Abstract
The commonly used automatic segmentation algorithms often have over-segmentation and error-segmentation problems when segmenting conglutinated bone fragments in computed tomography (CT) images. And the end face details of the fragments are lost in this process. At present, bone fragments are often manually segmented by doctors. This increases the workload of doctors and takes low efficiency and poor accuracy in bone segmentation. We propose an automatic bone fragment segmentation algorithm based on morphology to segment the conglutinated and separated bone fragments and reconstruct them in 2D and 3D spaces. The pixel classification and clustering method based on morphology are proposed in in the algorithm to identify the pixels in the fracture images. And a concave points detection method is proposed to segment the conglutination area of the fractured bone. This algorithm and two other commonly used algorithms are used in the experiment to segment the three typical fracture images. The segmentation results are quantitatively analyzed to demonstrate the advantages of the morphology algorithm. The morphology algorithm performs well in the metrics of Accuracy (.93 ± .07), Sensitivity (.92 ± .08), Dice coefficient (.92 ± .08) and Mean intersection over union (MIoU) (.92 ± .07). The segmentation results of this algorithm are closest to the manual segmentation results of doctors compared to the other two algorithms. We have demonstrated that the algorithm proposed in this paper has good segmentation accuracy for separated and conglutinated bone fragments. This algorithm significantly improves the segmentation accuracy and efficiency of bone fragments under complex fracture conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Recently, our team is developing a robot surgery system for the reduction of lower limb fractures. The fracture CT images segmentation and reconstruction are the foundation for preoperative planning in robotic surgery systems [1,2,3]. At present, bone fragment segmentation in CT images is usually segmented manually by doctors. This increases the workload of doctors and results in low efficiency of segmentation and reconstruction. The segmentation accuracy is also affected by subjective factors of doctors [4, 5]. Therefore, incorporating bone fragments segmentation and reconstruction algorithms into the surgical system to guide surgeons in selecting the optimal parameters to automatically complete image processing tasks is an effective method to solve these problems.
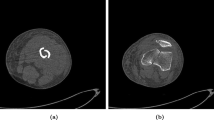
Using computer algorithms to segment bones has always been a research hotspot in the medical image processing field. Most of the fracture image segmentation algorithms currently studied focus on complete bone segmentation [6,7,8]. However, the position and posture of bone fragments demonstrate specificity and randomness under the muscle traction after fracture [9]. If the bone fragments are separated from each other, we could segment the bone fragments using these algorithms [10, 11]. If the bone fragments are staggered and conglutinated with each other, because the density of bone fragments is almost the same, these bone fragments in the CT images are mostly integrated, as shown in Fig. 1. Which poses a significant challenge to the segmentation algorithms.
At present, there are few studies on the segmentation of conglutinated bone fragments. For example, Liu et al. [12] introduced a system for assessing the degree of injury in tibia comminuted fractures. The system can segment the conglutinated bone fragments at the tibia joints and calculate the surface area of the bone fragments to determine the degree of fracture. Zhang et al. [13] proposed a bone fragments segmentation algorithm. The algorithm divides the bone fragments from the conglutination region based on erosion and expansion operations and improves computing efficiency through GPU acceleration technology. Shadid and Willis [14, 15] proposed an improved watershed algorithm. The algorithm segments cortical bone and cancellous bone from surrounding tissues by setting the prior pixel value threshold. The remaining bone pixels are classified into corresponding fragments according to the probability function. This algorithm can separate conglutinated bone fragments from most fracture images. However, for some special fracture states, such as the case where cortical bones of bone fragments conglutinate together, the segmentation results may be incorrect.
Through communication with orthopedic experts and literature review, we can find that orthopedic doctors often combine clinical experience with the shape of bone fragments in fracture images to determine different areas of bone fragment [16, 17]. This is similar to the target segmentation process of image recognition technology in the engineering field [18]. In this process, the pixels are labeled to segment targets in images by morphological methods: pixel classification and clustering [19, 20]. In addition, concave point detection is often used to segment conglutinated targets in many fields, such as biological field [21], mining field [22], and industrial manufacturing field [23]. In the medical field, this method is used to segment cells and organs [24, 25]. Therefore, pixels classification and clustering method as well as concave point detection method are the feasible approach to segment conglutinated bone fragments in fracture images.
Based on the study above, this paper proposes an automatic segmentation algorithm for bone fragments segmentation in lower limb fracture cases. In order to facilitate the system operation of fracture image processing by doctors, the algorithm guides users to obtain the optimal image processing parameters through several simple steps, thereby achieving automatic segmentation and reconstruction of preoperative images of patients. Therefore, this algorithm enables clinical doctors without image processing technology and computer programming foundation to complete complex image processing tasks.
2 Methods
The morphological segmentation algorithm proposed in this paper achieves the extraction of bone fragment targets by filtering and classifying pixel points in CT images. This overcomes the drawback of traditional segmentation algorithms relying solely on pixel values for object recognition. To achieve this goal, the algorithm processes the pixels in the image in four steps.
In the first step, the pixels in original images are classified into different point sets based on their pixel values and positions. In the second step, the bone fragments boundary is formed by detecting and connecting concave points in the bone fragments conglutination area. In the third step, the pixels within the boundary are divided into proximal bone fragment pixels and distal bone fragment pixels through the region filling. In the fourth step, the pixel points on the boundary are collected into the proximal and distal bone fragments through pixel clustering, and the bone fragments segmentation task is completed. The implementation process of this algorithm is shown in Fig. 2.
Several user-defined parameters are required in the four steps: (1) Bone pixel prior threshold Tbone, (2) Secondary boundary point classification threshold TS-boundary, (3) Concave point detection radius Rconcave, (4) Concave point detection threshold Tconcave, (5) Concave point filtering radius RS-concave.
2.1 Pixels classification
In this step, the pixels in the original CT image are classified into boundary point set (B), background point set (BG), secondary boundary point set (S) and interior point set (I) by the algorithm based on threshold Tbone and TS-boundary. This step includes three stages. Firstly, the bone pixels in the original CT image are filtered out based on the threshold Tbone. Then, the filtered bone pixels are classified into different pixels based on their pixel value and position. Finally, secondary boundary points are filtered out based on the threshold TS-boundary.
In the first stage, the value of Tbone is estimated by the prior distribution of bone pixel value. The prior distribution of human bone and non-bone tissue as shown in Fig. 3. Bone pixels have higher pixel values than non-bone tissue in CT images. Thus, bone pixels can be filtered out from non-bone tissue by setting an appropriate threshold value. If the value of the pixel is greater than Tbone, it is classified as a bone pixel. If the value of the pixel is less than Tbone, it is classified as a background pixel and the value of the pixel is set to 0.
In the second stage, the 8-neighborhood connection value is calculated to determine the position of the bone pixel as shown in Eq. (1). And classify the pixel into the following categories based on its connection value.
where \({N}_{c}^{\left(8\right)}\left(x\right)\) is the 8-neighborhood connection value of the x-position pixel, k is the number of pixels around the x-position pixel in 8-neighborhood, M = [0, 2, 4, 6], \(\overline{I }\left(x\right)=1-I\left(x\right)\).when k + 2 = 8, x8 = x0.
-
(1) Isolated point: If the connection value of a pixel \({N}_{c}^{\left(8\right)}\left(x\right)=0\) and pixels’ value in 8-neighborhood are all 0, the pixel should be called isolated point. Its pixel value is set to 0.
-
(2) Interior point (I): If the connection value of a pixel \({N}_{c}^{\left(8\right)}\left(x\right)=0\) and pixels’ value in 8-neighborhood are all 1, the pixel is classified as the interior point.
-
(3) Boundary point (B): If the connection value of a pixel \(1\le {N}_{c}^{\left(8\right)}\left(x\right)\le 4\), the pixel is classified as the boundary point.
In the third stage, secondary boundary points are filtered out from the point set I according to TS-boundary. The number of interior points in 8-neighborhood around the pixel is used to determine whether the pixel is a second boundary point. If the number of boundary points in the 8-neighborhood is greater than or equal to TS-boundary the point should be called second boundary point and classified into the point set S.
After this step, non-bone tissue pixels in the original CT image are removed, and bone pixels are classified into boundary points, secondary boundary points, and interior points. And the boundary lines formed by the boundary point set and the secondary boundary point set can segment bone fragments that are separated or slightly conglutinated (The width of the conglutination area is only four pixels) to each other. But for bone fragments with severe conglutination, the next steps of concave point detection and connection are required.
2.2 Concave point detection and connection
For CT images with severe bone fragment conglutination, only identifying the boundary points and secondary boundary points of the bone fragment is not sufficient to form the bone fragment boundary. Moreover, repeated boundary detection is not conducive to the recognition and segmentation of slender and small bone fragments. Through communication with orthopedic doctors, they judge the boundary of bone fragments through experience, and the boundary of bone fragments is often on the connecting line of concave points. Therefore, this algorithm is used to search for concave points on bone boundaries through concave point detection. And the concave point connection is used to connect two concave points to form the bone fragment boundary. This step includes three stages. Firstly, the concave points on the bone boundary (point set S) are detected according to the search radius Rconcave and threshold Tconcave. Then, correct concave points are filtered out based on the search radius RS-concave. Finally, the correct concave points are connected to form bone fragment boundaries.
Compared with the other points in the point set S, the number of background pixels in the area centered on the concave point is much smaller than that of other points, as shown in Fig. 4. Therefore, in the first stage, the number of background points is counted in the area with a radius of Rconcave and centered on the point in the point set S. If the number of background points is less than Tconcave, the pixel will be classified into the concave point set (C). On account of the broken edge of the bone fragments, there are much interference concave points are detected, as shown in Fig. 5. These interference points will hinder the correct concave points connection. Thus, these points should be filtered out.
The fracture image after concave point detection. The white points are concave points, the concave points marked by black circles are correct concave points, and the rest are interference concave points. Due to the unsmooth boundary of the bone fragment, some of the detected concave points are not on the boundary of the bone fragment conglutination area
In the second stage, any other concave points are detected in the area with radius RS-concave to determine whether the center point is a correct concave point. If there is, the point is correct concave point and retained in the point set C; If there is not, the point is classified to the point set S. Due to the different fracture situations of different patients, the value of RS-concave needs to be adjusted according to the specific circumstances.
In the third stage, the 4-path connecting the concave points are searched in the point set I by the 4-neighborhood search method, the pathfinding method as shown in Fig. 6. This method includes 8 search directions. Due to the long program, the figure only shows the starting point located at the bottom left of the target point as an example. The pixels on the path are classified into point set S.
After this step, boundaries have been formed between the conglutinated bone areas in the original CT image. But currently, the bone areas in the image are all interior points. These interior points need to be classified into proximal bone fragment and distal bone fragment.
2.3 Closed area filling
Through the above two steps, the bone area in the CT image can be divided into two enclosed areas. This step uses flood filling method to classify the pixels in the two enclosed areas into proximal fragment point set (P) and distal fragment point set (D) based on the proximal and distal bone fragment seed points.
This step starts from the seed point and searches for interior points in the 6-neighborhood around the pixel, as shown in Fig. 7, and classifies the searched interior points into point set P or point set D. Then use these points as the starting points to continue the search operation, until all the interior points in the images are classified into point set P or point set D.
After this step, the bone area in the original CT image has been preliminarily divided into proximal and distal bone fragments. Only the pixels in the boundary point set and secondary boundary point set in the image have not been classified.
2.4 Boundary points and secondary boundary points clustering
After the above three steps, the pixels in the image are classified into boundary point set, secondary boundary point set, proximal bone fragment point set and distal bone fragment point set. This step classifies the points in the boundary point set and secondary boundary point set into the proximal bone fragment point set and the distal bone fragment point set by calculating the distance between the points in the proximal and distal bone fragments separately.
In this step, the pixels in the point set B and point set S are classified according to the minimum Euclidean distance between the pixels and the bone fragment areas, as shown in Fig. 8. The calculation rule is shown in Eq. (2). If the minimum Euclidean distance between the pixel and the proximal bone fragment area is less than that of the distal bone fragment area, the pixel is classified into point set P. Conversely, the pixel is classified into point set D. After this step, the bone pixels in the original CT image are divided into proximal and distal bone fragments.
where L (x, P) is the minimum Euclidean distance between the pixel x and the proximal bone fragment area, L (x, D) is the minimum Euclidean distance between the pixel x and the distal bone fragment area. The calculation of the two is shown in Eqs. (3) and (4).
where xpj and ypj are the coordinate of pixel in the point set P, xdj and ydj are the coordinate of pixel in the point set D.
An example of how to classify the pixels in point set B and S. Calculate the distance between the point and all pixels in the area, and take the minimum value to represent the minimum distance between the point and the area. Determine the classification of the point by comparing its minimum distance from two areas
3 Results
The clinical datasets used in the experiment are provided by the Department of Traumatology and Orthopedics, Affiliated Hospital of Shandong University of Traditional Chinese Medicine. CT data was obtained using a GE Revolution CT scanner. Each volume dataset consisted of 657–809 axial slices, with a thickness of 0.5–0.625mm and a scanning interval of 0.625mm. Each case image included images of fractured and unfractured limbs. The dataset used in the experiment included three sets of fracture cases, which were from a 33-year-old male with tibial fractures, a 33-year-old male with fibular fractures, and a 66-year-old female with femoral fractures. These fracture cases are quite representative. In dataset 1, the fractured ends of adult male fibula fractures are relatively fragmented, and the small cross-sectional area of the fibula poses a great challenge for segmentation of fractured bone targets. In dataset 2, the tibia of adult males is relatively thick, and after a fracture, there is a different degree of conglutination between the two ends of the fractured bone, which can represent a typical adult male long bone fracture. In dataset 3, adult female femur fractures occurred near the trochanter. Due to the thin bone mass at the trochanter and the patient's age, the fractured bone image at the fracture site is very slender, with only two pixels at the thinnest area, which is very challenging for segmentation of fractured bone targets.
C + + program is designed for segmentation algorithm implementation and model visualization based on the Insight Segmentation and Registration Toolkit (ITK) and the Visualization Toolkit (VTK). The ‘ground truth’ data set is set to compare and evaluate the accuracy of segmentation results generated by segmentation algorithm and this dataset is segmented manually by orthopedic experts. As two commonly used segmentation algorithms, Otsu threshold segmentation algorithm and watershed segmentation algorithm are used as a comparison of morphology segmentation algorithms in experiments [26, 27]. The segmentation parameters of different segmentation algorithms for three datasets are shown in Table 1. The prior bone threshold Tbone is set to segment the bone areas according to the study reported by Inacio and Kranioti et al. [28, 29].
The segmentation results generated by different algorithms are shown in Figs. 9, 10 and 11. It is shown in these figures that the segmentation results generated by morphological algorithm are most similar to ‘ground truth’. In the segmentation results generated by the watershed algorithm, over-segmentation appears at the edge of the bone fragments. And in the segmentation result of the fibula image, error-segmentation appears at the distal bone fragment. In the segmentation results generated by the Otsu threshold algorithm, serious over-segmentation appears at the bone fragments. And in the segmentation result of the fibula image, error-segmentation appears at the proximal bone fragment.
The segmentation results of fibula fracture dataset using different algorithms. Taking the 426th, 442nd and 455th slice as examples. The segmentation results of morphological algorithms are closest to ground truth. The segmentation results of other two algorithms have corrosion and incorrect segmentation problems
The segmentation results of tibia fracture dataset using different algorithms. Taking the 247th, 256nd and 266th slice as examples. The segmentation results of morphological algorithms are closest to ground truth. The segmentation results of watershed algorithm have incorrect segmentation problem, and the segmentation results of threshold algorithm have severe corrosion problems in the cancellous bone area
The segmentation results of femur fracture dataset using different algorithms. Taking the 304th, 319nd and 354th slice as examples. The segmentation results of morphological algorithms are closest to ground truth. The segmentation results of watershed algorithm have incorrect segmentation and corrosion problems. The segmentation results of threshold algorithm have more serious corrosion problems, even resulting in bone area loss
Several evaluation metrics are used to evaluate the segmentation effect of different algorithms. The metrics include the Hausdorff Distance (HD) [30], Region Rank metric (RR) [15], and four evaluation metrics based on the error matrix: Accuracy, Sensitivity (Recall) Dice coefficient (F1 coefficient), MIoU and Precision [31]. The values of these evaluation metrics for different algorithms are shown in Tables 2, 3 and 4. By comparing the evaluation metric scores, the same conclusion can be drawn as above. The segmentation results generated by the morphological algorithm are closest to ‘ground truth’.
Figure 12 shows the relative error distribution of three CT image segmentation results using different segmentation algorithms. When segmenting the conglutination area and the two ends area of the fractured bone, the Otsu threshold algorithm and the watershed algorithm obviously have higher segmentation error. However, the morphological algorithm has less error in these two segmentation cases.
The relative error distribution of segmentation results for three CT datasets using different segmentation algorithms. Where (a) is data set 1, (b) is data set 2, (c) is data set 3. The accuracy of the three segmentation algorithms for images with fractured bone sections has decreased when segmenting bone fragments in the fracture area, especially in the bone fragments conglutination area. The difference between the segmentation results of morphological algorithms and ground truth is minimal, especially at the ends of bone fragment, the segmentation results of the other two algorithms show significant deviations
Figure 13 shows the reconstruction models of the segmentation results of three fracture image datasets by different segmentation algorithms. Through the 3D models, the advantages of morphological algorithm in bone segmentation can be further demonstrated compared with the other two algorithms. The models segmented by morphological algorithm have smooth surface and no bone pixels loss. However, the models segmented by the watershed algorithm have many surface defects and serious bone pixels loss in the joint head areas. And the models segmented by the threshold algorithm have serious bone pixels loss. This causes the bone fragment models to become transparent and even the joint head areas are lost.
3D reconstruction models of segmentation results of three CT datasets using different segmentation algorithms. Where (a) is the tibia data set, (b) is the fibula data set, (c) is the femur data set. The model segmented by morphological algorithms is smooth and complete. The watershed algorithm model shows partial bone pixels loss at the joint head and shaft. The threshold algorithm model has severe bone pixels loss
In addition, the morphological algorithm segmentation results and ‘ground truth’ are reconstructed in 3D space to compare the segmentation results of the algorithm comprehensively. Figures 14, 15 and 16 show the reconstruction model of the algorithm segmentation results and ‘ground truth’ in different directions. The bone fragments models generated based on the morphological algorithm segmentation results are displayed on top of the corresponding bone fragments in ‘ground truth’, respectively shown in Figure c and Figure d. And the RR metric values and HD metric values of the morphological algorithm segmentation are shown in Table 5. Comparing the values of HD metric, it can be found that the algorithm has the highest segmentation accuracy for tibia fracture images, followed by fibula, and femur is the worst. The same conclusion can be drawn by comparing the models of three fracture datasets. The segmentation result of the tibia fracture dataset is almost the same as ‘ground truth’. In the fibula fracture dataset, a little over-segmentation appears at the edge of the distal bone fragment. In the femur fracture dataset, some over-segmentation appears at the end of the proximal bone fragment. These problems will be discussed in detail in the next section.
3D reconstruction models based on the segmentation result of fibula dataset. Each model is observed from the sagittal, coronal and cross-sectional directions. (a) displays the morphological segmentation algorithm result. (b) displays the ‘ground truth’. (c) displays the proximal bone fragment. (d) displays the distal bone fragment
3D reconstruction models based on the segmentation result of tibia. Each model is observed from the sagittal, coronal and cross-sectional directions. (a) displays the morphological segmentation algorithm result. (b) displays the ‘ground truth’. (c) displays the proximal bone fragment. (d) displays the distal bone fragment. The segmentation results of the morphological algorithm are almost consistent with the ground truth
3D reconstruction models based on the segmentation result of femur fracture images. Each model is observed from the sagittal, coronal and cross-sectional directions. (a) displays the morphological segmentation algorithm result. (b) displays the ‘ground truth’. (c) displays the proximal bone fragment. (d) displays the distal bone fragment. The segmentation results of morphological algorithms are mostly consistent with ground truth. But there are slight corrosion problems at the end of the bone fragments
4 Discussion
The segmentation experimental results demonstrate that the morphological algorithm proposed in this paper has higher accuracy in segmenting fracture datasets compared to two traditional algorithms. The Otsu threshold segmentation algorithm has the lowest accuracy among the three automatic segmentation algorithms. This is because the gray value of the bone conglutination area is close to the gray value of the bone fragments. In order to segment the bone fragments from the conglutination area, the threshold must be increased to filter out the area.This causes areas with low trabecular bone displacement density such as cancellous bone and joint head to be filtered out. As a result, the segmentation result has a serious over-segmentation problem. Therefore, the threshold algorithm has better segmentation accuracy for fracture images with completely separated bone fragments. However, for the case images of bone congratulation or osteoporosis, the segmentation accuracy will be greatly reduced.
The watershed algorithm also segments bone fragment areas based on the gray value of pixels. The algorithm is prone to segmentation errors when segmenting targets with small difference in pixel values. In the experiment of this paper, the watershed algorithm has a wrong segmentation of the conglutination area when the conglutinated bone fragments are segmented. In order to segment the conglutinated bone fragments, it is necessary to increase the filtering threshold of the algorithm. Similar to the threshold algorithm, this results in areas with lower bone density being filtered out. Therefore, this algorithm has high segmentation accuracy for targets with significant differences in pixel values between the target boundary pixels and interior pixels. However, this algorithm is prone to error-segmentation and over-segmentation problems when segmenting fracture images with conglutinated bone fragments.
However, in the morphological algorithm, the identity of the pixel is only related to its own location and its relative position with the surrounding pixels. All bone pixels in the dataset, regardless of their gray values, can be classified and clustered. In addition, the concave point detection method is used to segment the conglutination areas between bone fragments. Avoiding error-segmentation problems caused by the same gray value of the bone fragments conglutination area as the bone area, as shown in Fig. 17. Therefore, this algorithm will not encounter the problems of error-segmentation and over-segmentation like threshold algorithms and watershed algorithms.
Segmentation results calculated using and not using the concave point detection method. (a) is the 278th slice of the original tibia fracture CT image. (b) is segmentation result without concave points detection. (c) is segmentation result with concave points detection. (d) and (e) are the corresponding 3D models. The use of concave point detection method can accurately generate the boundaries of conglutinated bone fragments, which ensures the accuracy of 3D reconstruction
By comparing the 3D models, it can also be found that the morphological algorithm has high segmentation accuracy. The segmentation result of the algorithm for tibia fracture images is almost consistent with ‘ground truth’. However, some over-segmentation appears in the segmentation results of fibula fracture images and femur fracture images.
This is because that the cross section of fibula is small. In the fibula fracture case, the end face of bone fragments is broken after fracture. There are several small bone fragment areas in the slice image. These areas only have boundary points and second boundary points after pixel classification step and cannot be filling into proximal and distal bone fragments in the closed area filling step. Therefore, there are little over-segmentation problems in the segmentation results. In the femur fracture case, the fracture is located at the distal part of the femur near the ankle. The bone in this part is thinner than that in the shaft. In addition, bone loss in elderly patient makes the bone fragments in this part thinner than the general bone. The bone area presents a slender shape in the slice image. Some boundary points and second boundary points are clustered into the wrong bone fragment in the pixel clustering step. Therefore, there are some over-segmentation and error-segmentation problems in the segmentation results.
Through analyzing the problems mentioned above, it can be found that the morphological algorithm is prone to problems when segmenting small fragments and slender fragments. But these problems can be solved by optimizing algorithm parameters. For example, for the over-segmentation problem when segmenting small bone fragments, this problem can be solved by optimizing the parameters in the concave point detection stage to avoid mistakenly segmenting the bone fragment area into many small closed areas. For the error-segmentation problem when segmenting slender bone fragments, this problem can be solved by adding seed points in areas where segmentation problems appeared.
Therefore, for the complex fracture images segmentation, the values of the parameters in the morphological algorithm should be optimized and adjusted according to the segmentation result. And the segmentation accuracy of the algorithm can be improved by optimizing parameters. Take the femur fracture images used in the segmentation experiment as an example. In the segmentation results before optimizing the algorithm parameters, some over-segmentation and error-segmentation problems appear at the end of the proximal bone fragment, as shown in Fig. 18 (a). After optimizing the parameters by adding seed points in areas where segmentation problems appeared, the over-segmentation and error-segmentation problems in the segmentation results are well resolved, as shown in Fig. 18 (b).
Comparison of segmentation results of proximal fibular bone fragment before and after optimization of algorithm parameters. (a) displays the segmentation result before optimizing parameters, where the green area is the ‘ground truth’ mask. (b) displays the segmentation result after optimizing the parameters. By optimizing algorithm parameters and adding seed points, the corrosion problem of the algorithm can be effectively solved
From this perspective, there is uncertainty in the algorithm when segmenting small and slender bone fragments. The main problem is that the algorithm needs to classify the boundary points and secondary boundary points of the bone fragments in bone fragment segmentation to form the boundary of the proximal and distal bone fragments. But for small and slender bone fragments, the area of their ends in the image is very small, and after calculation, there are no interior points in this area. Therefore, during the third step of calculation, these areas were not filled. As a result, when calculating the distance between pixels and two bone fragments in the fourth step, the pixels that should belong to the proximal bone fragment are more distant from the proximal bone fragment than from the distal bone fragment. The same problem also occurs on the pixels of the distal fractured bone fragment. This resulted in incorrect classification of pixels in the boundary point set and secondary boundary point set. Thus, when segmenting small and slender bone fragments, seed points are added to the end image slices of the bone fragments. This can increase the number of interior points in the area where segmentation errors occur during the third step of the algorithm. At the same time, considering canceling the algorithm's secondary point boundary point detection step when processing similar images can also increase the number of interior points at the ends of small and slender bone fragments. This can avoid errors when calculating the distance in the fourth step and reduce segmentation errors.
5 Conclusion
A novel automatic bone fragment segmentation algorithm based on morphology is proposed in this paper to segment the lower limb fracture CT images. In this algorithm, pixels classification and clustering methods are designed to segment the bone fragments that are separated from each other and slightly conglutinated to each other. In addition, the concave point detection method is designed to segment the bone fragments with the conglutination area. These methods improved the adaptability of the algorithm for segmenting complex fracture images. This method determines the boundaries of different bone fragments through their morphology, improving the traditional object segmentation algorithm's approach of simply segmenting objects based on their pixel values. This avoids the shortcomings of traditional segmentation algorithms in fracture image segmentation and provides new ideas and research directions for clinical image target segmentation. Through the description in this paper, the parameters in the algorithm are easy to understand and control in the process of image segmentation, which makes the algorithm can be intuitively applied to orthopedic clinical images segmentation.
In the segmentation experiment, the three segmentation algorithms are used to segment the typical fracture CT datasets. The segmentation results are presented and quantitatively evaluated in 2D and 3D spaces. Through comparative analysis of segmentation experiment results, it is demonstrated that the segmentation algorithm proposed in this paper has more advantages than commonly used segmentation algorithms. The experimental results show that the algorithm can not only correctly segment the bone fragments but also have good segmentation accuracy in conglutinated bone fragments segmentation.
However, the algorithm also exposes some problems in the segmentation experiment. In the future, we will optimize the algorithm to improve the segmentation accuracy of the algorithm for small and slender targets. Through communication with clinicians, we learned that orthopedic surgeons often judge the boundary of bone fragments through context and clinical experience. The machine learning method is also based on the existing segmented datasets to accumulate experience, and then complete the segmentation task. And machine learning algorithms have been widely used in target segmentation of medical images. [32,33,34] Therefore, we will study machine learning fracture image segmentation algorithms and use doctors' manual segmentation of images as training samples to improve the segmentation accuracy of the algorithm.
Data availability
Data will be made available on reasonable request.
References
Zhao JX, Li C, Ren H, Hao M, Zhang LC, Tang PF (2020) Evolution and current applications of robot-assisted fracture reduction: a comprehensive review. Ann Biomed Eng 48:203–224. https://doi.org/10.1007/s10439-019-02332-y
Dauwe J, Mys K, Putzeys G, Schader JF, Richards RG, Gueorguiev B et al (2020) Advanced CT visualization improves the accuracy of orthopaedic trauma surgeons and residents in classifying proximal humeral fractures: a feasibility study. Eur J Trauma Emerg Surg 48:4523–4529. https://doi.org/10.1007/s00068-020-01457-3
Pan MZ, Liao XL, Li Z, Deng YW, Chen Y, Bian GB (2023) Semi-Supervised Medical Image Segmentation Guided by Bi-Directional Constrained Dual-Task Consistency. Bioengineering 10(2):225. https://doi.org/10.3390/bioengineering10020225
Arabi H, Zaidi H (2017) Comparison of atlas-based techniques for whole-body bone segmentation. Med Image Anal 36:98–112. https://doi.org/10.1016/j.media.2016.11.003
Klein A, Warszawski J, Hillengaß J, Maier-Hein KH (2019) Automatic bone segmentation in whole-body CT images. Int J Comput Assisted Radiol Surg 14:21–29. https://doi.org/10.1007/s11548-018-1883-7
Rehman F, Ali SI, Riaz MN, Gilani SO (2020) A region-based deep level set formulation for vertebral bone segmentation of osteoporotic fractures. J Digital Imaging 33:191–203. https://doi.org/10.1007/s10278-019-00216-0
Van Eijnatten M, van Dijk R, Dobbe J, Streekstra G, Koivisto J, Wolff J (2018) CT image segmentation methods for bone used in medical additive manufacturing. Med Eng Phys 51:6–16. https://doi.org/10.1016/j.medengphy.2017.10.008
Wang M, Yao J, Zhang G, Guan B, Wang X, Zhang Y (2021) ParallelNet: Multiple backbone network for detection tasks on thigh bone fracture. Multimed Syst 27:1091–1100. https://doi.org/10.1007/s00530-021-00783-9
Xu H, Lei J, Hu L, Zhang L (2022) Constraint of musculoskeletal tissue and path planning of robot-assisted fracture reduction with collision avoidance. Int J Med Robot 18(2):e2361. https://doi.org/10.1002/rcs.2361
Paulano F, Jiménez JJ, Pulido R (2014) 3D segmentation and labeling of fractured bone from CT images. Vis Comput 30:939–948. https://doi.org/10.1007/s00371-014-0963-0
Ruikar DD, Santosh KC, Hegadi RS (2019) Automated fractured bone segmentation and labeling from CT images. J Med Syst 43:1–13. https://doi.org/10.1007/s10916-019-1176-x
Liu P, Hewitt N, Shadid W, Willis A (2021) A system for 3D reconstruction of comminuted tibial plafond bone fractures. Comput Med Imaging Graphics 89:101884. https://doi.org/10.1016/j.compmedimag.2021.101884
Zhang Y, Tong R, Song D, Yan X, Lin L, Wu J (2020) Joined fragment segmentation for fractured bones using GPU-accelerated shape-preserving erosion and dilation. Med Biol Eng Comput 58:155–170. https://doi.org/10.1007/s11517-019-02074-y
Shadid W, Willis A (2013) Bone fragment segmentation from 3D CT imagery using the Probabilistic Watershed Transform. In: 2013 Conf Proc IEEE SOUTHEASTCON, IEEE. 1–8
Shadid WG, Willis A (2018) Bone fragment segmentation from 3D CT imagery. Comput Med Imaging Graphics 66:14–27. https://doi.org/10.1016/j.compmedimag.2018.02.001
Mys K, Visscher L, van Knegsel KP, Gehweiler D, Pastor T, Bashardoust A et al (2023) Statistical Morphology and Fragment Mapping of Complex Proximal Humeral Fractures. Medicina 59(2):370. https://doi.org/10.3390/medicina59020370
Rakesh Y, Akilandeswari A (2022) Bone Fracture Detection Using Morphological and Comparing the Accuracy with Genetic Algorithm. J Pharm Negat Results 13(4): 270–276. https://doi.org/10.47750/pnr.2022.13.S03.030
Cai L, Gao J, Zhao D (2020) A review of the application of deep learning in medical image classification and segmentation. Ann Transl Med 8(11): 713. https://doi.org/10.21037/atm.2020.02.44
Shakeel PM, Burhanuddin MA, Desa MI (2019) Lung cancer detection from CT image using improved profuse clustering and deep learning instantaneously trained neural networks. Measurement 145:702–712. https://doi.org/10.1016/j.measurement.2019.05.027
Xie X, Zhang W, Wang H, Li L, Feng Z, Wang Z et al (2021) Dynamic adaptive residual network for liver CT image segmentation. Comput Electr Eng 91:107024. https://doi.org/10.1016/j.compeleceng.2021.107024
Chen Z, Fan W, Luo Z, Guo B (2022) Soybean seed counting and broken seed recognition based on image sequence of falling seeds. Comput Electr Agric 196:106870. https://doi.org/10.1016/j.compag.2022.106870
He L, Wang S, Guo Y, Cheng G, Hu K, Zhao Y et al (2022) Multi-scale coal and gangue dual-energy X-ray image concave point detection and segmentation algorithm. Measurement 196:111041. https://doi.org/10.1016/j.measurement.2022.111041
Wang Z, Wang E, Zhu Y (2020) Image segmentation evaluation: a survey of methods. Artif Intell Rev 53:5637–5674. https://doi.org/10.1007/s10462-020-09830-9
Zhang W, Li H (2017) Automated segmentation of overlapped nuclei using concave point detection and segment grouping. Pattern recognit 71:349–360. https://doi.org/10.1016/j.patcog.2017.06.021
Ranjbarzadeh R, Saadi SB (2020) Automated liver and tumor segmentation based on concave and convex points using fuzzy c-means and mean shift clustering. Measurement 150:107086. https://doi.org/10.1016/j.measurement.2019.107086
Li Y, Song S, Sun Y, Bao N, Yang B, Xu L (2022) Segmentation and volume quantification of epicardial adipose tissue in computed tomography images. Med Phys 49(10):6477–6490. https://doi.org/10.1002/mp.15965
Zhang Z, Li Y, Shin BS (2022) C2-GAN: Content-consistent generative adversarial networks for unsupervised domain adaptation in medical image segmentation. Med Phys 49(10):6491–6504. https://doi.org/10.1002/mp.15944
Inacio JV, Malige A, Schroeder JT, Nwachuku CO, Dailey HL (2019) Mechanical characterization of bone quality in distal femur fractures using pre-operative computed tomography scans. Clin Biomech 67:20–26. https://doi.org/10.1016/j.clinbiomech.2019.04.014
Kranioti EF, Bonicelli A, García-Donas JG (2019) Bone-mineral density: clinical significance, methods of quantification and forensic applications. Res Rep Forensic Med Sci 9:9–21. https://doi.org/10.2147/RRFMS.S164933
Aydin OU, Taha AA, Hilbert A, Khalil AA, Galinovic I, Fiebach JB, Frey D, Madai VI (2021) On the usage of average Hausdorff distance for segmentation performance assessment: hidden error when used for ranking. Eur Radiol Exp 5:1–7. https://doi.org/10.1186/s41747-020-00200-2
Wang J, Li Z, Chen Q, Ding K, Zhu T, Ni C (2021) Detection and Classification of Defective Hard Candies Based on Image Processing and Convolutional Neural Networks. Electronics 10(16):2017. https://doi.org/10.3390/electronics10162017
Hassan E, Mahmoud Y et al (2023) The effect of choosing optimizer algorithms to improve computer vision tasks: a comparative study. MULTIMED TOOLS APPL 82:16591–16633. https://doi.org/10.1007/s11042-022-13820-0
Hassan E, Mahmoud Y et al (2023) COVID-19 diagnosis-based deep learning approaches for COVIDx dataset: A preliminary survey. Artificial Intell Disease Diag Prognosis Smart Healthcare 16:107
Hassan E, Elmougy S, Ibraheem MR, AlMutib HMS, K, Ghoneim A, AlQahtani SA, Talaat FM, (2023) Enhanced Deep Learning Model for Classification of Retinal Optical Coherence Tomography Images. Sensors 23:5393. https://doi.org/10.3390/s23125393
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 52075301), the National Key R&D Program of China (No. 2019YFC0119200) and the Key R&D Program of Shandong Province (No. 2019GHZ002).
We thank all the team members for their support for this study.
Funding
Key Technologies Research and Development Program,2019YFC0119200,Qinhe Zhang,National Natural Science Foundation of China,52075301,Qinhe Zhang,Key Technology Research and Development Program of Shandong,2019GHZ002,Qinhe Zhang
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that there are no financial interests or personal relationships conflicts that influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Miao, G., Zheng, X., Han, Y. et al. An automatic segmentation algorithm for conglutinated bone fragments in 3D CT images of lower limb fractures based on morphology. Multimed Tools Appl 83, 67001–67022 (2024). https://doi.org/10.1007/s11042-023-18060-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-18060-4