Abstract
Introduction
Glioblastomas (GBMs) usually occur as a solitary lesion; however, about 0.5–35% present with multiple lesions (M-GBM). The genetic landscape of GBMs have been thoroughly investigated; nevertheless, differences between M-GBM and single-foci GBM (S-GBM) remains unclear. The present study aimed to determine differences in clinical and molecular characteristics between M-GBM and S-GBM.
Methods
A retrospective review of multifocal/multicentric infiltrative gliomas (M-IG) from our institutional database was performed. Demographics, clinical, radiological, and genetic features were obtained and compared between M-GBM IDH-wild type (IDH-WT) vs 193 S-GBM IDH-WT. Mutations were examined by a targeted next-generation sequencing assay interrogating 315 genes.
Results
33M-IG were identified from which 94% were diagnosed as M-GBM IDH-WT, the remaining 6% were diagnosed as astrocytomas IDH-mutant. M-GBM and S-GBM comparison revealed that EGFR alterations were more frequent in M-GBM (65% vs 42% p = 0.019). Furthermore, concomitant EGFR/PTEN alterations were more common in M-GBM vs. S-GBM (36% vs 19%) as well as compared to TCGA (21%). No statistically significant differences in overall survival were observed between M-GBM and S-GBM; however, within the M-GBM cohort, patients harboring KDR alterations had a worse survival (KDR-altered 6.7 vs KDR-WT 16.6 months, p = 0.038).
Conclusions
The results of the present study demonstrate that M-GBM genetically resembles S-GBM, however, M-GBM harbor higher frequency of EGFR alterations and co-occurrence of EGFR/PTEN alterations, which may account for their highly malignant and invasive phenotype. Further study of genetic alterations including differences between multifocal and multicentric GBMs are warranted, which may identify potential targets for this aggressive tumor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM) is the most common primary malignant tumor of the central nervous system (CNS) and is associated with poor prognosis despite current therapies [1,2,3]. GBMs usually occur as a solitary lesion; however, about 0.5 to 35% of all GBMs present with multiple lesions (M-GBM) [4,5,6,7,8]. Based on radiographic features, M-GBMs can be further divided into multifocal and multicentric GBMs. Multifocal GBMs have a clear contiguity pathway of spread between foci, which can be demonstrated by contiguous areas of T2-weighted signal on magnetic resonance imaging (MRI) of the brain. Meanwhile, multicentric GBMs are widely separated lesions that cannot be attributed to contiguous pathways [9]. The prognosis of multifocal and multicentric GBMs (M-GBM) have been historically reported to be more dismal than single foci GBM (S-GBM) [6,7,8].
The genetic landscape of GBMs has been thoroughly investigated revealing important genetic pathways that influence its clinicopathologic characteristics and outcomes [10, 11]. The exact mechanism of multifocality/multicentricity is still not fully comprehended, as studies defining genomic alterations of these tumors are scarce. The molecular characteristics of M-GBM might explain its inherent ability to migrate and invade, resulting in the poor outcome observed in these patients. It has been described that M-GBM lacks IDH1, ATRX, and PDGFRA mutations. Nevertheless, given the small sample sizes of prior studies, the true genomic differences between S-GBM and M-GBM remain unclear [8].
The goal of the present study is to determine from our institutional experience the molecular characteristics and clinical features between M-GBM and S-GBM, to allow a better understanding of genetic alterations in M-GBM that could explain their biology and behavior.
Material and methods
Patients and tumor samples
We performed a retrospective review of patients diagnosed with infiltrative glioma between January 2004 and December 2019. Patients were included if they had (1) histologic diagnosis of diffuse glioma; (2) multifocal or multicentric tumor; (3) mutation analysis by next-generation sequencing. Patients with multifocal tumors were defined as those having at least two distinct tumor foci, separated by at least 1 cm. Multifocal and multicentric GBM (M-GBM) were analyzed together and individually for a subanalysis. A cohort including 193 S-GBM, IDH-WT was used for comparison of demographic, clinical, and molecular characteristics (Online Resource 1). Patients diagnosed with diffuse astrocytic gliomas, IDH-WT, with molecular features of GBM, WHO grade IV were considered as GBM, IDH-WT according to the 3rd cIMPACT-NOW update [12].
Study data were collected from the electronic medical record of Memorial Hermann Hospital and managed using REDCap electronic data capture tools hosted at the University of Texas Health Science Center at Houston (UT Houston) [13, 14]. These included age, gender, Karnofsky performance status (KPS), histologic diagnosis, tumor location, radiographic extent of resection, treatment strategy, recurrence, and survival. Tumors were classified by a Board-certified neuropathologist following the 2016 WHO Classification of Tumors of the Central Nervous System [15]. This study was approved by the Institutional Review Board of the UT Houston and Memorial Hermann Hospital, Houston, TX.
Targeted sequencing
Tumor samples were analyzed for genomic alterations by a targeted next-generation sequencing assay (NGS) interrogating 315 genes and 28 gene rearrangements (FoundationOne CDx®, Foundation Medicine, Inc., Cambridge, MA, USA). The FoundationOne® assay was performed in a clinical laboratory improvement amendments certified laboratory, as previously described [16,17,18]. Telomerase reverse transcriptase promoter (TERTp) status was not available for four patients.
Co-occurrence of most common mutations
To evaluate the co-occurrence of EGFR/PTEN, EGFR/KDR, and EGFR/PTEN/TERTp alterations, we compared M-GBM vs S-GBM. In addition, to further investigate these differences we compared our results to the known incidence of these mutations in The Cancer Genome Atlas (TCGA) PanCancer Atlas for EGFR/PTEN and EGFR/KDR, and to a published study evaluating co-occurrence of EGFR/PTEN/TERTp alterations, as TCGA lacks TERTp information [19,20,21,22].
Statistical analyses
Clinical, demographic, and frequency of genomic alterations were evaluated by Fisher’s exact test or Mann–Whitney U test for categorical and continuous variables, respectively. Overall survival was calculated as the time in months from diagnosis to death or date of last available follow-up. Kaplan–Meier method was used to plot survival curves and statistical significance was examined by the univariable log-rank test. Cox proportional hazard regression models were utilized for univariable and multivariable analysis to calculate the hazard ratio (HR) estimates with 95% confidence interval (CI). Multivariable Cox proportional hazard regression model analysis was adjusted for the following set of covariates: age, male gender, KPS, Stupp protocol, salvage bevacizumab, multifocal vs multicentric, and KDR mutant gene, as these are factors well-known to affect survival [23], KDR was selected as it was found to be significant in univariable analysis. A p-value ≤ 0.05 was considered statistically significant. Statistical analyses were performed in EZR (1.40) [24] and Prism v.8.2.1 (GraphPad, La Jolla, California, USA).
Results
Clinico-demographic characteristics of multifocal infiltrative gliomas
A total of 564 patients with infiltrative gliomas (IG) were identified, from which 41 (7.3%) had a multifocal/multicentric profile (M-IG). There were 39/395 (9.9%) M-GBM IDH-WT patients, 2/69 (2.9%) astrocytomas IDH-mutant (IDH-MT) presented multiple lesions at the time of initial diagnosis. From the 41 patients, 33 met the inclusion criteria and were further analyzed (Online Resource 1).
The 33M-IG patients had a median age of 60 years (range 26–77). There were 17 males and 16 females, 24.2% had KPS scores ≥ 80, and 29/33 (88%) underwent resection, meanwhile, the remaining 12% underwent biopsy. Temozolomide was prescribed to 31 (93.9%) patients with concomitant radiotherapy to 30 (90.9%) according to the Stupp protocol [25]. Moreover, some patients were treated with 1st line bevacizumab (7/33, 21.2%), irinotecan (5/33, 15.2%), and tumor-treating fields (5/33, 15.2%). Histologically, 31 patients (94%) were diagnosed as GBM, IDH-WT, 1 (3%) GBM IDH-mutant (IDH-MT), and 1 (3%) diffuse astrocytoma (DA), IDH-MT. Furthermore, 22/33 (66.7%) patients could be further classified as multicentric and 30 (90.9%) patients had enhancing tumors (Fig. 1). The demographic and clinical characteristics are depicted in Table 1.
M-GBM MRI imaging. Patient 1, axial T1 post contrast (a) and axial T2 flair (b) demonstrating a multifocal GBM IDH-WT. Patient 27, sagittal T1 post contrast (c) and axial T2 Flair (d) demonstrating a multicentric GBM IDH-WT. Patient 20, axial T2 Flair (e), and axial T1 post contrast (f) demonstrating a multicentric partially enhancing GBM IDH-WT. Patient 32, axial T1 post contrast (g) demonstrating a multifocal GBM IDH-MT. Patient 33, sagittal (h) MRI demonstrating a multicentric diffuse astrocytoma IDH-MT
The comparison between S-GBM and M-GBM revealed that the latter was less likely to have gross total resection (GTR, 12.9% vs 33.5%, p = 0.02) but was treated more frequently with 1st line irinotecan (1.5% vs 12.9%, p = 0.007) and bevacizumab (9.3% vs 19.4%, p = 0.11). Re-resection was less likely to occur in M-GBM patients (39.8% vs 19.2%, p = 0.07). Furthermore, the analysis between multifocal (n = 10) and multicentric (n = 21) GBM IDH-WT showed that within this group, multifocal patients were more likely to undergo re-resection and salvage therapies at recurrence (Table 1).
Genetic alterations
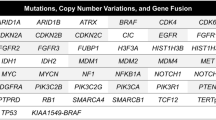
NGS analysis revealed that TERTp was mutated in 22/29 (75.9%) patients. CDKN2A/B loss was observed in 25/33 (75.7%), while EGFR alterations in 21/33 (63.6%) patients. PTEN was mutated in 18/33 (54.5%) patients. The other frequently altered genes were TP53 in 8/33 (24.4%), KDR, KIT, and PDGFRA, each mutated in 5/33 (15.2%) patients (Fig. 2; Table 1).
Mutations in cancer-related genes of 33 multicentric/multifocal infiltrative glioma patients. PFS progression-free survival (months), OS overall survival (months), WT wild type, Mut mutant, M male, F female. *Anaplastic astrocytoma IDH-WT with molecular features of GBM WHO grade IV according to cIMPACT-NOW Update 3
The comparison between S-GBM and M-GBM revealed that EGFR alterations were more frequently present in M-GBM (65% vs 42% p = 0.019, Table 1 and Online Resource 2). Moreover, we observed that concomitant mutations in EGFR and PTEN genes were more frequently observed in M-GBM (M-GBM 36% vs S-GBM 19%, p = 0.05). Also, the percentage of patients harboring the aforementioned mutation concomitantly with TERTp mutations was higher in M-GBM (M-GBM 25% vs S-GBM 18%, p = 0.43). Analysis of TCGA data revealed a co-occurrence of EGFR/PTEN alterations in 81/378 (21.4%) GBM IDH-WT cases. This was similar to the S-GBM group (19%) but lower than M-GBM patients in our study [19, 20, 22]. The co-occurrence of EGFR, PTEN, and TERTp also appeared to be higher in M-GBM (25%) compared to prior reports (5.6%) [21]. Also, we evaluated the co-occurrence of EGFR/KDR alterations in the entire cohort (n = 224) and M-GBM (n = 31), in which there was not an association between these genes (p = 0.34 and 0.12, respectively). These results were further validated by the lack of association in the TCGA database between EGFR/KDR alterations (p = 0.49) and EGFR amplification and KDR alterations (p = 0.17) [19, 20, 22].
Prognosis and survival
Univariable analysis of GBM IDH-WT patients (n = 224) demonstrated that patients with age < 55 years, KPS 80 or higher, and 1st line Stupp protocol had improved OS. However, only age and 1st line Stupp protocol were independent predictors of poor survival after multivariable analysis. Importantly, we observed a trend towards shorter survival (13.0 vs 17.9 months, p = 0.31) in M-GBM patients (Fig. 3a, Online Resource 3).
Overall survival in GBM IDH-WT. a Overall survival of GBM IDH-WT (n = 224) by multifocality, in which there was not statistically significant difference between the survival of M-GBM (n = 31) and S-GBM (n = 193, 13.0 vs 17.9 months, p = 0.31). b Overall survival within M-GBM (n = 31). There was not statistically significant difference between multifocal (n = 10) and multicentric (n = 21) patients (27.7 vs 12.5 months, p = 0.07). c Overall survival of M-GBM (n = 31) by KDR gene status, in which KDR mutant patients (n = 4) had worse survival compared to KDR WT patients (n = 27, 6.7 vs 16.6 months, p = 0.038). d Overall survival of M-GBM by EGFR gene status, in which there was not statistically significant difference between EGFR altered (n = 20) and EGFR WT (n = 10) patients (12.5 vs 16.5 months, p = 0.54). GBM glioblastoma, M-GBM multifocal/multicentric GBM, S-GBM single foci GBM, WT wild type. Kaplan–Meier curves were examined by the log-rank test, a p ≤ 0.05 was considered as statistically significant
No statistically significant differences were observed between multifocal and multicentric GBM. However, multifocal GBM showed a trend towards better outcomes (Fig. 3b). In addition, M-GBM patients with KDR alterations had a worse survival than KDR-WT patients in univariable analysis (6.7 vs 16.6 months, p = 0.038) and multivariable analysis (HR 9.3 [1.17–73.8], p = 0.035, Fig. 3c; Table 2). The effect of KDR alterations in the outcome of S-GBM was observed in the univariable log-rank test, in which KDR altered S-GBM had a worse survival than their KDR-WT counterparts (11.4 vs 18.5 months, p = 0.015). However, this difference was not observed after multivariable analysis (KDR altered HR 1.78 [0.74–4.31], p = 0.20). Survival differences in M-GBM patients were not observed between EGFR-altered (12.5 months) and EGFR-WT (16.5 months) tumors (Fig. 3d; Table 2).
Discussion
Several studies on M-GBM have reported decreased survival when compared to S-GBM; however, studies focusing on the molecular characteristics of M-GBM are scarce. It is still unclear whether M-GBM represents a distinct biologic variant of GBM or if multifocal progression is part of the natural history of the disease [7, 8]. The current study represents, the largest M-GBM cohort with comprehensive genomic characterization, in which subtle molecular differences were observed. Our data suggest that M-GBM genetically resembles typical GBM.
In the present cohort, we identified that 9.9% of GBM IDH-WT had multiple lesions at diagnosis, which is comparable to recent studies that have used similar criteria to define M-GBM [5, 7, 8]. Interestingly, we identified 2 cases (2.9%) of multifocal astrocytomas IDH-MT. A recent study reported that multifocality is an independent predictor for IDH1-WT status, as only 1/102 (1%) patients had an IDH1 mutation [26]. The multifocal tumor, in this case, was an oligodendroglioma, IDH-mutant and 1p/19q co-deleted. A prior study by Liu et al. [8] reported all multifocal patients to be IDH-WT. In our study, both patients (32 and 33) were astrocytomas, IDH-mutant (1p/19q intact). Thus, to the best of our knowledge, these cases represent the first reports of multifocal IDH-MT astrocytomas (Figs. 1g, h, 2).
Previous studies have described the differential expression of MAPK, PTEN, MGMT, and EGFR between S-GBM and M-GBM; however, these studies are limited by their small sample size and lack of comprehensive molecular characterization [5, 7, 27]. A recent study on 18M-GBM patients with mutation analysis identified the absence of IDH1, ATRX, or PDGFRA mutations in all patients [8]. These results contrast with our findings in which in addition to the observed IDH1 p.R132H mutation in two patients, we also observed mutations in ATRX and PDGFRA amplification in 2.9% and 15.2% of patients, respectively. ATRX mutations are widely distributed across the gene and are mostly truncating (including frame shift and nonsense variants) as seen in the current study (patient 32 with an ATRX N111fs*16 and patient 33 with an ATRX E1541*) [28]. ATRX mutations occur in 78% and 63% of GBM, IDH-MT and astrocytoma IDH-MT, respectively, and only in 3% of GBM IDH-WT [29]. Moreover, a Japanese study identified loss of ATRX expression in 28.6% of 14 multicentric astrocytomas, IDH-WT [27]. Thus, it is not surprising that ATRX mutations were not observed in previous smaller cohorts. PDGFRA discrepancies between our study and previous studies could be also explained by sample size, as we observed that PDGFRA is amplified in 15.2% of M-IG and 12.9% of M-GBM. This represents roughly the same known frequency of PDGFRA amplifications in GBM, as 13.5% of S-GBM and 14.7% of GBMs in TCGA harbored PDGFRA amplification [19]. Moreover, PDGFRA amplification has been reported in 16.7% of a small M-GBM study [30].
EGFR alterations are found in 30–50% of all GBMs and are associated with tumor development and progression [19, 31]. Therefore, EGFR has been proposed as a potential therapeutic target for GBMs; however, clinical trials targeting this gene have been unsuccessful [32, 33]. A previous study with six M-GBM found aberrations in EGFR in all patients [30]. In our study, we also identified that EGFR alterations were more common in M-GBM than in S-GBM (65% vs 42%, p = 0.019).
Importantly, EGFR alterations have been associated with an increased infiltrative and invasive phenotype [30, 34], and therefore with a multifocal appearance as seen in our study. Notably, we observed that co-occurrence of EGFR and PTEN alterations, as well as, EGFR, PTEN, and TERTp alterations, was increased in M-GBM compared to both S-GBMs in our cohort and published literature [19,20,21,22]. These findings are in accordance with the results of a previous study that described 67% of M-GBM showed alterations in EGFR, PTEN, and TERTp [30]. Therefore M-GBM genetically resembles typical GBM with an increased incidence of EGFR mutations and co-occurrence of EGFR, PTEN, and TERTp alterations.
A previous study has also identified a higher expression of CYB5R2 in M-GBM, which has been independently associated with worse OS and has been proposed as a potential prognostic and diagnostic marker [8]. Unfortunately, we did not examine the expression of this gene in our study.
Overall survival and multifocality
Multifocality has been consistently correlated with poor survival in GBM [4,5,6,7,8, 35, 36]. In our study, M-GBM trended towards a worse survival compared to S-GBM; however, it did not reach statistical significance. This could be explained due to the limited sample size, the relatively small percentage of biopsies performed in these cases (12.2%) compared to prior reports (28–100%) [5,6,7, 35,36,37,38], and the more aggressive adjuvant upfront approach demonstrated by increased utilization of chemotherapeutic agents in these patients (bevacizumab and irinotecan) compared to S-GBM (Online Resource 4). Prior centers have also described an aggressive approach for these cases, suggesting multiple craniotomies for maximal cytoreduction that could potentially improve survival [9]. Interestingly, we identified for the first time that alterations in KDR predicted worse survival within the M-GBM group. Even though our results should be taken with caution and validated in a larger cohort, KDR, also known as VEGFR2, is a tyrosine kinase receptor that plays an important role in GBM angiogenesis, aggressiveness, and progression [39,40,41,42]. Previous studies have demonstrated that the KDR CAGT haplotype increases GBM aggressiveness and that concomitant high mRNA expression of KDR, FLT1, and VEGFA has been associated with shorter survival [39, 40]. Bevacizumab, a monoclonal antibody towards VEGF, is one of the few drugs currently approved for GBM treatment. A recent study has demonstrated that EGFR-altered patients with recurrent GBM, have a significantly shorter time to progression when treated with bevacizumab [43]. Moreover, KDR activation through paracrine secretion of VEGF-C has been demonstrated to represent an escape mechanism employed by GBM to counteract bevacizumab therapy, which could potentially explain the worse survival seen in KDR-altered patients [44]. While KDR alterations have been demonstrated to predict bevacizumab response in other malignancies such as colon cancer [45], it is still unknown if alterations in KDR and its related genes FLT1 and VEGFA could predict response to bevacizumab in GBM.
Multifocal vs multicentric GBM
Finally, we investigated the differences between multifocal and multicentric GBM. While no demographic differences were observed, our study revealed that multicentric GBM was less likely to undergo re-resection, salvage therapies, and GTR. Prior studies have also reported worse outcomes in multicentric GBM compared to multifocal GBM (3 vs. 11 months) [6], which is in concordance with our results. Interestingly, various genetic differences were observed in the univariate analysis between these groups demonstrating differences in CDKN2A/B, CDK4, PTEN, and TP53 genes that have not been previously described and may warrant further study (Table 1).
Despite several limitations, such as the retrospective nature, limited sample size, and lack of sequencing information from distinct tumor foci, the present study represents the largest cohort of multifocal/multicentric gliomas undergoing comprehensive genetic characterization and demonstrates the unique molecular features of this aggressive type of GBM.
Conclusions
The results of the present study demonstrate that M-GBM genetically resembles S-GBM, however, M-GBM have a higher frequency of EGFR alterations and co-occurrence of EGFR/PTEN alterations, which may account for their highly malignant and invasive phenotype.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
References
Ostrom QT, Gittleman H, Truitt G et al (2018) CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-oncology 20:iv1–iv86. https://doi.org/10.1093/neuonc/noy131
Zhu P, Du XL, Zhu J-J, Esquenazi Y (2019) Improved survival of glioblastoma patients treated at academic and high-volume facilities: a hospital-based study from the National Cancer Database. J Neurosurg. https://doi.org/10.3171/2018.10.JNS182247
Ferlay J, Colombet M, Soerjomataram I et al (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144:1941–1953. https://doi.org/10.1002/ijc.31937
Showalter TN, Andrel J, Andrews DW et al (2007) Multifocal glioblastoma multiforme: prognostic factors and patterns of progression. Int J Radiat Oncol Biol Phys 69:820–824. https://doi.org/10.1016/j.ijrobp.2007.03.045
Patil CG, Yi A, Elramsisy A et al (2012) Prognosis of patients with multifocal glioblastoma: a case–control study. J Neurosurg 117:705–711. https://doi.org/10.3171/2012.7.JNS12147
Thomas RP, Xu LW, Lober RM et al (2013) The incidence and significance of multiple lesions in glioblastoma. J Neurooncol 112:91–97. https://doi.org/10.1007/s11060-012-1030-1
Paulsson AK, Holmes JA, Peiffer AM et al (2014) Comparison of clinical outcomes and genomic characteristics of single focus and multifocal glioblastoma. J Neurooncol 119:429–435. https://doi.org/10.1007/s11060-014-1515-1
Liu Q, Liu Y, Li W et al (2015) Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta Neuropathol 130:587–597. https://doi.org/10.1007/s00401-015-1470-8
Hassaneen W, Levine NB, Suki D et al (2011) Multiple craniotomies in the management of multifocal and multicentric glioblastoma. J Neurosurg 114:576–584. https://doi.org/10.3171/2010.6.JNS091326
Yan H, Parsons DW, Jin G et al (2009) Mutations in gliomas. N Engl J Med 360:765–773. https://doi.org/10.1056/NEJMoa0808710
Brennan CW, Verhaak RGW, McKenna A et al (2013) The somatic genomic landscape of glioblastoma. Cell 155:462. https://doi.org/10.1016/j.cell.2013.09.034
Brat DJ, Aldape K, Colman H et al (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol 136:805–810. https://doi.org/10.1007/s00401-018-1913-0
Harris PA, Taylor R, Thielke R et al (2009) Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–381. https://doi.org/10.1016/j.jbi.2008.08.010
Harris PA, Taylor R, Minor BL et al (2019) The REDCap Consortium: building an international community of software platform partners. J Biomed Inform 95:103208. https://doi.org/10.1016/j.jbi.2019.103208
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Zorofchian S, El-Achi H, Yan Y, et al (2018) Characterization of genomic alterations in primary central nervous system lymphomas. J Neurooncol 140:509–517. https://doi.org/10.1007/s11060-018-2990-6
Frampton GM, Fichtenholtz A, Otto GA et al (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023–1031. https://doi.org/10.1038/nbt.2696
Schwaederle M, Krishnamurthy N, Daniels GA et al (2018) Telomerase reverse transcriptase promoter alterations across cancer types as detected by next-generation sequencing: a clinical and molecular analysis of 423 patients. Cancer 124:1288–1296. https://doi.org/10.1002/cncr.31175
McLendon R, Friedman A, Bigner D et al (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068. https://doi.org/10.1038/nature07385
Gao J, Aksoy BA, Dogrusoz U et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:1–20. https://doi.org/10.1126/scisignal.2004088
Nonoguchi N, Ohta T, Eun J (2013) TERT promoter mutations in primary and secondary glioblastomas. 931–937. https://doi.org/10.1007/s00401-013-1163-0
Cerami E, Gao J, Dogrusoz U et al (2012) The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. https://doi.org/10.1158/2159-8290.CD-12-0095
Thakkar JP, Dolecek TA, Horbinski C et al (2014) Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomark Prev 23:1985–1996. https://doi.org/10.1158/1055-9965.EPI-14-0275
Kanda Y (2013) Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant 48:452–458. https://doi.org/10.1038/bmt.2012.244
Stupp R, Mason WP, van den Bent MJ et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. https://doi.org/10.1056/NEJMoa043330
Park YW, Han K, Ahn SS et al (2018) Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. Am J Neuroradiol 39:37–42. https://doi.org/10.3174/ajnr.A5421
Karlowee V, Amatya VJ, Hirano H et al (2017) Multicentric glioma develops via a mutant IDH1-independent pathway: immunohistochemical study of multicentric glioma. Pathobiology 84:99–107. https://doi.org/10.1159/000447951
Heaphy CM, De Wilde RF, Jiao Y et al (2011) Altered telomeres in tumors with ATRX and DAXX mutations. Science (80-) 333:425. https://doi.org/10.1126/science.1207313
Pekmezci M, Rice T, Molinaro AM et al (2017) Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol 133:1001–1016. https://doi.org/10.1007/s00401-017-1690-1
Abou-El-Ardat K, Seifert M, Becker K et al (2017) Comprehensive molecular characterization of multifocal glioblastoma proves its monoclonal origin and reveals novel insights into clonal evolution and heterogeneity of glioblastomas. Neuro-oncology 19:546–557. https://doi.org/10.1093/neuonc/now231
Parsons DW, Jones S, Zhang X et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science (80-) 321:1807–1812. https://doi.org/10.1126/science.1164382
Lassman AB, Van Den Bent MJ, Gan HK et al (2019) Safety and efficacy of depatuxizumab mafodotin + temozolomide in patients with EGFR-amplified, recurrent glioblastoma: results from an international phase I multicenter trial. Neuro-oncology 21:106–114. https://doi.org/10.1093/neuonc/noy091
Lassman AB, Pugh SL, Wang TJC, Aldape K, Gan HK, Preusser M, Vogelbaum MA, Sulman E, Won M, Zhang P, Moazami G, Macsai MS, Gilbert MR, Bain E, Blot V, Ansell PJ, Samanta S, Kundu MG, Seidel C, de Vos FY, Hsu S, Cardona AF, Lombardi G, Bentsion D, Peterson R, Gedye C, Lebrun-Frenay C, Wick A, Curran WJ, Mehta M (2019) Epidermal Growth Factor Receptor (EGFR) amplified (amp) newly diagnosed glioblastoma (nGBM). In: Paper presented at the annual meeting of Society of Neuro-Oncology, Phoenix, AZ
Talasila KM, Soentgerath A, Euskirchen P, et al (2013) EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. 683–698. https://doi.org/10.1007/s00401-013-1101-1
Syed M, Liermann J, Verma V et al (2018) Survival and recurrence patterns of multifocal glioblastoma after radiation therapy. Cancer Manag Res 10:4229–4235. https://doi.org/10.2147/CMAR.S165956
Pérez-Beteta J, Molina-García D, Villena M et al (2019) Morphologic features on MR imaging classify multifocal glioblastomas in different prognostic groups. Am J Neuroradiol 40:634–640. https://doi.org/10.3174/ajnr.A6019
Singh G, Mehrotra A, Das K et al (2015) Multiple glioblastomas: are they different from their solitary counterparts? Asian J Neurosurg 10:266. https://doi.org/10.4103/1793-5482.162685
Burger MC, Breuer S, Cieplik HC et al (2017) Bevacizumab for patients with recurrent multifocal glioblastomas. Int J Mol Sci 18:1–11. https://doi.org/10.3390/ijms18112469
Vasconcelos VCA, Lourenço GJ, Brito ABC et al (2019) Associations of VEGFA and KDR single-nucleotide polymorphisms and increased risk and aggressiveness of high-grade gliomas. Tumor Biol 41:1–10. https://doi.org/10.1177/1010428319872092
Zhang SD, Leung KL, McCrudden CM, Kwok HF (2015) The prognostic significance of combining VEGFA, FLT1 and KDR mRNA expressions in brain tumors. J Cancer 6:812–818. https://doi.org/10.7150/jca.11975
Sjöström S, Wibom C, Andersson U et al (2011) Genetic variations in VEGF and VEGFR2 and glioblastoma outcome. J Neurooncol 104:523–527. https://doi.org/10.1007/s11060-010-0504-2
Wu HB, Yang S, Weng HY et al (2017) Autophagy-induced KDR/VEGFR-2 activation promotes the formation of vasculogenic mimicry by glioma stem cells. Autophagy 13:1528–1542. https://doi.org/10.1080/15548627.2017.1336277
Hovinga KE, McCrea HJ, Brennan C et al (2019) EGFR amplification and classical subtype are associated with a poor response to bevacizumab in recurrent glioblastoma. J Neurooncol 142:337–345. https://doi.org/10.1007/s11060-019-03102-5
Michaelsen SR, Staberg M, Pedersen H et al (2018) VEGF-C sustains VEGFR2 activation under bevacizumab therapy and promotes glioblastoma maintenance. Neuro-oncology 20:1462–1474. https://doi.org/10.1093/neuonc/noy103
Zhang SD, McCrudden CM, Meng C et al (2015) The significance of combining VEGFA, FLT1, and KDR expressions in colon cancer patient prognosis and predicting response to bevacizumab. Oncotargets Ther 8:835–843. https://doi.org/10.2147/OTT.S80518
Acknowledgements
None.
Funding
No funding to disclose.
Author information
Authors and Affiliations
Contributions
Study design: AD, LYB, and YE. Data recollection: AD, EW, and AR Data analysis: AD and VLR. Manuscript writing: AD, EW, and YE. Manuscript revision and editing: AD, NT, LYB, and YE. Study supervision: LYB and YE. Approved final manuscript: all authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared no conflict of interest.
Ethical approval
This retrospective study was approved by the Institutional Review Board of The University of Texas Health Science Center at Houston and Memorial Hermann Hospital, Houston, TX following the 1964 Helsinki declaration and its later amendments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dono, A., Wang, E., Lopez-Rivera, V. et al. Molecular characteristics and clinical features of multifocal glioblastoma. J Neurooncol 148, 389–397 (2020). https://doi.org/10.1007/s11060-020-03539-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-020-03539-z