Abstract
Skin cancer is a major public health concern and the most common type of cancer among the other types. Reliable automated classification systems will provide clinicians with great help to detect malignant skin lesions as quickly as possible. Recently, deep learning-based approaches have efficiently outperformed other conventional machine learning models in medical image classification tasks. In this study, a novel computer-aided approach is designed for Skin Lesion Detection by creating an Ensemble of Deep (SLDED) models. More specifically, we initially performed a modified faster R-CNN using VGGNet feature extractor on ISIC archive database, including 4668 skin lesion images for lesion localization, and we obtained a mean average precision (mAP) of 0.96. Then we fused four different convolutional neural networks (CNNs) into one framework to obtain high classification accuracy. Moreover, a weighted majority voting method is proposed to aggregate the final decision of each individual voter. We evaluate our experimental classification results on 934 and 200 images from ISIC and PH2 test data. We achieved the average accuracy of 97.1% and 96%, Area under receiver operating characteristics curve (AUC) of 98.6% and 98.1%, precision of 87.1% and 90.2%, recall of 86.7% and 85.4% for ISIC and PH2 test data, respectively. As another objective evaluation, we have tested our proposed procedure on official test set of 2016 and 2017 International Symposium on Biomedical Imaging (ISIB) challenges. It outperforms the results of other proposed frameworks that have been published in those challenges. The results demonstrate that our proposed SLDED method is a meaningful approach to classify four different skin lesions with a high accuracy despite the lack of access to expensive computational equipment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Skin cancer is one of the main public health problems that each year detects about 123 million new cases worldwide [29]. Melanoma is the deadliest type among various skin cancers, with a dramatic increasing incidence rate [61]. In 2019, approximately 96,480 people with melanoma were expected in the United States, and one estimated in five people being diagnosed with skin cancer [17, 59].
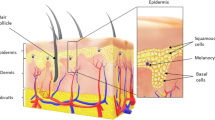
The human skin is a structured tissue, including the epidermis, dermis, and hypoderm. The epidermis has melanocytes that can produce melanin, and under certain conditions, such as ultraviolet radiation, it generates melanin at an extremely abnormal rate [29]. Malignant tumors are caused by an atypical growth of melanocyte, which is known as melanoma. Melanoma begins in melanocytes to make a pigment as melanin, but despite other skin cancers, it spreads rapidly among other tissues (metastasize) [58].
The annual cost of skin cancer treatment in the USA is estimated at $ 8.1 billion, and it is still rising. In order to enhance diagnostic accuracy and diminish health care expenses, there is a high amount of stimulus in developing the diagnosis of skin cancer, especially for melanoma [47]. A common diagnosis examination of a skin cancer diagnosis is a Biopsy method, an invasive and unpleasant procedure. It has a great deal of time for the patient as well as a physician [37]. Without extra technological support, dermatologists have a 65–80% accuracy in melanoma detection [3].
A non-invasive method that can aid in diagnosing skin cancer by providing high-resolution skin images is Dermoscopy. It is a physical checkup method based on light radiation and immersion in oil, providing a beneficial solution for visual examination of the underlying skin structure [36]. Dermoscopy provides an excellent opportunity for dermatologists to collect magnified images with a high resolution. This technology improves the visual quality of the collected data drastically [52].
In 1992, the potential advantages of employing digital imaging to detect skin-related diseases were pointed out [64]. In the work of Moss et al., an expert system has been proposed, which has been based on analysis of the texture features extracted from Fourier transform [55]. Chang et al. have proposed a pipeline including pre-processing the images, extracting 91 features describing the tissue shape, color, and texture; and finally using a Support Vector Machine (SVM) to classify the images [18]. Some studies [1, 4, 15, 35] have classified skin cancer images according to ABCD rules. The features have described the asymmetry (A), border(B), color(C), and differential structure(D). They have been computed total dermoscopy score (TDS) from A, B, C, and D features and classified thembased on their TDS. All of the mentioned studies in this paragraph are based on extracting hand-crafted features from a skin lesion. Additionally, some other studies [16, 65] have extracted other traditional hand-crafted features from the images in order to classify the skin lesions. However, the discriminative power of hand-crafted features is low, and they are computationally intensive.
The first step in skin lesion images’ classification is lesion localization and segmentation of the image [54]. Due to a wide variety of lesions, detecting and segmenting them is still a challenging issue, and therefore many studies have been conducted in this area [24, 27, 57, 62]. In computer aided diagnosis (CAD) system, a better quality can be achieved if the classification task is done on some areas of the images, including the lesions, known as a region of interest (ROI). This is due to the extraction of a set of features that can be a great indicator of the lesion [7, 14]. In deep learning methods to prevent feature maps’ saturation, preprocessing of images is prior to the classification task [5]. In a study by Badrinarayan et al., they used a SegNet autoencoder-based approach to preprocessing their images [6]. In another study by Bi et al., a fully convolutional network (FCN) method was used for lesion detection [10]. Also, Attia et al. proposed a combination of CNNs and recurrent neural networks (RNNs) for lesion detection and segmentation [5].
Classification based on computer vision pipelines and feature engineering is very complicated and time-consuming. It requires specialized knowledge to choose and design the most appropriate feature extraction methods. Moreover, the development of these models should be robust against the diversity of lesions as well as intra-classes variations and inter-class similarities [68]. The previously proposed automated image processing techniques for skin cancer diagnosis achieved a high classification owing to the employment of recent emerging deep learning models [11,12,13, 57].
Deep learning comprises a collection of machine learning algorithms called deep neural networks (DNNs), which had enormous achievements in the processing of real data, such as image, text, as well as sound in the past decade [41]. In 2012, Krizhevsky proposed a convolution neural network (CNN) to make a significant leap in the accuracy of image detection tasks [40]. The overall success and excellence of CNNs have been shown in a wide range of computer vision tasks [9]. In recent years, CNN architectures, such as GoogleNet [66], ResNet [30], ResNeXt [69] VGGNet [56], which are the most popular pre-trained models, have been proposed for the classification of natural images.
After building and deploying GPU cards with high computing power at affordable prices in recent years, several methods based on CNNs have been eased for the processing of skin cancer images [24, 29, 32, 57, 68, 70]. Some of the most recent studies, which have applied CNNs to classify skin lesions are shown in Table 1. In the study by [44], they have used pre-trained VGGNet architecture and transfer learning paradigm to classify skin lesion images. One of the limitations to the transfer learning technique is that they can achieve good performance when a target problem’s data content is similar to the pre-trained model’s trained data. To the best of our knowledge, the existing pre-trained models do not include sufficient skin lesion images. They can be trained from scratch to address this issue while it is time-consuming and suffers from high computational volume.
According to the results of studies mentioned above, deep models, if train with enough data, can show better accuracy and can aid the dermatologists in decision making with higher confidence and accuracy, such as the study by Esteva et al. [24]. However, one of the main limitations of deep learning methods in medical imaging is the lack of sufficient training data required to provide the model with high accuracy, especially in images with soft tissues. Therefore, regarding the small number of available medical images, an individual CNN possibly cannot extract all the discriminative features to obtain a high classification accuracy.
In this work to tackle the mentioned problems, we propose a novel computer-assisted approach by creating an ensemble of four different CNNs. Our main contributions lie in four-folds, including:
The main contributions of this study lie in four-folds, including:
-
Designing a novel ensemble-based method (SLDED) by inspiring from the most popular pre-trained architectures which were especially used in skin cancer detection.
-
Proposing a new VGG-based faster R-CNN approach and using Inception-ResNet in region proposal network (RPN) for skin lesion segmentation.
-
Improving the classification results by designing a new weighted majority voting approach to aggregate each individual CNN’s vote.
-
Introducing a deep-based approach for classification of skin lesions with the applicability of training in a short time, achieving a high accuracy.
This paper is organized as follows. In Section 2, a description of the dataset as well as the main steps of our proposed SLDED method for skin image classification, are presented. Experimental results are illustrated in Section 3. Section 4 discusses the main findings. Finally, concluding remarks are shown in Section 5.
2 Materials and methods
In this study, we aim to classify skin images of four different skin lesions by building an ensemble of deep neural networks. The class labels are Basal cell carcinoma (BCC), malignant melanoma (MM), nevus lesions (NV), and Seborrheic keratosis (SK).
BCC is the most commonly diagnosed skin cancer worldwide Fig. 1a This lesion type is a typical non-aggressive cancer, and its corresponding tumors grow slowly with rarely metastasize (metastatic rate < 0.1%) [63]. MM lesions have an uppermost mortality rate compared to other skin disorders Fig. 1b Given the aggressive growth of invasive MM lesions, its early diagnosis is a critical issue [51]. The nevus lesions Fig. 1c refers to occurring several conditions such as neoplasm and hyperplasia in melanocytes [26]. Moreover, the lesions labeled as SK class Fig. 1d have the highest occurrence rate among benign skin lesions, affecting almost 83 million Americans [8].
Figure 2a describes the main steps of our proposed SLDED method. After data collection and augmentation prior to the classification task, the lesions were segmented using the proposed VGG-based faster R-CNN model, shown in Fig. 2b Afterward, the segmented lesions were fed to each module of the SLDED method as input data to do feature map extraction, illustrated in Fig. 2c Finally, by performing a weighted majority voting approach on each module’s predicted probabilities, which were obtained from each module’s fully connected layer, a final decision has been made to classify the lesion types, presented in Fig. 2d.
More details about the proposed SLDED method are described in the following subsections.
2.1 Dataset
The images analyzed in this study are collected from the International Skin Imaging Collaboration (ISIC) Archive[20], which has been gathered from different melanoma detection challenges in recent years. The total number of these images is 4668, and the number of images labeled as BCC, MM, nevus, and SK are 583, 2131, 1535, 419, respectively. To train and evaluate our proposed SLDED method, the data by a ratio of 8:2 has been randomly divided into training and test sets. Therefore, the total number of training and test images are 3734 and 934 images, respectively. Additionally, 10% of the training data as a validation set is considered to provide the model’s training process unbiased. The number of images per class for training and test datasets is illustrated in the second and the last columns of Table 2 respectively.
Moreover, for the sake of completeness evaluation, another dermoscopic image dataset (PH2) with a total number of 200 skin lesion images, including 160 Nevus and 40 melanoma images are used as another test set.
2.2 Data preprocessing
The steps of preprocessing data are described in the following subsections.
2.2.1 Data augmentation
Data augmentation is a strategy to increase the training data volume significantly. It can be helpful to prevent deep models from being overfitted [42, 60]. Additionally, the data augmentation approach is advantageous for CNNs to lead them to be more able in the extraction of general features, especially when the data set is imbalanced [40]. Since the amount of data in our study is insufficient to train the proposed SLDED approach, the data augmentation is applied using different methods. The original images have been rotated with \({45}^{^\circ }\) to \({45}^{^\circ }\) (\({45}^{^\circ }\), \({90}^{^\circ }\), \({135}^{^\circ }\), \({180}^{^\circ }\), \({210}^{^\circ }\)) and flipped horizontally and vertically. The second column of Table 2 illustrates the training dataset’s size after augmenting per class.
2.2.2 Lesion detection and localization
Region-based CNN (R-CNN) was first introduced by Girshick et al. [31]. R-CNN performs object detection in two stages. In the first step, it generates independent object proposals using a selective search method [67]. Then in the second step, after wrapping each object proposal into constant sizes, the features are extracted to be used by a classifier and a regressor for object detection. Despite the high accuracy of R-CNN, it also has a high computational volume, so then, fast R-CNN was introduced to solve this problem. In fast R-CNN, instead of convolving each wrapped area, only the whole image is convolved. As a result, a fixed feature vector is extracted for each object proposal. Moreover, the Region of Interest (ROI) enables fast R-CNN to use some pre-trained models as well.
R-CNN and fast R-CNN use hand-crafted models, such as selective search, to generate object proposals. These hand-crafted methods are time-consuming, and they suffer from high computational volume. To tackle those problems and achieve a greater accuracy, faster R-CNN model is introduced, including two parts. This model is robust against the noises and performs well applying on benchmark dataset. The first part includes an RPN that do the task of generating objects and the second part is a fast R-CNN to refine the proposals. Faster R-CNN has the ability to share the convolutional layers between RPN and fast R-CNN. As a result, it is sufficient for the image to be passed through the convolutional layer only once. Therefore, faster R-CNN can generate proposal objects and also refine them more quickly. This enables us to use very deep learning networks, such as ResNet 50 and VGGNet 19, to achieve high accuracy in object detection tasks. In the entire faster R-CNN system, the input of fast R-CNN is completely dependent on the output of RPN, and these two modules must share their convolutional layers with each other. As a result, in the optimization phase, the optimizer in fast R-CNN must consider ROI according to the coordinates of predicted proposals of RPN.
After extracting the features from VGGNet, two steps have to be taken in order to form bounding boxes. Initially, 9 anchor boxes with different sizes were generated on 3⋅3 non-overlapped patches of each image’s feature map. Then the RPN model consisting of an Inception-ResNet module, with 6 convolutional layers with different kernel sizes was designed to predict the coordinates and the probability of the mentioned anchor boxes labeling them as a lesion or normal area. This was done by labeling the anchor boxes based on the intersection over union (IOU) threshold of 0.5. In the second step, as shown in Fig. 3 each feature map of the proposed fixed-size regions is given as fast R-CNN input; this was done by ROI pooling.
In this work, our lesion detection faster R-CNN method is based on a pre-trained model, VGGNet 19. For the training process, we were first trained the VGGNet with our images, and its weights were fine-tuned in order to learn the specific features of these images. The VGGNet weights were kept constant in the next step, and the RPN and fast R-CNN weights were fine-tuned. Then we ran the model with 100 iterations using Adam optimizer [39].
The SOFTMAX activation function in the last layer of R-CNN can predict any image area, either a normal skin region or a lesion. Then, the greedy suppression algorithm was used to generate the bounding box around the lesion.
2.3 Classification by an ensemble of different CNNs
2.3.1 Convolution neural network
CNN is a subset of deep learning methods that has fascinated much attention in recent years and has been used in image recognition, such as analyzing skin medical images [38]. In general, the main parts of CNNs are convolutional layers and subsampling parts that extract the features’ hierarchy from input images. These layers are usually followed by fully connected layers (dense layers) and a SOFTMAX classifier. Therefore, CNNs are used for the classification of images. The CNN architecture mainly encompasses (1) Convolutional layers, (2) Pooling layers, as well as one or multiple (3) Fully connected layers [25, 34]:
-
(1)
Convolutional layer: The essential capability of deep learning, notably for image recognition, is due to its convolutional layers. These layers convolve the whole image using various kernels and generate different feature maps [25]. These layers take an input volume of size \({W}_{1}\)×\({H}_{1}\)×\({D}_{1}\) and produce output volume of size \({W}_{2}\)×\({H}_{2}\)×\({D}_{2}\) based on the following Eq. (1) by adjusting the parameters K, E, S, and P:
$$\begin{array}{*{20}c}W_2:{(W}_1+2P-F)/S+1\\ H_2:(H_1+2P-F)/S+1\\ \kern -7pc D_2:K\end{array}$$(1) -
(2)
Pooling layer: Pooling operation is used to reduce the dimensions of the output neurons from the convolutional layer, reducing the required computational time and memory and preventing the overfitting of the model.
-
(3)
Fully connected layer: A fully connected layer that uses the convolutional layer’s output to predict an image’s class.
Activity functions, which are used to get output from neurons, play an essential role in training deep neural networks. Nowadays, one of the most frequently and most successful activation functions is the Rectified Linear Unit (ReLU) [28], which has fast convergence ability and prevents from exploding and vanishing gradient problem. In this work, two activation functions are used: (1) Rectified linear unit and (2) SOFTMAX for the last layer.
-
(1)
Rectified linear unit: After all convolution layers as well as fully connected layers, the RELU activation function is used. Equation (2) shows this function.
$$f\left(x\right)=\left\{\begin{array}{*{20}c}0, \; for \; x \; < \; 0 \\ x, \; for \; x \geq 0\end{array}\right.$$(2)Where f (x) is zero if x is less than zero, and f (x) is equal to x when x is greater than or equals to zero.
-
(2)
SOFTMAX: The SOFTMAX function is a more generalized logistic activation function used for multi-class classification problems. The SOFTMAX function calculates the probability distribution for the k output classes. Therefore, the last layer (the third layer of fully connected) employs this function to predict the class label of the input images. Equation (3) shows this function mathematically.
$$\sigma{\left(x\right)}_i=\frac{e^{x_i}}{\sum_{j=1}^ke^{x_j}},\;for\;i=1,\;\dots,\;k\;and\;x=\left(x_1\;\dots\;x_k\right)\in\mathbb{R}^k$$(3)Where x is a vector of the inputs to the output layer, and i is the index of the output neurons. Output values of \(\sigma {\left(x\right)}_{i}\) lie between 0 and 1, and their sum equals 1.
2.3.2 The architecture of the proposed SLDED method
The existing models that are pre-trained on a large natural ImageNet dataset have a considerable performance in a classification task. But to use them in areas that are not well trained, we require to train them from scratch. However, due to their very deep architecture training them from scratch is a very time consuming and expensive task. Consequently, on our own task, inspiring pre-trained models, including VGGNet, GoogleNet, ResNet, and ResNeXt we developed some modules that are a light sample of the original networks. Each of these networks has different advantages with various architectures; therefore, combining them provides our model with extracting more varied features. In the following, we will describe how we used them in this work.
-
(a)
GoogleNet module
It is first introduced by Szegedi et al. in 2015. Their proposed V1 and V2 Inception models include 9 inception modules. The following year, they presented Inception-V4 and Inception-ResNet with more changes compared to their previous versions. Instead of adding up more deep layers, the GoogleNet is based on employing different kernel sizes in each layer. This is because that the large kernels lead the model to identify global features and the smaller ones resulting in extracting local features. In our first network, which is inspired by Google Net, we use 5 modules with kernel sizes of 1 ⋅ 1, 3 ⋅ 3, and 5 ⋅ 5 for convolutional layers as well as 3 ⋅ 3 for the pulling layer. For simplicity, we call it CNN1.
-
(b)
VGGNet module
In a study by Simonyan et al., they proposed a deep CNN architecture called VGG. Different versions of VGG model, including VGG16 and VGG19 differ only in total number of layers with 16 and 19 convolutional layers, respectively. The main attribute of this network is using fixed-size kernels, and the idea behind this is to reduce the number of parameters and to improve the network training time. Accordingly, kernels with different sizes can be replaced by several convolutional layers with the same kernel size in one block. In this work, our VGG-based network called CNN2 includes 14 convolutional layers with kernel size of 3⋅3, 6 max-pooling layers with kernel size of 2⋅2 and stride two.
-
(c)
ResNet module
One of the problems with CNNs is vanishing of gradients. It is because that the CNNs lack the ability to identify identical and straightforward feature maps, especially when the training iteration number is large. He et al. introduced the ResNet architecture to address the mentioned problem by presenting a shortcut connection between the next layers’ input and output. As a result, the model can also be trained on an input with simpler feature maps. ResNet has various versions that are named based on the total number of layers. In our ResNet-based network called CNN3 we employed 24 convolutional layers with a 3 ⋅ 3 filter size and stride 1 or 2 without max pulling layer. Also, we put a shortcut connection between every two or three layers.
-
(d)
ResNeXt module
ResNeXt is a simple, highly modularized architecture for image classification tasks, first introduced by Xie et al. and ranked first in the ILSVRC classification competition task 2017. ResNeXt’s architecture includes aggregated residual transformation blocks (ARTB). It achieved better results training on ImageNet dataset compared to its ResNet counterpart. Using ARTBs, we designed a network called CNN4 that is a light sample of ResNeXt model. Each path in ARTBs includes three convolutional layers with kernel sizes of 1⋅1, 3⋅3, and 1⋅1.
Figure 4 depicts the architecture of our proposed SLDED method. Each network’s parameters, including depth, kernel size, stride, dimensions, and cardinality (for CNN4), are illustrated.
Fig. 4 Architecture of the SLDED method. The networks from left to right represents GoogleNet (CNN1), VGGNet (CNN2), ResNet (CNN3), and ResNeXt (CNN4) modules, respectively. The numbers insides the boxes from left to the right denote the number of filters, kernel size, and stride, respectively. i.e., 3 × 3 × 2 means the convolutional layer with the number of 3 filters, kernel size of 3, and stride 2. C = 32 means the cardinality of 32
2.3.3 Weighted majority voting of CNNs
A better decision can be made when the information is derived from several experts. Aggregating the multiple opinions by a decision-maker can improve prediction accuracy [43]. In this study, we elaborate on an automated approach employing an ensemble of four different CNNs to achieve considerable accuracy in our image classification task.
In the first step, each of the proposed CNNs classifies the skin lesions. To do so, in each CNN, we employed two fully connected layers with sizes of 1024 and 4, respectively. Additionally, soft max activation function is used in the last layer in order to calculate the predicted probabilities. The maximum probability of classes calculated in Eq. (4) is considered as an image’s label and its assigned vote associating to that individual CNN.
Where \({c}_{i}\), \({p}_{i}\), and \({s}_{x}\) represents class i, the value of predicted probability for class i, and output of the SOFTMAX function, respectively.
Secondly, a weighted majority voting method illustrated in Eq. (5) is applied to lead the model to make the final decision for every input image. According to Eq. (6) if \({CNN}_{j}\) allocates label i to the input image x, the vote of \( {CNN}_j \) equals to 1 for that label and 0 for other classes.
Where \({p}_{ij}\) is the probability that \({CNN}_{j}\) assigns label i to x. \(V \left({p}_{ij}\right)\) represents the vote that \({CNN}_{j}\) assigns label i to x and \({w}_{j}\) is the weight of \({CNN}_{j}\)’s vote
Since the number of aggregated votes for two classes may be the same, we determine a weight for each network’s vote. To do this, we first determined an initial weight for the individual models according to their calculated mean AUC score, then optimized the weights using genetic (GA) algorithm [21].
2.3.4 Training the SLDED method
Each member CNN of the weighted voting ensemble model are trained end-to-end. In training, categorical cross-entropy was used as a loss function for calculating the weights of the voters. Moreover, an RMSProp [50] optimizer with a learning rate of 0.01 was used to minimize the loss. The data were fed to the CNNs in a batch of 16 images through 300 training epochs per network. The training has been performed in a computer equipped with NVIDIA GeForce GTX 1070 SLI, CPU @ 2.6 GHz, with 20 cores. The implementations were performed in Python using the Keras library.
2.4 Evaluation metrics
To evaluate the accuracy of faster R-CNN, creating bounding boxes around the lesions, we used IOU criterion. The generated bounding box is defined as the area of bl (a, b, l, w), where (a, b) denotes the center coordinates and l, w are the length and width of the bounding box. RCNN performs the lesion area detection using a greedy overlapping or IOU criterion of the actual and the predicted box. The area is known as a lesion when IOU is between 0.5 and 1, and also, the region will be labeled as a normal one if the IOU is between 0 and 0.5. According to Eq. (7) IOU is written as:
Where area of overlap is calculated by finding the overlapped area between predicted bounding box and the ground truth. Also, area of union is obtained by getting the sum of areas of predicted and ground truth bounding box.
We used the mAP, illustrated in Eq. (8) to evaluate the accuracy of RCNN lesion localization. mAP is a good metric to investigate the accuracy of a bounding box prediction model. The higher the mAP value is, the greater the accuracy of the model is in lesion localization.
For calculating mAP, we first calculate IOU, if the value of IOU is greater than 0.5 and less than 1 (0.5 < IOU ≤ 1), the predicted bounding box is labeled as TP. This means that the most area of predicted bounding box contains the most of lesion area. On the other hand, if IOU is greater than 0 and less than 0.5 (0 ≤ IOU ≤ 0.5), the most area of predicted bounding box includes normal area, so it is considered as FP.
To evaluate the performance of the SLDED method, several measures are calculated for comparison. Common measures for assessing the performance of a classification model include Accuracy, AUC, Precision, Recall, and F1-Score.
Since our problem is multi-class classification, it is required to calculate the average of AUC, F1-score, precision, and recall measures. For each measure, there are micro and macro averages that will calculate slightly different values. Since the micro-average of the performance measures for imbalance data is preferred to macro-average ones [46], the micro-average of F1-score, precision, recall, and AUC for the models are reported. A micro average will sum up the contributions of all categories to calculate the average of the measures. Equations (9–11) indicate how to calculate the mentioned measures.
3 Experimental results
This section will describe the results of lesion localization achieved by a VGG-based faster R-CNN method. We will also use two ISIC and PH2 test sets to compare and analyze the results of the proposed SLDED approach with other previous state-of-the-art models.
3.1 Lesion localization results
Faster R-CNN can distinguish areas in the image that are as lesions from the ones which are normal. To do so, faster R-CNN assigns the features extracted from the last convolutional layer to the SOFTMAX function and sets them a probability to consider extracted features, either a lesion or a normal skin area. The confined pixels relating to the lesion area are labeled as positive specimens and the rest of the regions as negative ones to train the faster R-CNN as a binary classifier for lesion localization. The detected overlapped lesion area is labeled based on a presumed IOU threshold value. In this work, we assumed a threshold of 0.5, and if IOU exceeded it, we consider it as a lesion area. The accuracy of lesion localization by faster R-CNN for ten randomly selected images from the ISIC test set is shown in Table 3. Also, the mAP criteria for total images is 0.958.
Figure 5 shows 12 selected images from ISIC test set that had the highest confidence score. As shown in the Fig. 5, the proposed faster R-CNN method was able to detect the lesions very meticulously regardless of the rotation of the images, their differed lesion sizes, and the presence of hair and/or other noises in them.
3.2 Comparison of the models
We have investigated the SLDED method with the initial weights deriving from the average AUC score and with optimal weights adjusted by the GA algorithm for evaluation and validation. Additionally, for the sake of completeness evaluation, each member CNN has also been assessed. Table 3 demonstrates the calculated results in terms of micro-average on the ISIC as well as PH2 test sets.
As illustrated by Table 3 SLDED with w2 derived from the GA algorithm outperforms the other skin lesion classification methods while evaluated on both ISIC and PH2 test dataset.
Moreover, for complete visual comparison, the four CNNs and their ensemble’s ROC curves are depicted in Fig. 6 regarding the classification of four skin lesion types, including BCC, MM, SK, and Nevus.
Figure 7 presents the confusion matrix of the proposed SLDED method for skin cancer diagnosis.
As shown in Fig. 7, the SLDED-w2 method on ISIC test set can correctly classify 98/117 of BCC, 396/426 of MM, 273/307 of NV, and 72/84 of SK classes. Moreover, on PH2 test data, including only two classes (MM and Nevus), SLDED-w2 can classify 36/40 of MM and 157/160 Nevus correctly.
“As an additional evaluation, we compared our approach with other state-of-the-art methods that have been selected as the best-developed models during 2016 and 2017 ISIB challenges towards skin lesion classification. In the 2016 challenge, the aim was developing an automated system to classify MM and Nevus lesions while in the 2017 competition, SK lesion images have also been added to the dataset. We note that the same data was considered for assessment of the models in terms of average AUC and accuracy and using an extended data has been permitted for the learning process. For this purpose, we used a transfer learning technique. Therefore, in our model we replaced the last dense layer including four neurons with a dense layer with two neurons so as to compare our results with the challenge 2016. Also, we replaced the dense layer with three neurons to make a fair comparison with the results of the challenge 2017. It is worth mentioning that we have frozen the weights for the rest of the layers. Also, we did not fine-tune the model because the images in these challenges were available in our data set, so there was no need to fine-tune the parameters. In order to have a fair comparison, we used the images in the test set of these data sets for the testing. Table 4 illustrates the comparison results; and the SLDED method performing on the ISIB challenges’ official test data outperforms the other proposed methods.
4 Discussion
Combining different networks will lead to the extraction of various feature maps, some of which can lead the classification model with high accuracy. Considering the same weights for the candidates results in integrating both weak and strong classifiers’ votes with the same weight. Therefore, we used a weighted majority voting approach to take the individual classifier’s accuracy into account. On one hand, the average AUC of each CNN is determined as its initial weight. For CNN1 to CNN4, we have set the weights 0.901, 0.898, 0.866, and 0.912, respectively. Then the weights have been optimized by the GA algorithm.
The genetic algorithm inspired by the natural selection process is a heuristic search and an optimization method. It is commonly used to find an approximate optimal solution for large-parameter space optimization problems. The evolution process of species (weights in this study) is mimicked according to biologically inspired components. Finally, the algorithm finds the optimal weights to minimize the cost (average AUC in our case). The optimal weights obtained from the GA algorithm for CNN1 to CNN4 equal to 0.883, 0.806, 0.656, and 0.954, respectively. As shown in Table 3 in Section 3.2 the SLDED-w2 method, whose weights are obtained by the GA algorithm, outperforms the others.
During the training process, the classification results of ensemble member CNNs on the validation and training set after each epoch using top-1 error rate are plotted, see Fig. 8a It can be pointed out that the training and validation curves move together in a descending way, which shows addressing the overfitting problem by applying data augmentation methods. Additionally, we fine-tuned and trained VGGNet, ResNet, GoggletNet, and ResNeXt pre-trained models to compare time-consuming trends. Comparing the gradient convergence of the pre-trained models and our designed approach in Fig. 8b displays the time-consuming trend. In general, our approach’s training time was 8 h and ResNet, ResNeXt, GoggletNet, and VGGNet were 11, 12, 15, and 19 h, respectively.
5 Conclusion
Despite the increasing trend in the employment of CNNs for diagnosis of skin lesions, the lack of large annotated images to train these networks is still a promising challenge in medical image analysis. In this paper, we have investigated the possibility of combining deep neural networks which have had excellent performance in medical image classification to improve the classification accuracy.
The main motivation is the development of an automated skin lesion detection approach. In this study, we exploited a collection of 3361 ISIC archive’s images for training, 373 for evaluation, and 934 images for testing. Moreover, 200 images of PH2 dataset have been employed as another additional test set. It is noteworthy that if we set the CNNs’ weights appropriately using the weighted majority voting approach, our proposed fusion method outperforms the individual CNNs according to the classification accuracy.
We note that our proposed ensemble approach is modular, and adding additional CNNs to the framework can improve the classification accuracy while also increasing the computational complexity. Moreover, other image segmentation methods rather than deep-based ones, such as clustering or threshold-based approaches, can be exploited, which can reduce the implementation time.
Data availability
The datasets analyzed during the current study are available in the International Skin Imaging Collaboration (ISIC) Archive repository, https://challenge.isic-archive.com/data/.
References
Abbes W, Sellami D (2017) Automatic skin lesions classification using ontology-based semantic analysis of optical standard images. Procedia Comput Sci 112:2096–2105
Agarwal M, Damaraju N, Chaieb S Skin lesion analysis toward melanoma detection
Argenziano G, Soyer HP (2001) Dermoscopy of pigmented skin lesions–a valuable tool for early. Lancet Oncol 2(7):443–449
Argenziano G et al (2006) Dermoscopy improves accuracy of primary care physicians to triage lesions suggestive of skin cancer. J Clin Oncol 24(12):1877–1882
Attia M et al (2017) Skin melanoma segmentation using recurrent and convolutional neural networks. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
Barata C, Celebi ME, Marques JS (2017) Development of a clinically oriented system for melanoma diagnosis. Pattern Recogn 69:270–285
Baumann LS et al (2018) Safety and efficacy of hydrogen peroxide topical solution, 40%(w/w), in patients with seborrheic keratoses: results from 2 identical, randomized, double-blind, placebo-controlled, phase 3 studies (A-101-SEBK-301/302). J Am Acad Dermatol 79(5):869–877
Bengio Y (2009) Learning deep architectures for AI. Found Trends Mach Learn 2(1):1–127
Bi L et al (2017) Semi-automatic skin lesion segmentation via fully convolutional networks. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE
Brinker TJ et al (2019) A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer 111:148–154
Brinker TJ et al (2019) Comparing artificial intelligence algorithms to 157 German dermatologists: the melanoma classification benchmark. Eur J Cancer 111:30–37
Brinker TJ et al (2019) Deep neural networks are superior to dermatologists in melanoma image classification. Eur J Cancer 119:11–17
Burdick J et al (2017) The impact of segmentation on the accuracy and sensitivity of a melanoma classifier based on skin lesion images. In: SIIM 2017 scientific program: Pittsburgh, PA, June 1-June 3, 2017, David L. Lawrence Convention Center
Carli P et al (2000) Preoperative assessment of melanoma thickness by ABCD score of dermatoscopy. J Am Acad Dermatol 43(3):459–466
Celebi ME et al (2007) A methodological approach to the classification of dermoscopy images. Comput Med Imaging Graph 31(6):362–373
center, c.s. Estimated new cases, 2019. Available from: https://cancerstatisticscenter.cancer.org/#!/
Chang W-Y et al (2013) Computer-aided diagnosis of skin lesions using conventional digital photography: a reliability and feasibility study. PLoS ONE 8(11):e76212
Codella NC et al (2017) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE
Collaboration, I.S.I. (2020) ISIC archive. Available from: https://www.isic-archive.com/#!/topWithHeader/wideContentTop/main
Conn AR, Gould NI, Toint P (1991) A globally convergent augmented Lagrangian algorithm for optimization with general constraints and simple bounds. SIAM J Numer Anal 28(2):545–572
Dascalu A, David E (2019) Skin cancer detection by deep learning and sound analysis algorithms: A prospective clinical study of an elementary dermoscope. EBioMedicine 43:107–113
Díaz IG (2017) Incorporating the knowledge of dermatologists to convolutional neural networks for the diagnosis of skin lesions. arXiv preprint arXiv:1703.01976
Esteva A et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT press
Grichnik JM, Rhodes AR, Sober AJ (2008) Benign neoplasias and hyperplasias of melanocytes. Fitzpatrick’s dermatology in general medicine, 7th edn, pp 1099–103
Haenssle HA et al (2018) Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol 29(8):1836–1842
Hahnloser RH et al (2000) Digital selection and analogue amplification coexist in a cortex-inspired silicon circuit. Nature 405(6789):947
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32
He K et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition
He K et al (2017) Mask r-cnn. In: Proceedings of the IEEE international conference on computer vision
Hekler A et al (2019) Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur J Cancer 115:79–83
Hekler A et al (2019) Superior skin cancer classification by the combination of human and artificial intelligence. Eur J Cancer 120:114–121
Hu H et al (2018) CNNAuth: continuous authentication via two-stream convolutional neural networks. In: 2018 IEEE international conference on networking, architecture and storage (NAS). IEEE
Isasi AG, Zapirain BG, Zorrilla AM (2011) Melanomas non-invasive diagnosis application based on the ABCD rule and pattern recognition image processing algorithms. Comput Biol Med 41(9):742–755
Jain S, Pise N (2015) Computer aided melanoma skin cancer detection using image processing. Procedia Comput Sci 48:735–740
Jaleel JA, Salim S, Aswin R (2013) Computer aided detection of skin cancer. In: 2013 International Conference on Circuits, Power and Computing Technologies (ICCPCT). IEEE
Kallenberg M et al (2016) Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging 35(5):1322–1331
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems
Levine AB et al (2019) Rise of the machines: advances in deep learning for cancer diagnosis. Trends Cancer 5:157–169
Li Y, Hu H, Zhou G (2018) Using data augmentation in continuous authentication on smartphones. IEEE Internet Things J 6(1):628–640
Li Y et al (2020) Using feature fusion strategies in continuous authentication on smartphones. IEEE Internet Comput 24(2):49–56
Lopez AR et al (2017) Skin lesion classification from dermoscopic images using deep learning techniques. In: 2017 13th IASTED international conference on biomedical engineering (BioMed). IEEE
Majtner T, Yildirim-Yayilgan S, Hardeberg JY (2016) Combining deep learning and hand-crafted features for skin lesion classification. In: 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA). IEEE
Manning C, Raghavan P, Schütze H (2010) Introduction to information retrieval. Nat Lang Eng 16(1):100–103
Mar VJ, Scolyer RA, Long GV (2017) Computer-assisted diagnosis for skin cancer: have we been outsmarted? Lancet 389(10083):1962–1964
Marchetti MA et al (2018) Results of the 2016 international skin imaging collaboration international symposium on biomedical imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol 78(2):270-277. e1
Maron RC et al (2019) Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks. Eur J Cancer 119:57–65
Matsunaga K et al (2017) Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. arXiv preprint arXiv:1703.03108
Megahed M et al (2002) Reliability of diagnosis of melanoma in situ. Lancet 359(9321):1921–1922
Mendonca T et al (2015) PH2: A public database for the analysis of dermoscopic images. In: Dermoscopy image analysis. CRC Press
Menegola A et al (2017) RECOD titans at ISIC challenge 2017. arXiv preprint arXiv:1703.04819
Mirzaalian-Dastjerdi H et al (2018) Detecting and measuring surface area of skin lesions, in Bildverarbeitung für die Medizin 2018. Springer, pp 29–34
Moss RH et al (1989) Skin cancer recognition by computer vision. Comput Med Imaging Graph 13(1):31–36
Mueller SA et al (2019) Mutational patterns in metastatic cutaneous squamous cell carcinoma. J Invest Dermatol 139(7):1449-1458.e1
Nida N et al (2019) Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering. Int J Med Informatics 124:37–48
Okur E, Turkan M (2018) A survey on automated melanoma detection. Eng Appl Artif Intell 73:50–67
Renzi M et al (2019) Management of skin cancer in the elderly. Dermatol Clin 37(3):279–286
Sarıgül M, Avci BMOM (2019) Differential convolutional neural network. Neural Netw 116:279–287
Schaefer G et al (2014) An ensemble classification approach for melanoma diagnosis. Memetic Comput 6(4):233–240
Soudani A, Barhoumi W (2019) An image-based segmentation recommender using crowdsourcing and transfer learning for skin lesion extraction. Expert Syst Appl 118:400–410
Sreekantaswamy S et al (2019) Aging and the treatment of basal cell carcinoma. Clin Dermatol 37:373–378
Stoecker WV, Moss RH (1992) Digital imaging in dermatology. Elsevier
Stoecker WV et al (2005) Detection of asymmetric blotches (asymmetric structureless areas) in dermoscopy images of malignant melanoma using relative color. Skin Res Technol 11(3):179–184
Szegedy C et al (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition
Uijlings JR et al (2013) Selective search for object recognition. Int J Comput Vision 104(2):154–171
Vasconcelos CN, Vasconcelos BN (2017) Experiments using deep learning for dermoscopy image analysis. Pattern Recognit Lett 139:95–103
Xie S et al (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition
Yu L et al (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36(4):994–1004
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shahsavari, A., Khatibi, T. & Ranjbari, S. Skin lesion detection using an ensemble of deep models: SLDED. Multimed Tools Appl 82, 10575–10594 (2023). https://doi.org/10.1007/s11042-022-13666-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13666-6