RR fruits) from three biological replicates was pulverized to homogenate powder. The homogenate was used for isolation of total RNA with the TriZol reagent following manufacturer’s guidelines (Invitrogen, http://www.invitogen.com). Total RNA integrity was verified by agarose gel electrophoresis and their purity (A 260/A 280 > 1.95) with a NanoDrop ND-1000 spectrophotometer (Thermofischer, USA). This was followed by RNase free DNase (Fermentas, www.fermentas.com) treatment to remove contaminating genomic DNA. For northern blot analyses, total RNA (15 µg) was fractionated using formaldehyde agarose gel (Sambrook et al. 1989) and blotted onto the positively charged nylon membrane (Hybond N, Pharmacia). The membrane was incubated for 6 h at 42 °C in a prehybridization buffer. Hybridization was carried out using 32P-labeled m-ODC and SlyACTIN probes at 42 °C for 16 h (Sambrook et al. 1989).
cDNA was generated from total RNA using Superscript reverse transcriptase (Invitrogen, USA) according to manufacturer’s instructions. qRT-PCR was performed using SYBR Green I technology in Realplex2 thermal cycler (Eppendorf, Germany). A master mix was prepared by using MESA Blue qPCR mix as a PCR core reagent (Eurogentec, USA). 1.5 ng of cDNA and 750 nM each of the specific primers were added in a final volume of 10 µl (Table S1). The amplification reactions were carried out at 95 °C 4 min, 40 cycles at 95 °C 15 s followed by 60 °C for 1 min. Specificity of amplification was assessed by disassociation or melt curve analysis at 60–95 °C after 40 cycles. For all the qPCR experiments, three independent biological replicates were included (each with two technical replicates). Relative expression was calculated using comparative D cycle threshold (CT) method and an average of three biological replicates X each with two technical replicates, was plotted along with calculated standard deviation. The constitutively expressed SlACTIN gene was used as reference gene for normalization of the expression. Primer pairs used for each gene with their respective accession numbers are provided in Supplementary Table S1.
Initially, semi-Quantitative RT-PCR was also performed to check the transcript level of the few ripening specific genes. DNA-free RNA (50 ng) was used for the one-step RT-PCR reaction as specified in the kit manual (Taurus-Scientific, India, www.taurusscientific.com). The reaction conditions were as follow: 45 °C for 50 min, followed by 94 °C for 5 min, n cycles of 94 °C for 30 s, annealing temperature for 30 s and 72 °C for 30 s and finally 72 °C for 15 min. The products were analyzed on ethidium bromide stained 1.5 % agarose gel. All semi-quantitative-RT-PCR experiments were performed in triplicates, with RNA from three fruits.
Accession numbers
The accession numbers for ripening-associated genes selected for transcript analysis are: SlODC (NM 001247687.1), SlADC1 (NM 001247135.1), SlSAMDC1(EU196515.1), SlSPDSYN (NM 001247564.1), ACS6 (AF179249), SlACS1A (U18056.2), SlACS2 (NM 001247249.1), SlACS4 (NM 001247351), SlACO1 (NM 001247095), SlE8 (X13437.1), 1-deoxy-d-xylulose-5-phosphate synthase (SlDXS1, AF143812), (SlPSY1, EF157835.1), lycopene-epsilon cyclase (SlLES, Y14387), SlTAGL1 (AY098735.2), polygalacturonase (SlPG, X05656), expansin (SlEXP1, U82123), β-galactosidase 4 (SlTBG4, AF020390), α-xyloglucan endotransglucosylase/hydrolase (SlXTH5, AY497475), and SlACTIN, (BT012695).
Quantification of polyamine levels
PAs were estimated by TLC method using 100 mg of tissue each from leaf, floral bud (FB) and pericarp of MG, BR, PR and RR fruits. Tissue was homogenized in 1 mL of 10 % perchloric acid and was centrifuged at 18,000×g for 20 min at 4 °C. The resultant supernatant formed the source of free and conjugated fractions of PAs and the pellet as the bound fraction. PA fractions thus obtained, were dansylated, chromatographed and quantified as described by Bajaj and Rajam (1996) using dual wavelength fluorometer (Bio-Rad, Versa Fluor, USA) with an excitation wavelength of 350 nm and emission wavelength of 495 nm. PA analysis was performed in three independent experiments.
Ethylene measurements
In order to determine ethylene levels, three RR fruits (pre-weighed) were enclosed in 100 mL of air-tight container for about 1 h at room temperature. Headspace atmosphere (3 mL) of the container was withdrawn and injected into a gas chromatograph (Model HP 5890, Hewlett Packard, USA) (Singh and Pal 2008). The ethylene estimation was carried out in four generations each having ten independent experiments with three experimental replicates.
Estimation of respiration rate
Five pre-weighed RR fruits were sealed in an air-tight container for 1 h and their respiration rate was determined by measuring the head space carbon dioxide (CO2) concentration using CO2/O2 analyzer (Model Checkmate 9900 O2/CO2, PBI Dansensor, Denmark) (Singh and Pal 2008). The respiratory activity was measured in ten independent experiments with three experimental replicates (repeated for four generations; T0–T3 of transgenic tomato plants). Respiration rate was calculated taking into account the weight of the fruit, residual volume of the container and the incubation time (1 h). CO2 evolution rate was expressed as mL kg−1 h−1.
On-vine ripening period determination
Flowers were tagged at anthesis and days were noted for the fruit formation. Mature fruits displaying first sign of colour change were identified as BR stage. On-vine ripening period was expressed as the number of days taken to reach RR state from BR stage. The experiment was carried out with nine fruits as biological replicates and repeated with ten independent sets. All ten sets were repeated for four generations (T0–T3) of transgenic tomato plants.
Determination of fruit shelf life
RR fruits were kept at room temperature and time was noted for the first visual sign of shrivelling after being detached. The shelf life of fruits was noted for nine fruits as biological replicates each in ten independent sets for four generations of transgenic tomato plants.
Determination of physiological loss of water
PLW from tomato plants was determined as loss in weight of tomato fruits. The loss in weight of individual fruit was calculated daily as a percentage of the original weight. A mean of nine fruits in each of the ten independent sampling periods in each generation (repeated for four generations; T0-T3 of transgenic tomato plants).
Measurement of total soluble solids
TSS concentration in RR homogenate was measured by a hand refractrometer (Model: Fisher, Japan). Results were expressed as °Brix and were corrected to 20 °C temperature. TSS content was measured in nine fruits as biological replicates in ten independent experiments in each of the four generations; T0–T3 of transgenic tomato plants.
Quantification of titratable acids
TAs content was determined by the method of Srivastava and Kumar (1993). A known volume of filtered juice was titrated with 0.1 N NaOH using phenolphthalein indicator and light pink colour as an end point. Acidity was computed and expressed as percent citric acid. The estimation was done in nine biological replicates in each of the ten independent experiments followed for four generations (T0–T3) of transgenic tomato plants.
Determination of sugars
Total fruit sugar content was determined following the method of Ranganna (1977). Sugar content was measured in nine fruits as biological replicates in each of the ten independent experiments carried out for four generations (T0–T3) of transgenic tomato plants.
Estimation of ascorbic acid content
AsA content in the red fruits was estimated titrimetrically using 2,6-dichlorophenol indophenol as the indicator dye (Singh and Pal 2008). AsA standard was prepared by dissolving 100 mg of l-ascorbic acid in 100 mL of 1 % HPO3 (Singh and Pal 2008). AsA content was measured in nine fruits in each of the ten independent experiments followed for four generations (T0–T3) of transgenic tomato plants.
Quantification of lycopene content
Lycopene fraction of RR fruit pericarp tissue (~2 g) was determined by spectrophotometric method as described by Ranganna (1977). Lycopene fractions were estimated in nine biological replicates in each of the ten independent experiments followed for four generations (T0–T3)of transgenic tomato plants.
Determination of total antioxidant activity
The total antioxidant activity was measured in 2 g of fruit pericarp tissue using total radical-trapping antioxidant parameters (TRAP) assay. The total antioxidant extraction and assay was performed according to the procedure reported by Fanasca et al. (2006). The total antioxidant activity was assayed in nine fruits as biological replicates in each of the ten independent experiments carried out for four generations (T0–T3) of transgenic tomato plants.
Data collection and analysis
For each of the parameter, data from four consecutive generations, with their replicates was collected. Data are the means of 10 experiments (9 replicates in each experiment). The data presented are the mean with the standard error from all the experiments. Significant differences were determined by Student’s t test (p < 0.05).
References
Alcázar R, Altabella T, Marco F, Bortolotii C, Reymond M, Koncz C, Carrasco P, Tiburcio AF (2010) Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231:1237–1249
Apelbaum A, Burgoon AC, Anderson JD, Lieberman M, Benarie R, Mattoo AK (1981) Polyamines inhibit biosynthesis of ethylene in higher plant tissue and fruit protoplast. Plant Physiol 68:453–456
Bajaj S, Rajam MV (1996) Polyamine accumulation and near loss of morphogenesis in long term Callus cultures of rice: restoration of plant regeneration by manipulation of cellular polyamine levels. Plant Physiol 112:1343–1348
Bassie L, Zhu C, Romagosa I, Christou P, Capell T (2007) Transgenic wheat plants expressing an oat arginine decarboxylase cDNA exhibit increases in polyamine content in vegetative tissue and seeds. Mol Breed. doi:10.1007/s11032-007-9154-2
Berta G, Altamura M, Fusconi A, Cerruti F, Capitani F, Bagni N (1997) The plant cell wall is altered by inhibition of polyamine biosynthesis. New Phytol 137:569–577
Brummell DA (2006) Cell wall disassembly in ripening fruit. Funct Plant Biol 33:103–119
Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P (1999) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11:2203–2216
Cassol T, Mattoo AK (2002) Do polyamines and ethylene interact to regulate plant growth, development and senescence? In: Nath P, Mattoo A, Ranade SR, Weil JH (eds) Molecular insights in plant biology. Science Publishers, Enfield, pp 121–132
Defilippi BG, Dandekar AM, Kader AA (2004) Impact of suppression of ethylene action or biosynthesis on flavor metabolites in apple (Malus domestica Borkh) fruits. J Agric Food Chem 52:5694–5701
D’Orazi D, Bagni N (1987) In vitro interactions between polyamines and pectic substances. Biochem Biophys Res Commun 148:1259–1263
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fanasca S, Colla G, Maiani G, Venneria E, Rouphael Y, Azzini E, Saccardo F (2006) Changes in antioxidant content of tomato fruits in response to cultivar and nutrient solution composition. J Agric Food Chem 54:4319–4325
Ferguson IB, Boyd LM (2002) Inorganic nutrients and fruit quality. In: Knee M (ed) Fruit quality and its biological basis. Sheffield Academic Press, Sheffield, pp 14–45
Giovannoni JJ (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52:725–749
Guillén F, Castillo S, Zapata PJ, Martínez-Romero D, Serrano M, Valero D (2007) Efficacy of 1-MCP treatment in tomato fruit. 1. Duration and concentration of 1-MCP treatment to gain an effective delay of postharvest ripening. Postharvest Biol Technol 43:23–27
Gupta A, Pal RK, Rajam MV (2013) Delayed ripening and improved fruit processing quality in tomato by RNAi-mediated silencing of three homologs of 1-aminopropane-1-carboxylate synthase gene. J Plant Physiol 170:987–995
Hamilton AJ, Lycett GW, Grierson D (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346:284–287
Handa AK, Mattoo AK (2010) Differential and functional interactions emphasize the multiple roles of polyamines in plants. Plant Physiol Biochem 48:540–546
Hobbs SLA, Warkentin TD, DeLong CMO (1993) Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol 21:17–26
Igarashi K, Kashiwagi K (2010) Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42:39–51
Kameji T, Pegg AE (1987) Effect of putrescine on the synthesis of S-adenosylmethioine decarboxylase. Biochem J 243:285–288
Khan AS, Singh Z, Abbasi NA, Swinny EE (2008) Pre- or postharvest applications of putrescine and low temperature storage affect fruit ripening and quality of ‘Angelino’ plum. J Sci Food Agric 88:1686–1695
Knee M, Bartley IM (1981) In: Friend J, Rhodes MJC (eds) Recent advances in the biochemistry of fruit and vegetables. Academic Press, New York, pp 131–148
Kumar SV, Rajam MV (2004) Polyamine-ethylene nexus: a potential target for postharvest biotechnology. Indian J Biotech 3:299–304
Kumar A, Taylor MA, Arif SAM, Davies HV (1996) Potato plants expressing antisense and sense S-adenosylmethionine decarboxylase (SAMDC) transgenes show altered levels of polyamines and ethylene: antisense plants display abnormal phenotypes. Plant J 9:147–158
Kusano T, Berberich T, Tateda C, Takahashi Y (2008) Polyamines: essential factors for growth and survival. Planta 228:367–381
Kushad MM, Dumbroff EB (1991) Metabolic and physiological relationship between the polyamine and ethylene biosynthetic pathways. In: Slocum RD, Flores HE (eds) Biochemistry and physiology of polyamines in plants. CRC Press, Boca Raton, pp 77–92
Lasanajak Y, Minocha R, Minocha SC, Goyal R, Fatima T, Handa AK, Mattoo AK (2014) Enhanced flux of substrates into polyamine biosynthesis but not ethylene in tomato fruit engineered with yeast S-adenosylmethionine decarboxylase gene. Amino Acids 46:729–742
Lashbrook CC, Giovannoni JJ, Hall BD, Fischer RL, Bennett AB (1998) Transgenic analysis of tomato endo-beta-1,4-glucanase gene function. Role of cel1 in floral abscission. Plant J 13:303–310
Li N, Parsons B, Liu D, Mattoo AK (1992) Accumulation of wound-inducible ACC synthase transcript in tomato fruit is inhibited by salicylic acid and polyamines. Plant Mol Biol 18:477–487
Madhulatha P, Pandey R, Hazarika P, Rajam MV (2007) High transformation frequency in Agrobacterium mediated genetic transformation of tomato by using polyamines and maltose in shoot regeneration medium. Physiol Mol Biol Plants 13:191–198
Madhulatha P, Gupta A, Gupta S, Kumar A, Pal RK, Rajam MV (2014) Fruit-specific over-expression of human S-adenosylmethionine decarboxylase gene results in polyamine accumulation and affects diverse aspects of tomato fruit development and quality. J Plant Biochem Biotechnol 23:151–160
Mehta R, Cassol T, Li N, Ali N, Handa AK, Mattoo AK (2002) Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nat Biotech 20:613–618
Mirdehghan SH, Rahemi M, Castillo S, Martínez-Romero D, Serrano M, Valero D (2007) Pre-storage application of polyamines by pressure or immersion improves shelf-life of pomegranate stored at chilling temperature by increasing endogenous polyamine levels. Postharvest Biol Technol 44:26–33
Nambeesan S, Datsenka T, Ferruzzi MG, Malladi A, Mattoo AK, Handa AK (2010) Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato. Plant J 63:836–847
Neily MH, Matsukura C, Maucourt M, Bernillon S, Deborde C, Moing A, Yina Y, Saitoa T, Moria K, Asamizua E, Rolinb D, Moriguchie T, Ezuraa H (2011) Enhanced polyamine accumulation alters carotenoid metabolism at the transcriptional level in tomato fruit over-expressing spermidine synthase. J Plant Physiol 168:242–252
Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254:437–439
Oke M, Pinhero RG, Paliyath G (2003) The effects of genetic transformation of tomato with antisense phospholipase D cDNA on the quality characteristics of fruits and their processed products. Food Biotechnol 17:163–182
Parra-Lobato MC, Gomez-Jimenez MC (2011) Polyamine-induced modulation of genes involved in ethylene biosynthesis and signaling pathways and nitric oxide production during olive mature fruit abscission. J Exp Bot 62:4447–4465
Payasi A, Sanwal GG (2010) Ripening of climacteric fruits and their control. J Food Biochem 34:679–710
Pear JR, Ridge N, Rasmussen R, Rose RE, Houck CM (1989) Isolation and characterization of a fruit-specific cDNA and the corresponding clone from tomato. Plant Mol Biol 13:639–651
Pirrello J, Regad F, Latché A, Pech JC, Bouzayen M (2009) Regulation of tomato fruit ripening. In: Perspectives in agriculture, veterinary science, nutrition and natural resources. CAB Rev 4:1–14
Ranganna S (1977) Manual of analysis of fruit and vegetable products. Tata McGraw Hill, New Delhi
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbour, New York
Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M (2013) Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451:145–154
Schwartz CE, Wang X, Stevenson RE, Pegg AE (2011) Spermine synthase deficiency resulting in X-linked intellectual disability (Snyder–Robinson syndrome). Methods Mol Biol 720:437–445
Serrano M, Martinez-Romero D, Guillén F, Valero D (2003) Effects of exogenous putrescine on improving shelf life of four plum cultivars. Postharvest Biol Technol 30:259–271
Singh SP, Pal RK (2008) Controlled atmosphere storage of guava (Psidium guajava L.) fruit. Postharvest Biol Technol 47:296–306
Sinha R, Rajam MV (2013) RNAi silencing of three homologues of S-adenosylmethionine decarboxylase gene in tapetal tissue of tomato results in male sterility. Plant Mol Biol 82:169–180
Srivastava RP, Kumar S (1993) Fruit and vegetable preservation—principles and practices. International book distribution company, New Delhi
Takahashi T, Kakehi J-I (2010) Polyamines: ubiquitous polycations with unique roles in growth and stress Responses. Ann Bot 105:1–6
Torrigiani P, Bressanin D, Beatriz RK, Tadiello A, Trainotti L, Bonghi C, Ziosi V, Costa G (2012) Spermidine application to young developing peach fruits leads to a slowing down of ripening by impairing ripening-related ethylene and auxin metabolism and signalling. Plant Physiol 146:86–98
Valero D, Martínez-Romero D, Serrano M (2002) The role of polyamines in the improvement of the shelf life of fruit. Trends Food Sci Technol 13:228–232
Wang T-W, Zhang C-G, Wu W, Nowack LM, Madey E, Thompson JE (2005) Antisense suppression of deoxyhypusine synthase in tomato delays fruit softening and alters growth and development. Plant Physiol 138:1372–1382
Watada AE, Aulenbach BB, Worthington JT (1976) Vitamins A and C in ripe tomatoes as affected by stage of ripeness at harvest and by supplementary ethylene. J Food Sci 41:856–858
Winer L, Apelbaum A (1986) Involvement of polyamines in the development and ripening of avocado fruits. J Plant Physiol 126:223–456
Xie Q, Hu Z, Zhu Z, Dong T, Zhao Z, Cui B, Chen G (2014) Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci Rep. doi:10.1038/srep04367
Xiong AS, Yao QH, Peng RH, Li X, Han PL, Fan HQ (2005) Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep 23:639–646
Yahia EM, Contreras-Padilla M, Gonzalez-Aguilar G (2001) Ascorbic acid content in relation to ascorbic acid oxidase and polyamine content in tomato and bell pepper fruits during development, maturation and senescence. Lebensm-Wiss Technol 34:452–457
Ziosi V, Bregoli AM, Bonghi C, Fossati T, Biondi S, Costa G, Torrigiani P (2006) Transcription of ethylene perception and biosynthesis genes is altered by putrescine, spermidine and aminoethoxyvinylglycine (AVG) during ripening in peach fruit (Prunus persica). New Phytol 172:229–238
Acknowledgments
We express our gratitude to Prof. Tony Pegg (Hershey, USA) for providing the m-ODC cDNA and Dr. V. S. Reddy (International Centre for Genetic Engineering and Biotechnology, New Delhi) for providing the 2A11 tomato promoter. This work was generously supported by Grants from the Department of Biotechnology (Govt. of India), New Delhi (Grant Nos. BT/PR/2990/Agr/16/232/2002 and BT/PR8657/PBD/16/738/2007). We also thank the University Grants Commission, New Delhi for Special Assistance Programme and Department of Science and Technology, New Delhi for FIST and DU-DST PURSE programmes. Research fellowships to Roopali Pandey and Aarti Gupta by the Council of Scientific and Industrial Research are acknowledged.
Conflict of interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Roopali Pandey and Aarti Gupta have contributed equally to the article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2014_273_MOESM2_ESM.jpg
Supplementary Fig. 2 Molecular characterization and segregation analysis of tomato transformants. (a) PCR analysis with primers specific to NPT-II gene in primary transformants: L - 1 kb ladder; PC - Plasmid DNA; UT - DNA from untransformed control; LeODC 22-72 - DNA from different transgenic tomato lines. (b) Southern blot analysis of T0 generation of LeODC transgenic plants, revealing transgene integration and copy number using m-ODC gene radio-labeled probe. UT - DNA from untransformed control; LeODC 11-60- DNA from different transgenic lines digested with Xba I enzyme. (c) The leaves of T1 transgenic line LeODC22 showing segregation of the m-ODC transgene on kanamycin (50 mg/l) amended MS basal medium, and this was based on the bleaching of leaves. The untransformed leaves (UT) showed complete bleaching on antibiotic medium. (d1 and d2). PCR analysis with primers specific to m-ODC gene showing segregation pattern of transgene in T2 and T3 generations respectively. L - 1 kb ladder; PC - Plasmid DNA, UT-DNA from untransformed control; B - blank, 1-12 - DNA from different progenies of LeODC27 line. (e) Southern blot analysis of T3 generation of LeODC transgenic plants revealing homozygous status of integrated transgene, using m-ODC gene radio-labeled probe. LeODC22-LeODC32 - DNA from T3 progenies of different transgenic lines digested with XbaI enzyme. (JPEG 90 kb)
11103_2014_273_MOESM3_ESM.jpg
Supplementary Fig. 3 A lineage chart illustrating analysis of segregation patterns across different generations. Leaves from 100 T1 seedlings (named as e.g., LeODC22-1, LeODC22-2……. LeODC22-100) of each positive T0 plant (e.g. LeODC22) were screened by kanamycin resistant assay and 40 positive segregants were further confirmed by m-ODC gene specific PCR. All the homozygous lines followed 3:1 transgene segregation pattern in T1 generation. Ten positive segregants were taken to raise T2 generation. Hundred T2 seedlings (further named as e.g., LeODC22-1.1, LeODC22-1.2……. LeODC22-1.100.) from each of 10 such parents were further screened by kanamycin resistance assay and confirmed by PCR. (JPEG 72 kb)
11103_2014_273_MOESM4_ESM.jpg
Supplementary Fig. 4 Pollen viability UT and transgenic tomato lines. Pollen viability was scored based on pollen germination after 2 h incubation in germination media in dark. Data is the representation of T3 generation showing means of 500 pollen counts. Each experiment was repeated thrice in four generations (T0-T3) with reproducible results. (JPEG 12 kb)
11103_2014_273_MOESM5_ESM.jpg
Supplementary Fig. 5 Semi-quantitative RT-PCR analysis showing expression patterns of fruit specific genes. (a) Expression pattern of PA biosynthesis genes; SlODC, SlADC, SlSAMDC1 and SlSPDSYN, (b) Expression pattern of ethylene biosynthesis genes; SlACS2, SlACS4 and SlACO1, (c) Expression pattern of cell wall hydrolyzing genes; SlEXP1, SlTBG4, SlPG2a and SlXTH (d) Expression pattern of lycopene biosynthesis genes; SlPSY1, SlDXS1 and SlLES; in transgenic (T3 generation) and UT fruits at different stages (MG, BR, PR and RR) of ripening. SlACTIN gene expression was maintained as internal control. Expression analysis was done thrice using 50 ng of total RNA, with ‘n’ no. of cycles utilizing three biological replicates. (JPEG 69 kb)
Rights and permissions
About this article
Cite this article
Pandey, R., Gupta, A., Chowdhary, A. et al. Over-expression of mouse ornithine decarboxylase gene under the control of fruit-specific promoter enhances fruit quality in tomato. Plant Mol Biol 87, 249–260 (2015). https://doi.org/10.1007/s11103-014-0273-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0273-y