Abstract
Sleep disorder diseases have one of the major health issues across the world. To handle this issue the primary step taken by most of the sleep experts is the sleep staging classification. The whole visual inspection process is carried out manually by the sleep experts, which can be a highly time-consumed task and creates a lot of annotation errors due to more human interventions. In this study, we introduce an efficient and robust approach to improve the sleep staging accuracy. In this paper, we proposed an automated deep nine-layer one-dimensional convolution neural network for multi-class sleep staging classification (9 L-1D-CNN-SSC) using polysomnography (PSG) signals. The proposed 9 L-1D-CNN-SSC model comprises eleven layers with learnable parameters: nine convolution layers and two fully connected layers. The main objective of designing such a model is to achieve higher classification accuracy for multiclass sleep stages classifications with reduced learnable parameters. The proposed network architecture is tested on two different subgroups recordings of ISRUC-Sleep datasets namely ISRUC-Sleep subgroup1 (ISR-SG-I), and ISRUC-Sleep subgroup3 (ISR-SG-III). The proposed model is compiled with eight different individual experiments based on a single-channel electroencephalogram (EEG), electrooculogram (EOG), electromyogram (EMG), and combinations of EEG + EOG+ EMG signals. The proposed 9 L-1D-CNN-SSC model achieved the highest classification accuracy of 99.03%, 99.50%, and 99.03% for three to five sleep stages classification, respectively with single-channel of EEG signals, similarly, the model achieved 98.93% for two-state sleep stage classification with EMG signals using the ISR-SG-I dataset. The same model achieved the highest classification accuracy of 98.88%, 98.76%, and 98.67% for three-five sleep stages classification with a single-channel EMG signal, and 99.24% for two-state sleep classification with single-channel EOG using ISR-SG-III dataset. It has been observed that the obtained results from the proposed 9 L-1D-CNN-SSC model give the best classification accuracy performance on multiclass sleep stages classification incomparable to the existing literature works. The developed 9 L-1D-CNN-SSC deep learning architecture is ready for clinical usage with high PSG data.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nowadays it has been observed that the neurocognitive system directly decides the mental and cognitive performance in a particular task. It’s very difficult to determine the subject’s sleep behavior very accurately either numerically or any standard evaluation procedures [8]. Currently, sleep related diseases are an open challenge in the medical domain concerning different diseases such as neurology disorder, rehabilitation, and psychology-related disorders. It’s also very difficult to assess with a scenario of changes sleep characteristics in a known predictable manner. These types of diseases are more challenging concerning analysis and getting proper diagnosis solutions [23]. Sleep is one of the important ingredients for good human health and also responsible for maintaining the fitness and functioning of the different core systems of our body. It also put an impact on our proper functioning of mental and cognitive systems [12].
For human life, a total of one-third of its duration is constituted of the sleep cycle. It has been observed from several studies that sleep deficiency causes so many consequences like inability to solve the problem, not able to make proper decisions, not controlling the emotions, and reflected several changes in people [11, 70]. Sometimes the improper quality of sleep influenced different types of sleep-related disorders such as sleep apnea, insomnia, depression, narcolepsy, hypersomnia, breathing-related disorders, and circadian rhythm disorders [77]. Sometimes it has been seen that sleep deprivation is considered a stress-related disorder or sleep pathology, which causes high risk in performing some common cognitive risks such as workplace incidents, road accidents happened [25]. According to a report of the National Highway Traffic Administration of USA, due to drowsiness around one lakh, car accidents happened, as consequence more than 1500 death cases resulted and injuries cases reported around 71,000 annually [22].In this scenario, proper analysis of sleep stages is very important for identifying sleep-related irregularities. So that it is very essential to analyze the sleep stages behavior and accurate scoring of sleep states is a very crucial segment of the sleep staging process [27].
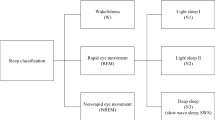
The polysomnography test is the primary step for any type of sleep-related disorder. It is a combination of different physiological signal which is useful during analyzing the sleep patterns of an individual subject. Several polysomnographic recording is recorded for sleep scoring: the EEG signal are used for monitoring the brain-behavior, the EOG signal used for monitoring eye movements and the EMG signals are tracked the changes behavior muscle tone. The entire sleep staging process is generally conducted through visualizing the sleep patterns of the subject during sleep periods by well-trained sleep experts according to two available standard sleep guidelines: the Rechtschaffen and Kales (R&K) [36] and the American Academy of Sleep Medicine (AASM) standards [78].As per AASM rules, the whole sleep stages are divided into five sleep stages: wake stage (W), non-rapid eye movement (NREM) sleep stage1 (N1), NREM sleep stage2 (N2), NREM sleep stage3 (N3), and rapid eye movement (REM) sleep stage. The main revision that occurred with AASM is, the merge of two R&K-defined sleep stages S3 and S4 into one single sleep stage, called N3.
Each defined sleep stage in AASM behaves differently during sleep periods. In this study, we are using an automated sleep stage classification system for these five sleep stages. Generally, one subject can go through all these sleep stages during its sleep cycle periods. One person can cover 3 to 5 sleep cycles, each of the time duration around 90–110 minutes during a full sleep [41]. During the first phase of the sleep cycle, the period of NREM stages is more and later part of the sleep cycles, periods REM stage increases. Earlier the sleep staging is done through visual inspection for a period of 2 to 5 hrs. in 8 hrs. of sleep for one subject. Traditionally, the sleep experts have segmented the entire sleep recordings into the 30-s interval, called one epoch, and each epoch labeled with one of the sleep stages through visualizing of its frequency and amplitude ranges, the characteristics of EEG waveforms, blinking of eye movements (EOG) and muscle movements (EMG) [76].
This traditional way of monitoring sleep stages methods has so many disadvantages such as requires more sleep experts to monitor the sleep recordings, time-consuming, and erroneous [10]. Due to more human interpretations during recording and it may not report good classification accuracy in the diagnosis of sleep stage classification [72]. Based on the above-mentioned drawbacks, automated classification of sleep stages is introduced, which ultimately gives benefits for quick diagnosis and also reported with increases of high classification accuracy [52, 55].
Sleep staging analysis and its scoring is a complicated procedure because of changes in sleep characteristics related to different sleep stages and also the non-stationary nature of the signal information [57].
The rest of the paper is presented as follows: In Section 2, the authors present the research contributions with related to sleep staging. Section 3 describes a brief overview on CNN model and its parameters. Section 4 describes briefly on the proposed methodology, which includes descriptions of the dataset used, proposed 9 L-1D-CNN-SSC model, model training and testing of 9 L-1D-CNN-SSC model are discussed. Section 5 presents the brief descriptions of experimental results of the proposed model. Section 6 discusses about the results and compares them with those by the state-of-the-art methods.Finally in Section 7 the concludes the paper and present the future directions.
2 Related work
Most of the authors are proposed an automatic sleep stage classification system for identifying the sleep patterns and diagnosis of several types of sleep-related disorders [29, 33, 34, 44]. In general sleep, staging procedures are conducted mainly on two strategies, one with single-channel input recording, and the other is multi-channel input recordings [60, 62, 63]. In the first approach, only one channel is considered for extracting the informative features about the sleep characteristics of the subjects. Similarly in a multi-channel system of recordings, a number PSG signal is used that is more than one EEG channel, EOG channel, and EMG channel [65, 66]. There is a standard procedure obtained for an automated sleep stage classification by most of the authors to their sleep staging experiments through five basic stages: 1) Signal the acquisition, 2) Pre-processing, 3) feature extraction,4) feature reduction, and 5) classification [79, 82]. The 3)feature extraction step is used for extracting the different characteristics parameter from preprocessed signal stage 2).These feature values can be extracted in frequency,time,time-frequency and non-linear domains [2].It has been seen that some of the s, one additional step used by authors that is feature reduction or dimensionality reduction stage. It is very helpful in screening the relevant features for the classification model. From our survey, it has reported that some of the feature selection algorithms used by different authors in their ASSC study are: principal component analysis (PCA) [46], relief algorithm [50], linear discriminate analysis (LDA) [58], minimum redundancy maximal relevance (mRMR) [71], and sequential forward and backward selection method [73, 74].Similarly in case of classification model used in recent automated sleep stage classification includes: support vector machine (SVM) [38, 53, 80], k-nearest neighbor (KNN) [104], k-means clustering [84], decision tree (DT) [96], bootstrap aggregating [7],random forest (RF) [20],naïve bayes [3],Gaussian mixture model (GMM) [97],adaboost [6], sparse auto encoders (SAE) [61],and artificial neural networks (ANNs) [93].
In recent research developments, deep learning techniques are becoming more popular in machine learning research applications and also used so many applications such as human-brain computing, computer vision, natural language processing, and speech recognition. Recently it has been found that deep learning concepts such as CNN [87, 99], RNN [28, 37], and LSTM [35] applied to the sleep staging approach. Currently, research on sleep staging plays an important role in NCP and Human-Machine interaction (HMI). There are several studies related to automated sleep staging using various physiological datasets and multimedia data, such as EEG, EMG, EOG, ECG, and audio, etc. One of the most popular contributions of sleep stage classification is the study of sleep behavior through human brain-computer- interaction (BCI) [102, 105].Till now also traditional machine learning techniques used for sleep staging, and recently in this research deep learning methods used in several contributions of automated recognition of sleep stages. We now look upon some of the recent contributions presented by different authors related to sleep staging using machine learning and deep learning concepts.
2.1 Polysomnography (PSG) based sleep staging using machine learning approaches
Most of the research contributions until now depend on the machine learning techniques for the recognition of sleep stages in an automated system. Krakovska et al. [35] used 6 EEG channels, 2 EOG channels, and 1 EMG channel used for recordings of the sleep behaviour and obtained features like variance, average amplitude, and spectral power. For classification of sleep stages obtained using quadratic discriminate analysis and the accuracy result reported about 74%.
In [54] the author considered multiple signals such as EEG, EOG, and EMG for the automated sleep scoring through the extraction of features like skewness, kurtosis, variance, entropy and used a dendrogram-based SVM (DSVM) classifier for classifying the sleep stages and reported accuracy for the model as 88%.
Zhu et al. [106] obtained graph-oriented features from single-channel EEG and used SVM classification techniques and the accuracy result was reported for six-state classification as 87.5%.
Hassan et al. [32] applied the EEMD algorithm for signal enhancement from single-channel EEG signal and extracted statistical features are forwarded into boosting techniques and the reported accuracy for two-six sleep stages is reported as 98.15%, 94.23%, 92.66%, 83.49%, and 88.07% respectively.
Silveria et al. [85] presented a six-state sleep staging approach using a discrete wavelet concept and obtained a random forest classifier, the model achieved 90% accuracy.
Rahman et al. [75] introduced a single-channel EOG sleep scoring approach and extracted statistical features by applying discrete wavelet transform techniques. The average accuracy reported for six state classifications through RUSBoost, RF and SVM is 90%, 91%, and 91.7%.
Memar et al. [59] proposed two-state sleep staging and the acquired signal decomposed into eight sub-bands, finally 13 features are extracted from each sub-band epoch. The suitable features are identified through the mRMR feature selection algorithm. The model achieved an overall accuracy of 95.31% through a random forest classifier.
Imtiaz et al. [42] presented automated sleep staging through home-based polysomnography signal and the model reported accuracy for training and testing dataset are 89% and 72% respectively through decision tree classification algorithm.
Dimitriadis et al. [16] proposed one channel EEG sensor ASSC techniques and estimated cross-coupling frequency (CFC) from each epoch and the system achieved an overall accuracy of 94% through multi-class Naïve Bayes classification techniques.
Sen et al. [81] proposed a sleep staging system using a single-channel EEG signal and obtained 41 multiple feature parameters. The relevant features were selected through different feature selection algorithms such as minimal redundancy maximal relevance (mRMR), ReliefF feature selection algorithm, fast correlation-based feature selection algorithm (FCBF), and Fisher score algorithm. Finally, the selected features were fed into five different classification algorithms such as support vector machine(SVM), decision tree(DT), random forest(RF), feed-forward neural network (FFNN), and radial basis function neural network (RBF) and the RF classification model reported accuracy of 97.03% for six sleep states classification problems.
Dikhya et al. [17] presented automated sleep staging system by obtaining the statistical time-domain features, structural graph similarity feature based on single-channel of EEG signal under R&K sleep scoring rules and the proposed model performed best using SVM classifier, an average accuracy of 95.53%.
T. Zhang et al. [103] proposed a novel mechanism for feature selection using the filter method with pairwise constraints. The author has obtained two different categories, one section of data having affected with mild sleep problem and the other category section, healthy controlled subjects. The whole recordings were collected from the S-EDF dataset. Finally, the model reported an accuracy of 97.66% and 93.57% with consideration of category-1 and category-2 data respectively.
Basha, A. J et al. [9] obtained the fuzzy-kernel SVM for classifying the sleep stages and extracted the statistical features and the selected features were fed into the recurrent neural network and the model reported an overall accuracy of 90.2%.
Shen, H et al. [83] proposed an improved model based on features with a combination of locality energy and state-space model for automated classifying the sleep stages based on single-channel electroencephalogram signals under the R&K and AASM sleep scoring guidelines. The model reached an overall classification accuracy of 92.04% and 78.92% using the S-EDF and Dreams dataset respectively. Similarly, the same model reported accuracy of 79.90% and 81.65% using Dreams and ISRUC-Sleep dataset.
Wang, Q. et al. [98] proposed a high-accuracy and high-efficiency automated sleep staging system using single-channel EEG data and extracted 30 features of types time, frequency, time-frequency features, and non-linear parameters. The selected features were classified through the ensemble learning stacking model and reported an overall accuracy of 96.67% for the five-class sleep stages classification task. It has been also seen that the multi-modal signals analysis also takes an important role during the diagnosis of different types of sleep-related diseases. Therefore several sleep studies were conducted by the different researchers using multi-channel signals.
Yan, R. et al. [100] develop an automated sleep staging system based on the eight combinations of the four multi-modality channels of PSG signals and obtained a total of 232 features of statistical, time, frequency, time-frequency entropy, fractal and non-linear parameters were extracted. The model reported an accuracy of 86.24% using a random forest classifier with the ReliefF selected features.
Ghimatgar, H. et al. [24] introduced a multi-modality approach using a deep learn-ing model and Hidden Markov Model (HMM) to improve the sleep staging performance using multi-channel EEG data. The experimental data collected from sixteen neonates having an age range of 38–40 weeks. The relevant features were screened using the MGCACO algorithm. The model is trained using bi-directional long-short time memory and post-processing done using HMM model. The proposed model reported an accuracy of 78.9% and 82.4% using the K-Fold cross-validation and LOOCV techniques respectively.
Cooray, N. et al. [14] presented a sleep staging system followed by Rapid Eye Movement sleep behavior disorder (RBD) detection. The extracted 156 features from EEG, EOG and EMG channels and forwarded them into the RF classifier. The model reported an accuracy of 92%.
Diykh, M et al. [17] presented the new sleep staging system based on the statistical features and weighted brain networks using multiple-channel of EEG signals under both R&K and AASM sleep scoring guidelines. The proposed model has been per-formed on the two most popular public datasets namely ISRUC-Sleep and S-EDF dataset. The model reported an average accuracy of 96.74% with C3-A2 channel under the AASM scoring standards and 96% with Pz-Oz channel under the R&K standards.
H. Korkalainen et al. [51] presented the deep learning approach for automatic sleep staging system and analyzed the severity of obstructive sleep apnea (OSA). The overnight polysomnography recordings obtained from S-EDF public dataset, both healthy and sleep apnea subjects were considered for analysis of the sleep behavior. The model reported an overall accuracy of 83.7% with single-channel EEG and 83.9% with single-channel EOG. Similarly, for the clinical data, the model achieved an accuracy of 82.9 with EEG signal and 83.8% with combinations of EEG and EOG data.
2.2 Polysomnography (PSG) based sleep staging using deep learning approaches
Nowadays the researchers are majorly focused on deep learning techniques for sleep staging because of its robustness, scalability, and adaptability with related to handle large amounts of signal recordings and it’s processing. Another important advantage related to deep learning models is, no need to require any explicit features for discriminating the subject’s sleep behaviour [30]. It has witnessed that deep learning techniques working well in different applications like image segmentation, recognition, detection, and natural language processing. It has also been observed that deep neural models are widely used in different fields of the biomedical research area. In recent research developments, it has found that notable increases happened with the use of the deep neural network in the field of biomedical signals (EEG, ECG, EMG, and EOG) [18].
Recently deep learning concepts proposed in many challenging applications by the different researchers with the input of biomedical signal data which includes epileptic seizures [18, 39, 45, 89], neurological disorders using CNN models [21, 40, 67, 94] and heart diseases using ECH channel [18, 30, 39, 51].some of the recent contributions conducted by different researchers using deep learning models for classifying sleep stages are described below here.
In [45], the author used a deep convolution neural network for automated sleep staging with the input of single-channel EEG. The model achieved an overall accuracy of 74%.
Sors A et al. [89] presented automatic sleep stages scoring for five-sleep states based on one-channel of EEG using the CNN model and the results reported for the proposed model are 87%.
Chambon et al. [94] introduced a deep learning model with the concept of multivariate signal analysis such as EEG, EOG, and EMG using KNN. The proposed model reached an overall accuracy of 80% with combinations of EEG + EOG + EMG.
In [40] the authors obtained five-layer convolution layers for classifying the sleep stages based on two-channels of EEG and EOG signal and one-channel of EMG signal and achieved result for the model is 83%.
Tripathy et al. [67] introduced a novel approach of sleep scoring based on coupling features of EEG data and RR time-series information using deep neural networks. The model resulted in an average accuracy of 95.71%, 94.03%, and 85.51% for the classification in between NREM vs REM, deep sleep vs light sleep, and sleep vs wake respectively.
Zhihong Cui et al. [21] proposed a sleep scoring system with input of 30s multi-channel signal information based on CNN and fine-grained properties, the model reported an average accuracy of 92.2% with the ISRUC-Sleep public dataset.
Supra Tk A et al. [56] designed a system of sleep scoring through extracted time-invariant information using CNN and find sleep stages transition information from the bidirectional LSTM network. The reported classification accuracy performance reached to 86.2%.
Akyol, K. et al. [5] presented a stacking ensemble learning model for analysis of the single-channel EEG signal and identifying the epileptic seizure detection. The clinical dataset was collected from the Bonn University. Finally, the author has com-pared the performance of the proposed model with the deep neural network (DNN) model. The model achieved an average accuracy value of 97.17%.
Yildirim, O. et al. [101] developed a deep learning model using a one-dimensional convolutional neural network (1D-CNN) using combinations of EEG, EOG, and EMG signal for classifying the six-two sleep stages classification problems. The model reported an overall accuracy of 91.00%, 91.22%, 92.36%, 94.64%, and 98.06% using the S-EDF dataset and similarly, the same model achieved an accuracy of 89.54%, 90.98%, 92.33%, 94.34%, and 97.62% using SE-EDF dataset for classifica-tion of six-two sleep classes problems.
Zhu, T. et al. [107] proposed a sleep staging system using a neural network with the implementation of the CNN concept and obtained inter and intra epoch features from the input signal. The model reached overall accuracies of 93.7% with S-EDF and 82.8% with SE-EDF datasets respectively.
Fernandez-Blanco, E et al. [19] proposed an ensemble technique for an automatic sleep scoring system using multiple-channel of EEG signals, and the model was trained using the convolutional neural network. The entire experiment work was done through a widely accepted bench-mark dataset as SE-EDF dataset and the test was carried out with help of leave-one-out cross-validation technique. The model achieved an accuracy of 92.67%.
C. Sun et al. [90] presented multi-class sleep stages classification using PSG signals based on a hierarchical neural network model. The model functioned in two phases, in the first phase using through feature learning stage and the second phase is the sequence learning stage. This study performed on 147 patients’ sleep recordings, which were obtained from the MASS dataset. Finally, the model reported an overall accuracy of 87.8% and an F1score of 81.8 respectively.
Chenglu Sun et al. [91] developed an automated sleep staging system based on a two-stage neural network model. During the first stage, the model learning hand-crafted features, and in the second stage the model is learned. The author also intro-duced the data augmentation techniques to resolve the class imbalance problem. The whole work was executed through two public datasets such as S-EDF and Sleep apnea dataset. The proposed model reported the result of F1score and Kappa score as 80.6% and 80% with healthy subjects, and 79% and 74% with sleep-disordered subjects.
Antoine Guillot et al. [26] proposed an automated sleep staging system using a deep learning model called as SimpleSleepNet, where the author retrieved two differ-ent categories of data from the Dreams dataset, one completely healthy controlled and other section of data are collected from the subjects who were affected by the sleep apnea subjects. The required data prepared by the five different sleep experts in the different sleep centers. Finally, the SimpleSleepNet framework achieved an average F1Score 89.9% with healthy controlled subjects and 88.3% with apnea subjects.
Mehdi Abdollahpour et al. [1] proposed the automated sleep staging system using the combinations of EEG and EOG signals. The extracted features from both the signals were separated into two different sets, one set contained the EEG features and the other set contained the fused feature of EEG + EOG. Each feature set is transformed into a horizontal visibility graph (HVG). The images of the HVG are classified by the convolutional neural network with the concept of transfer learning. The model has been performed on the two most popular datasets such as S-EDF and SE-EDF datasets. The model achieved an overall accuracy of 93.58% using the S-EDF dataset.
According to the existing contribution to sleep scoring, major challenges found that choosing the correct features which helps to distinguish the sleep stages. It has found that the maximum researchers extracted the time, frequency, and time-frequency features, then after finalizing the relevant features either manually or applied some conventional feature selection algorithm. In some cases, this selection algorithm increases the complexity factor and consumes more time. Another challenge related to feature selection is that some features are well fitted for some of the subjects but the same may not apply for another one.
The next challenge with earlier contribution is that, improper distributions of sleep epochs for all the sleep stages. This imbalance of sleep information may produce biased results with conventional machine learning algorithms. From the literature survey, it has found that the maximum researchers extracted the time, frequency, and time-frequency features, then the relevant features are selected either in manually or using some conventional feature selection algorithm, which takes the computational time and also increases the complexity factor, Another limitation regarding selected features, some of the features well suitable for classification for some of the subject cases may be the same many not applicable for other categories of subjects. This may create a problem to achieve higher classification accuracy.
Another challenge with subjects to sleep staging is that sometimes in the recorded data, it may see that the sleep epochs are not distributed equally for all the sleep stages. This imbalance of information may produce biased results with traditional machine learning algorithms. In most of the previous studies, these may not be properly addressed, so that reported classification accuracy performance is not up to the mark level. With consideration of all these issues, the authors obtained deep learning techniques for automated analysis and classification of sleep stages using polysomnography signals. Though our input data is in the form one-dimensional size. In this study, we propose a 9 L-1D-CNN-SSC for automated sleep stages classification. To recognize the sleep behavior, here the authors have proposed an end-to-end structure, without use any type of handcrafted features for learning. The proposed model learning the features automatically from the obtained layers of the model. Apart from this, other advantages of this architecture and addressing the multi-class sleep stage classification problems without changing any of its layers and its parameters for two to five sleep classes.
2.3 Contribution
The main contributions of our proposed research works are explained below:
1. The authors propose a 9 L-1D-CNN-SSC architecture for classifying multiple sleep classes based on multi-modality signal fusions under the AASM sleep scoring rules using two different categories of subjects’ sleep recordings.
2. The proposed architecture of 9 L-1D-CNN-SSC consists of a convolutional layer, pooling layer, batch normalization layer, and fully connected layer. The performance of the proposed model is also compared with the existing pre-trained model. The obtained features from the last fully connected layers of the 9 L-1D-CNN-SSC model were fed into the softmax activation function.
3. The complete sleep staging process was analyzed with the three different combinations of signals and each one executed in the individual experiment. The first three experiments were executed with the input of single-channel EEG, EMG, and EOG signal, and the final experiment is performed with combinations of the three signals, EEG + EMG + EOG.
4. The proposed methodology uses fewer parameters to train the model and extracting the prominent features from the input signal data automatically, which supports achieving the high classification accuracy incomparable to the earlier contributions. Concerning the earlier contribution of sleep staging using the CNN model by different researchers, our proposed model is well competitive with the results of existing sleep studies, which use even complex structure CNN architecture.
3 Background of CNN model
In recent years, deep learning techniques attempt excellent performance to learn the highly complicated behavior from the input biomedical signals through a designed hierarchical architecture model. Among the different modes in deep learning methods, the CNN model is a more effective technique in biomedical signals and image analysis and classification problems compared to traditional machine learning techniques. The network styles of the CNN model are quite similar to the conventional ANN model structure, a CNN model framed with compositions o the input and output layers, and a set of hidden layers. The hidden layers of the CNN model comprise a set of convolution, pooling, and fully connected layers, which extracts the highly commendable features from the input data automatically, which are more feasible in concerns to each and individual neuron representing in each layer. Like ANN, CNN also depends on the previous layer’s weight and bias information to get the final result. The typical structure of the CNN model for a one-dimensional signal is shown in Fig. 1.
The entire working procedures in CNN are implemented through two basic stages 1.feedforward stage, 2.backpropagation stage.
In the feedforward stage, the given input data are fed into the designed model, each input data are multiplied with the layer’s parameters of each layer and finally, the achieved feature map values are forwarded into the network output. During the backpropagation stage, the values of the weights are adjusted in each successive layer in the network to control and reduce the error values in the obtained model by implementing the proper loss function. Finally, the error rate determines by comparing actual and desired outcomes.
The CNN model consists of several network layers such as convolution layers (CONV), rectified linear unit (ReLU), batch normalization layers (BN), pooling layers, and fully connected layer, each layers description is described in detail below:
3.1 Convolution layer
In the CNN model, the convolution (CONV) layer is the basic core section of a CNN model. Generally, in one CNN architecture, there are more than convolutions layers are there, in this layer the given inputs are processed through the set of learnable parameters and with the number of filters with different dimension sizes. From each filter as an output, we generate a set of feature maps values, which are computed from dot product small unit of given input data and its weight values. It helps to learn the features from the input signal by learning the weight parameters of the filters. The given size of input data for a convolution layer is ShXSwXSd, the required output volume size of the layer is ShnewXSwnewXSdnew.The required output computed using four hyper parameters 1. Number of filters (NF), 2. Size of filter (Fs), 3.Padding size ( Ps), 4.Stride information (SI).The general form of computation the output volume from convolution layers as follows:
3.2 Batch normalization layer
The importance uses of batch normalization (BN) layer in the CNN model are normalizing the data present inside the network [4].It also supports to increase the training speed and reducing the internal changes of covariance values. The main intention to include BN layer in the model is to make confirm that how best the activation function distributes in a stable manner throughout the training procedure.
BNn = {B1, B2, B3……………Bm} Presents a minibatch size of m.
The batch normalization of BNn is computed using this set of mathematical equations.
Majorly BNn layer deployed in between convolution layers and ReLU layers, which permits the users to fix the higher learning rates. The most important advantage with batch normalization layer is, it mainly controls over fit issue and increase the training speed.
3.3 ReLU layer
It is a general trend in CNN model to deploy ReLU layer after each successive convolution layers, the main intention to use of this activation function is establishes the nonlinear impact in the network. In the proposed work, we have obtained two different convolution functions are used 1.ReLU and 2.Softmax.
In general, the ReLU activation function converts the negative map values into zeros and maintains the positive values. The mathematically form of ReLU function defined as
The role of softmax activation function is decides the probable classification of the output classes. So that softmax function used in the final fully connected layer, for predicting which input signal is belong to wake, N1, N2, N3 and REM sleep stages. The mathematically the equation defined as:
Where S is the input to the network model.
Pi is represents the output value.
The output values are lies in between 0 to 1.
3.4 Pooling layer
The main purposes of using pooling techniques in CNN model is reducing the number of trainable parameters which helps indirectly reduce in complexity in execution. Finally it helps to control the overfitting problem. After each layer of convolution, we applied the max-pooling techniques for down sampling the feature map volume size and during this layer, no parameters are to be trained. In general architecture of CNN model, the pooling layer placed in between two convolution layers, for the purpose of down sampling the feature map size. From many of studies, it has found that maxpooling techniques are more effective with concern to CNN model. So that in our proposed model obtained max-pooling techniques.
3.5 Fully connected layer
Generally in a CNN model, there is one or more fully connected layers (FC) are used after successive convolution, ReLU and max-pooling layers. The operational procedure of FC layer is same with accordance to conventional neural network, where each neuron is associated to all the preceding layer neurons. The most important concern with related to FC layer is, it holds number of learnable parameters which indirectly leading computational overhead during training. In the current research work, used only one FC layer in our proposed model.
4 Methodology
The main intention of the proposed research work is to develop an artificial intelligence-based deep learning classification model to automatically classifying the sleep stages, which alternatively help for diagnosing the major types of sleep diseases This paper proposed a novel nine-layer one-dimensional convolutional neural network-based automated classification model for multi-class sleep staging classification (9 L-1D-CNN-SSC) using polysomnography (PSG) signals. The main intention is to make use of the effective information of multi-modality combinations of the signals to improve the sleep staging classification performance. The concept of the proposed research work is shown in Figure 2 by block diagram. The proposed methodology involves signal processing concepts and proposed 9 L-1D-CNN-SSC architecture followed by finding the discriminatory features to distinguish these multi-modal signals for training and testing. Mainly this research work executed through these four parts (i) input module, which takes PSG signals,(ii) pre-processing is performed for eliminating the irrelevant noise compositions which are contaminated in the recorded signals and passes them into the 9 L-1D-CNN-SSC model (iii) final decision, the output of the 9 L-1D-CNN-SSC model are fed into the fully connected layer to take the final predictions,(iv) validated the results of the proposed 9 L-1D-CNN-SSC architecture for two to five sleep stages classification problems with the existing state-of-the-art works.
4.1 Sleep stages classes
According to the AASM sleep standards, the sleep classes can be divided into two to five sleep stage classes. The only changes with the AASM sleep standards are N3 and N4 stages of the R&K standards are merged into one stage called as N3 stage. The proposed sleep staging procedure is executed under the AASM sleep scoring rules. The brief description of sleep classes considered in this proposed research work is shown in Table 1.
4.2 Experimental data
In this proposed study, we have obtained ISRUC-Sleep (ISR) datasets to evaluating the proposed sleep staging method. It contained the sleep recordings from different medical conditioned subjects who were affected by different types of sleep problems. The whole database is divided into three sections such as ISR-SG-I, ISR-SG-II, and ISR-SG-III [5].In ISR-SG-I, 100 subjects one-session sleep recordings were available, all were affected with different types of sleep-related disorder and the average age of subjects in this section is 51 years, similarly, in the ISR-SG-II, 8 subjects sleep recordings were contained, among them 6 were male and 2 were female gender. In this section, two different recordings were collected from subjects on two different dates; maximum subjects in this section were affected with sleep apnea events. It has been found that the average age of participated subjects for this section was 47. Finally, the ISR-SG-III section collected 10 subject’s sleep records, which were completely healthy, controlled, and no prior symptoms with any type of sleep problems in earlier periods. All these sleep recordings are done by the set of sleep exports at the center of sleep in the Hospital of Coimbra University. Each sleep recordings were a collection of whole-night PSG recording containing 6 EEG channel, 2 EOG channel, 3 EMG channel, 1 ECG channel, airflow, abdominal efforts, pulse oximetry, and body position information. Each recorded signal is segmented into 30-s fragments called epochs, and each epoch is represented to one particular sleep stage and labeled according to AASM sleep manuals.
In the present work, two different categories of subjects, one set belong affected with sleep-related disorders and other groups are utterly healthy category. Both the subjects’ recordings were collected from ISRUC-Sleep public repository, which was used explicitly for sleep scoring. Sleep-disordered subjects were obtained from the ISR-SG-I section; similarly, healthy subjects were acquired from ISR-SG-III. In this experiment, the authors have randomly selected five sleep-disordered subjects and five healthy controlled subjects from ISR-SG-I and ISR-SG-III respectively. The subjects’ sleep recordings concerning different stages of sleep are briefly described in Table 2.
The proposed model used three different channels of EEG, EMG, and EOG signals as inputs for measuring the sleep stages scoring. In Figure 2, the 30s sample of C3-A2 channel of EEG signal, ROC-A2 channel of EOG signal, and X1(Chin) channel of EMG signal sample of the sleep-disordered subject-77 record is shown for five sleep stages, similarly, the sleep behavior sample of the healthy subject-2 are shown in Figure 3. Each collection of records from the subjects was different from other individuals due to different sleep patterns. The resulting distribution of sleep stages also distinct differences in their observed sleep behavior, and it may sometimes make the model challenging to train the model.
4.3 Preprocessing
In this section, the author has to discard non-sleep epochs from each PSG signal recordings, because sleep stages classifications of certain epochs depend on information about its neighboring epochs. Sometimes it has been observed that recorded signals are contaminated with different types of artifacts and noise information. To remove these irrelevant portions from the raw signal by implementing a Butterworth bandpass filter with the order of 10 at the frequency ranges of 0.1–45 Hz. After that, all polysomnographic signals are segmented into 30s epochs, and each epoch is labeled into a particular sleep stage. Sometimes it has been seen that more than one sleep stage information represented to one epoch, in that case, the author assigned the sleep stages to that particular epoch, which had maximum periods of appearance. Similarly scoring for N2 sleep stages, considered the previous epochs last part and current epochs first part, if there are containing more than one sleep spindles and k-complexes then that stage to be considered as N2. Similarly, if the same trends occurred in between N2 and REM sleep stages, then the middle of the epochs are labeled as N2 otherwise it is annotated as REM stage. Sleep spindles are appeared in between N2 and REM stages. Finally, we have applied normalization techniques and here we use zero mean and unit variance and a normalized feature vector is generated. Generally, it supports increasing the performance of the system.
4.4 Proposed 9 L-1D-CNN-SSC model architecture
The CNN concepts are frequently used for two-dimensional image processing [68].But it is not restricted to two or three dimensional image classifications problems only, but it also applicable for one-dimensional input data according to recent research developments.
1D-CNN model shares the same properties like other CNN models, but only the difference of 1D-CNN is with subject to convolution operation, only 1D convolution operation is performed.The known input data in the form of 1D-CNN is biomedical signal data [69].In a 1D-CNN model, with input of input signal SI, and kernel Kw, then the convolution operation is defined in Eq. (10):
From the above equation, the * operator determines the convolution operations in between the kernel (weights) which slides over the given input data. The resulted outcome from the convolution operation is called as feature map. The general formula for computing the output from the convolution operation is defined in Eq. (11):
The operation of the convolution layer is similar to the feature extraction stage, as the outcome from this layer generated a feature map of the input matrix. The resulted feature map values down-sampled further in the network using pooling layers, further, the values are processed with a set of convolution operations. The final layer of the CNN model contains the neural network layer, which is also called as a fully connected layer, mainly it performs classification operation. Figure 4 presents the graphical representation of the proposed 9 L-1D-CNN-SSC model for the automated classification of sleep stages.
The proposed 9 L-1D-CNN-SSC model comprises twenty layers: nine convolution layers (CONV-1 to CONV-9), nine max-pooling layers (Polling-1 to Polling-9), and one fully connected layer. Apart from these layers, the proposed model considered nine batch normalization and ReLU layers. The proposed network extracted hierarchical feature information automatically using a set of hidden layers. As we have mentioned earlier the use of more FC layers may increase the computational overhead. Therefore in this research work, we have only considered one FC layer, and finally, the model resulted in a vector of size five corresponding to five different classes of sleep stages, and at last, a softmax activation function is applied in the FC layer to determine the final class label.
The main objective of designing such a custom model is to increase the classification accuracy is compared to the preexisting trained model. The first convolution layer of the proposed model takes an input of preprocessed polysomnography signals of size 3000 × 1 sample points and the first layer model convolves it with 16 × 8 filters and with four stride ratios to produce the resulting feature map of size 750 × 16.The second layer of the proposed model is the pooling layer, in this work the authors have considered max-pooling techniques with filter size 2 × 2 and a stride 1 to produce a lower dimension size of output volume of size 375 × 16. The BN layer and ReLU activation applied over the output of each max-pooling layer of the models. Next, to the pooling layer, we have again applied the second convolution layer and it carried the previous convolution layer output information and convolves it with 32 × 3 kernel of size with two stride ratios, it provides the output volume of size 352 × 32.
The same procedure is repeated for seven times again in this proposed model. In the proposed model, the first convolution layer kernel of size 8 × 1 and the stride ratio set as four, further convolution layer kernel size is 3 × 1 and for the remaining layers, the stride size is 1.In this proposed 9 L-1D-CNN-SSC model the total nine max-pooling operations performed for down sampling the feature map volume size.BN and ReLU activation layers are performed over the output of the max-pooling layer.
Finally, the output result of the convolution layer is fully connected to five neurons of a fully connected layer and applied softmax function to determine the probability distribution over each class label and finally, the output decided upon the neurons having the maximum probability score. All the layers of the proposed model and its parameter description representations are given in Table 3. Algorithm 1 shows the pseudo code for basic block of the proposed model with input vector V and number of neurons N. Similarly the Algorithm 2 presents the pseudo code of the proposed 9 L-1D-CNN-SSC model with input of PSG signals for multi-class sleep stages classification.


The final parameterized structure of the -SSC model for multi-classification sleep stages is described as follows:
1. H1: The first hidden layers contained 16 filters with a kernel size of 8. This is followed by 4 strides with valid padding, and also obtained a rectified linear unit(ReLU) activation function and batch normalization with kernel size 2.Finally the output shape of the layer is (750,16).
2. H2: 32 filters with the kernel of size 3. This is followed by 2 strides, a ReLU activation, and batch normalization with kernel size 2. The output shape is (375, 32).
3. H3: 64 filters with the kernel of size 3. This is followed by 1 stride, a ReLU activation, and batch normalization with kernel size 2. The output shape is (187, 64).
4. H4: 64 filters with the kernel of size 3. This is followed by 1 stride, a ReLU activation, and batch normalization with kernel size 2. The output shape is (93, 64).
5. H5: 64 filters with kernel size 3. This is followed by 1 stride, a ReLU activation, and batch normalization with kernel size 2. The output shape is (46, 64).
6. H6:64 filters with kernel size 3. This is followed by1 strides, a ReLU activation, and batch normalization with kernel size 2. The output shape is (23, 64).
7. H7:64 filters with kernel size 3. This is followed by 1 stride, a ReLU activation, and batch normalization with kernel size 2. The output shape is (11, 64).
8. H8:64 filters with kernel size 3. This is followed by 1 stride, a ReLU activation, and batch normalization with kernel size 2. The output shape is (5, 64).
9. H9:64 filters with kernel size 3. This is followed by 1 stride, a ReLU activation, and batch normalization with kernel size 2. The output shape is (2, 64).
Output: 5 output classes followed by a softmax activation function. The output stage is 5 i.e., 5 classes of 5 sleep stages (Wake, NREM N1 Stage, N2 Stage, N3 Stage, and REM stage).
4.5 Model training and testing
The main important step in the deep learning structure is the training of the 9 L-1D-CNN-SSC model because it directly enhances the impact on the resulting accuracy of the entire sleep staging system. The proposed research work has used two different percentages of data for training, and testing. For experimental purposes, the author considered 70% training data, and the rest of 30% is using testing data. The model performance is validated with unseen test data. First of all, the PSG signal is presented to the semantic vector of 3000 bins. The training process is optimized using categorical cross-entropy loss. For proper memory management, split the training samples into several mini-batches that are processed during the train of the 9 L-1D-CNN-SSC model in each iteration. In each batch, 100 signals are selected randomly from the training samples (without repetition). The proposed model is trained for 100 epochs. Finally, the model is tested by using unseen test data. The hyperparameter used for training the proposed 9 L-1D-CNN-SSC model is described in Table 4 for the multi-class classification of the sleep stages. The proposed 9 L-1D-CNN-SSC model is trained by using 70% training data of the database, and finally, the 30% unused samples are used for testing the model performances. The trained 9 L-1D-CNN-SSC model classifying the sleep stages by studying the behavior of PSG signals for two to five sleep state classification problems (wake, NREM (N1), NREM (N2), NREM (N3), and REM). Those classification results are compared with the ground truth values, which provided by the sleep experts.
5 Experiments and results
The whole experiments were executed on the most popular and widely used ISR-SG-I and ISR-SG-III datasets. In this study,the authors have considered both subjects affected with different types of sleep-related disorders and healthy controlled subjects.In both the datasets five sleep stages recordings information are available. The detailed information with regarding to sleep staging analysis and classification are presented in this section. Besides, both the subgroups datasets are split into 70% training data and 30% testing data.
5.1 Experimental setup
The proposed model considered PSG signals as input, which is a combination of the EEG, EMG and EOG signals. Collected PSG signal are segmented into 30-s time duration. The acquired signals are sampled with the sampling rate of 100 Hz. Each epoch of PSG data hold 3000 sample points. We have conducted eight individual experiments for both the dataset ISR-SG-I and ISR-SG-III respectively and the brief details of the experiments are presented in Table 5. The complete illustration of the experimental steps is presented in Figure 5.
Hardware Requirements: To execute the entire experiments and analyze the performance of the model, the authors establish an experimental set-up on a personal computer system with an Intel Quad-Core i7 4th generation processor with a Level1 cache memory of 32 KB, Level2 cache memory of 256 KB, and 4 MB size of cache memory, and with 1 TB hard disk.
Software Requirements: This research work executed Microsoft Windows 10 as the base operating system. MAT LAB 2015b version.
5.2 Performance evaluation metrics
To evaluate the performance of the proposed 9 L-1D-CNN-SSC architecture, the authors considered confusion matrix outcome with subject to sleep scoring using PSG signals.The confusion matrix four terms True Positive (TP),False Positive (FP),True Negative (TN), and False Negative (FN) are used for measuring the sensitivity [19], precision [90], F1Score [91] and classification accuracy [26]. The mathematical exp- ression of these statistical indices is defined in Eqs. (12)–(15).
5.3 Results with the input of ISRUC-sleep subgroup-I dataset
5.3.1 Experiment-1
Using EEG signal
In this experiment, the authors considered with single-channel C3-A2 of EEG signal re-cordings of five sleep-disordered subjects. For this experiment also, the ratio for training and testing is 70:30 with same number features are used. Table 6 presents the confusion-matrix generated from the training set and testing set for five-state sleep staging.
The average accuracy resulted for training set data as 98.67% and for testing it re-ported as 98.49%. Similarly, the performances of sensitivity, precision and F1-score reported more than 96% for all the five sleep stages. The same performance results for both training and testing data are presented in Table 7.The achieved results from both the data are closely near.
Similarly, with the same layer structure and hyper parameters values the proposed 9 L-1D-CNN-SSC model applied for two to four classes of sleep stages classification. The reported classification accuracy results for two-five sleep classes using single-channel EEG data are presented in Table 8.The training and testing accuracy for two-five sleep classes for 100 epochs iterations are presents in Figure 6.
5.3.2 Experiment-2
Using EOG signal
It has observed sometimes that rapid eye movements occurred during different stages of sleep, it is also one of the sleep characteristics which may create disturb-ances in sleep patterns of the subjects. Here we extracted ROC-A1 channel of EOG signal. To explore the sleep staging behaviour with selected features, the author considered single-channel EOG information from subjects for being analysis the sleep behaviour. The ratio of training data and testing data are same kept like for other two experiment.The confusion matrix for this experiment is presented in Table 9.It can be observed that the average classification accuracy achieved with training set as 98.32% and for testing test, the result reported as 98.31%. The evaluation performance results are presented in Table 10.
Figure 7 presents the classification accuracy performance results with 70% training and 30% testing data sets using input of single-channel EOG. It has observed from the accuracy results, there is no overfitting issue occurred found from all the sleep classes. The performance graph signifies that both training and testing accuracy gain positive learning. The highest sleep staging accuracy achieved from two class and lowest from five sleep class classification.The training and resting stages achieved accuracy for two-five sleep class are presented in Table 11.
5.3.3 Experiment-3
Using EMG signal
The third experiment carried with input of X1(Chin) channel of EMG signal.The reported results of confusion matrix from both training and testing input data are presented in Table 12.
The average accuracy resulted for training set data as 98.67% and for testing it re-ported as 98.49%. Similarly, the performances of sensitivity, precision and F1-score reported more than 96% for all the five sleep stages. The same performance results for both training and testing data are presented in Table 13.The accuracy performance graph representation using training and testing data with input of single-channel EMG are shown in Figure 8 and the same information present in Table 14 for two-five sleep stages classification.
5.3.4 Experiment-4
Using combinations of EEG + EMG + EOG signals
In our final experiment of this proposed study was conducted with combinations of EEG, EMG and EOG channels. These combinations provide sleep characteristics information which may give more accurate identification of the sleep irregularities and the results of the confusion matrix are tabulated in Table 15 and the reported performance results using EEG+EMG+EOG signals are presented in Table 16.
The average accuracy achieved with training set as 98.44%, similarly the results reported with testing data as 98.52%.The results exceeded above 95% for sensitivity, precision and F1-Score. For these combinations of multi-variate signals, the performance of classification accuracy achieved using the proposed 9 L-1D-CNN-SSC model from both training and testing data are presented in Table 17 and the same result information are presented in graphical forms in Figure 9.
5.4 Results with the input of ISR-SG-III data
To analysis the effectiveness of the proposed methodology, the authors applied with the ISR-SG-III data, which contained only healthy controlled subjects.The experimental setup and input channel information is same as Experiment-1 to Experiment-4.The same channel C3-A2 of EEG, ROC-A1 of EOG, Chin(X1) of EMG signal used for sleep staging classification with input of healthy controlled subjects sleep recordings. Experiment-5 to Experiment-8 is executed with input of single-channel EEG, EOG, EMG signal and combinations of the EEG + EOG + EMG signals respectively. The detailed accuracy results for all the four experiments using ISRUC-Sleep (subgroup-III) dataset are given in Table 18.The graphical representation of resulted accuracy with both training and testing data using single-channel EEG,EOG,EMG, and EEG + EOG + EMG are shown in Fig. 10, Fig. 11, Fig. 12, Fig. 13 and Fig. 14 respectively.
5.5 Summary of experimental results
In the current research work, we have conducted sleep staging studies with considering two subgroups sleep recordings (ISR-SG-I/ISR-SG-III).For all the individual experiments we used only proposed 9 L-1D-CNN-SSC model and with same training and testing dataset size (70:30).The proposed model is an effective with subjects to sleep scoring without any manual feature extraction or feature screening. The reported summary results obtained from both the datasets with input of single and multi-variate signals using the proposed 9 L-1D-CNN-SSC model presents in Table 19.
6 Discussion
Many similar research works have been conducted by different researchers on multiple sleep staging using different methodologies through machine learning techniques. The proposed 9 L-1D-CNN-SSC architecture can automatically be learning high-level features from the input signals directly. The obtained results indicate that the proposed scheme achieved improved sleep stages classification accuracy compared to earlier similar contributed works using the DL approach. Table 20 presents the performance comparisons results between the proposed methodology and the existing similar multiple sleep staging studies using different deep learning techniques. Similarly the performance of the proposed model also compared with the recent contributions with subject to sleep staging using different traditional machine learning classification models and the same achieved results are presented in Table 21. It has been found that the proposed framework achieved higher classification accuracy in comparison to the other state art of the contributions.
The performance of the proposed model is evaluated from two different subgroups of ISR dataset. The proposed model yielded the higher classification accuracy of 99.03%,99.50%, and 99.03% for three to five sleep states classification using single-channel EEG signals, similarly, the reported results for two-state sleep stages classification as 98.93% with single-channel EMG and combinations of single-channel EEG + EMG + EOG respectively using SG-I dataset. Further, the proposed model obtained a higher classification accuracy of 98.88%, 98.76%, and 98.67% using single-channel EMG for the same three to five sleep classes, similarly, the highest classification accuracy performance for two-state sleep stages as 99.26% with single-channel EOG using ISR-SG-III dataset. Furthermore, it has been observed that the obtained results from the proposed 9 L-1D-CNN-SSC model give the best classification accuracy performance for multiple sleep stages classification incomparable to the existing literature works.
The proposed 9 L-1D-CNN-SSC model results indicated that the model is performed excellent with concern to two-five sleep stages classifications incomparable to the existing literature works. It has been seen that several similar works are completely based on handcrafted features and shallow classifiers. From this point, the proposed model completely differs from the rest of the work and provides good potential for sleep staging analysis. Despite the improved performance on classification accuracy, the proposed model reported some more advantages related to the other works: (i) the proposed model eliminates the traditional techniques for classification using multi-stage pipeline architecture, which creates a lot of complexity during execution and raises a lot of errors. (ii) the proposed model learned the features automatically from the polysomnography signals during the model training, therefore it does not require any types of handcrafted features (iii) the proposed model used less number of learnable parameters for training model incomparable to the existing some of the pre-trained DL models, (iv) with the same hyperparameter values, the sleep staging performance is significantly improved for two to five sleep states classification using both the subgroups of a dataset and (v) the proposed architecture obtains higher classification accuracy performance in comparisons with the existing state-of-the-artworks.
7 Conclusion
In this paper, the authors proposed a 9 L-1D-CNN-SSC model for automated sleep stages classification using PSG signals. The proposed architecture contains 11 learnable layers, which helps to learn the features automatically from the multi-modality signals. The main objective of designing such architecture is to improve the sleep staging classification accuracy results with better learnable parameters compared to the traditional shallow learning models.The entire experiments executed in eight individual experiments based on single-channel EEG, EMG, and EOG signals and the final experiments are combinations of EEG, EMG, and EOG signals with the ISR-SG-I and ISR-SG-III data respectively.The proposed model obtained excellent performance for two to five sleep states classification for both the subgroups of the ISRUC-Sleep dataset. The results obtained from the 9 L-1D-CNN-SSC model exhibited the superiority of other existing CNN models. The proposed model architecture achieved the highest classification accuracy of 99.07%,99.50%, and 99.03% with single-channel EEG signals using the SG-I dataset, similarly, the same model reported accuracy using the SG-III dataset of 98.88%,98.76%, and 98.67% using single-channel EMG for three-five sleep stages classification. The same architecture performed accuracy of 98.91% with ISR-SG-I and 99.24% with ISR-SG-III using single-channel EOG signals for two sleep states classification tasks.
The main difference between the performance of the proposed model and state-of-the-art works is, the existing sleep staging systems are dependent upon the hand-engineered feature extraction approaches, which require tuning of parameters, their performance highly depends upon the selection of the hyperparameter and the sleep data. As such the model does not generalize well across the different types of sleep-related datasets. On the other hand, the existing models also involve laborious designs such as first extracted the features and selected the suitable features, and then input into a classifier, all these steps need hyper-parameters whose joint effort is quite laborious. On the other hand, the proposed 9 L-1D-CNN-SSC model is an end-to-end system, which is completely based on the deep learning concept, which takes input signals and gives the decision without any kind of feature extraction, selection, and laborious parameter tuning. The model learns the discriminative information automatically from the input signal data and the learning and classification process is completed automatically.
The proposed model not required any types of handcrafted features and shallow learning classification models for the classification of polysomnography signals and it can assist clinicians during the sleep scoring. To date, there is no pre-trained CNN models exist for biomedical signals. Therefore it is one of the major challenges lies here to get large amounts of the biomedical signal dataset. In the future; the authors will collect the biomedical signal data from different medical institutes and test the efficiency of the model. Furthermore, the proposed work to be extended for different types of sleep related diseases. The authors will also include the data augmentation techniques to overcome the data imbalance issues. Finally, it has been observed that the proposed framework improves the existing state of the art and achieves better classification results for the two-five sleep classification tasks. The proposed fully automated sleep staging classification systems could replace the traditional error-prone classification models.
Data availability
All EEG files are available from the ISRUC-SLEEP database (https://sleeptight.isr.uc.pt/ISRUC_Sleep/).
References
Abdollahpour M, Rezaii TY, Farzamnia A, Saad I (2020) Transfer learning convolutional neural network for sleep stage classification using two-stage data fusion framework, in IEEE access 8, 180618-180632, https://doi.org/10.1109/ACCESS.2020.3027289
Acharya UR, Faust O, Kannathal N, Chua T, Laxminarayan S (2005) Non-linear analysis of EEG signals at various sleep stages. Comput Methods Prog Biomed 80(1):37–45. https://doi.org/10.1016/j.cmpb.2005.06.011
Acharya UR, Chua ECP, Chua KC, Min LC, Tamura T (2010) Analysis and automatic identification of sleep stages using higher order spectra. Int J Neural Syst 20(06):509–521. https://doi.org/10.1142/s0129065710002589
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adam M, Gertych A, Tan RS (2017) A deep convolutional neural network model to classify heartbeats. Comput Biol Med 89:389–396. https://doi.org/10.1016/j.compbiomed.2017.08.022
Akyol K. (2020) Stacking ensemble based deep neural networks modeling for effective epileptic seizure detection Expert Systems with Applications, 113239. https://doi.org/10.1016/j.eswa.2020.113239
Tharwat Alaa (2018) AdaBoost classifier: an overview. https://doi.org/10.13140/RG.2.2.19929.01122.
Awujoola O, Francisca O, Odion P (2020) Effective and Accurate Bootstrap Aggregating (Bagging) Ensemble Algorithm Model for Prediction and Classification of Hypothyroid Disease. International Journal of Computer Applications 176:40–48. https://doi.org/10.5120/ijca2020920542
Bajaj V, Pachori RB (2013) Automatic classification of sleep stages based on the time-frequency image of EEG signals. Comput Methods Prog Biomed 112(3):320–328. https://doi.org/10.1016/j.cmpb.2013.07.006
Basha AJ, Balaji BS, Poornima S, Prathilothamai M,Venkatachalam K (2020) Support vector machine and simple recurrent network based automatic sleep stage classification of fuzzy kernel. Journal of ambient intelligence and humanized computing. 10.1007/s12652-020-02188-4
Boashash B, Ouelha S (2016) Automatic signal abnormality detection using time-frequency features and machine learning: a newborn EEG seizure case study. Knowl Based Syst 106:38–50
Carskadon MA, Dement WC (2005) Normal human sleep: an overview. Principles Pract Sleep Med 4:13–23. https://doi.org/10.1016/j.mcna.2004.01.001
Chambon S, Galtier MN, Arnal PJ, Wainrib G, Gramfort A (2018) A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Transactions on Neural Systems and Rehabilitation Engineering 26(4):758–769. https://doi.org/10.1109/tnsre.2018.2813138
Chen Z (2020) Effective and efficient batch normalization using a few uncorrelated data for statistics estimation. IEEE Transactions on Neural Networks and Learning Systems 32:348–362. https://doi.org/10.1109/TNNLS.2020.2978753
Cooray N, Andreotti F, Lo C, Symmonds M, Hu MTM, De Vos M (2019) Detection of REM sleep behaviour disorder by automated polysomnography analysis. Clin Neurophysiol 130:505–514. https://doi.org/10.1016/j.clinph.2019.01.011
Cui Z, Zheng X, Shao X, Cui L (2018) Automatic sleep stage classification based on convolutional neural network and finegrained segments. Hindawi Complex 2018:9248410. https://doi.org/10.1155/2018/9248410
Dimitriadis SI, Salis C, Linden D (2018) A novel, fast and efficient single-sensor automatic sleep-stage classification based on complementary cross-frequency coupling estimates. Clin Neurophysiol 129(4):815–828. https://doi.org/10.1016/j.clinph.2017.12.039
Diykh M, Li Y, Abdulla S (2019) EEG sleep stages identification based on weighted undirected complex networks. Computer methods and programs in biomedicine, 105116. 10.1016/j.cmpb.2019.105116
Faust O, Hagiwara Y, Hong TJ, Lih OS, Acharya UR (2018) Deep learning for healthcare applications based on physiological signals: a review. Comput Methods Prog Biomed 161:1–13. https://doi.org/10.1016/j.cmpb.2018.04.005
Fernandez-Blanco E, Rivero D, Pazos A (2019) Convolutional neural networks for sleep stage scoring on a two-channel EEG signal. Soft Comput 24:4067–4079. https://doi.org/10.1007/s00500-019-04174-1
Fraiwan L, Lweesy K, Khasawneh N, Wenz H, Dickhaus H (2012) Automated sleep stage identification system based on time–frequency analysis of a single EEG channel and random forest classifier, Comput. Methods Progr Biomed 108, 10–19. https://doi.org/10.1016/j.cmpb.2011.11.005u.r.
Fraiwan L, Hassanin O, Fraiwan M, Khassawneh B, Ibnian AM, Alkhodari M (2020). Automatic identification of respiratory diseases from stethoscopic lung sound signals using ensemble classifiers. Biocybernetics and Biomedical Engineering. https://doi.org/10.1016/j.bbe.2020.11.003
Garcés Correa A, Orosco L, Laciar E (2014) Automatic detection of drowsiness in EEG records based on multimodal analysis. Med Eng Phys 36(2):244–249. https://doi.org/10.1016/j.medengphy.2013.07.011
Gevins Alan S (1994) Non-invasive human neurocognitive performance capability testing method and system, U.S. Pat
Ghimatgar H, Kazemi K, Helfroush MS, Aarabi A (2019) An automatic single-channel EEG-based sleep stage scoring method based on hidden Markov model Journal of Neuroscience Methods, 108320. https://doi.org/10.1016/j.jneumeth.2019.108320
Goel N, Rao H, Durmer J, Dinges D (2009) Neurocognitive consequences of sleep deprivation. Semin Neurol 29(04):320–339. https://doi.org/10.1055/s-0029-123711
Guillot A, Sauvet F, During EH, Thorey V (2020) Dreem open datasets: multi-scored sleep datasets to compare human and automated sleep staging. IEEE transactions on neural systems and rehabilitation engineering, 1–1. https://doi.org/10.1109/tnsre.2020.3011181
Güler I, Ubeyli ED (2005) Adaptive neuro-fuzzy inference system for classification of EEG signals using wavelet coefficients. J Neurosci Methods 148(2):113–121. https://doi.org/10.1016/j.jneumeth.2005.04.013
Güler NF, Übeyli ED, Güler İ (2005) Recurrent neural networks employing Lyapunov exponents for EEG signals classification. Expert Syst Appl 29:506–514. https://doi.org/10.1016/j.eswa.2005.04.011
Hassan AR, Bhuiyan MI (2016) A decision support system for automatic sleep staging from EEG signals using tunable Q-factor wavelet transform and spectral features. J Neurosci Methods 271:107–118. https://doi.org/10.1016/j.jneumeth.2016.07.012
Hassan AR, Bhuiyan MIH (2017) An automated method for sleep staging from EEG signals using normal inverse Gaussian parameters and adaptive boosting. Neurocomputing 219:76–87. https://doi.org/10.1016/j.neucom.2016.09.011
Hassan AR, Bhuiyan MIH (2017 Mar) Automated identification of sleep states from EEG signals by means of ensemble empirical mode decomposition and random under sampling boosting. Comput Methods Prog Biomed 140:201–210. https://doi.org/10.1016/j.cmpb.2016.12.015
Hassan AR, Hassan Bhuiyan MI (2016) Automatic sleep scoring using statistical features in the EMD domain and ensemble methods. Biocybernetics and Biomedical Engineering 36(1):248–255
Hassan AR, Subasi A (2017) A decision support system for automated identification of sleep stages from single-channel EEG signals. Knowl-Based Syst 128:115–124. https://doi.org/10.1016/j.knosys.2017.05.005
Hassan AR, Bashar SK, Bhuiyan, MIH (2015) On the classification of sleep states by means of statistical and spectral features from single channel Electroencephalogram. 2015 International conference on advances in computing, Communications and Informatics (ICACCI) https://doi.org/10.1109/icacci.2015.7275950
Hochreiter S, Schmidhuber J (1997 Nov 15) Long short-term memory. Neural Comput 9(8):1735–1780. https://doi.org/10.1162/neco.1997.9.8.1735
Hori T, Sugita Y, Koga E, Shirakawa S, Inoue K, Uchida S, Kuwahara H, Kousaka M, Kobayashi T, Tsuji Y, Terashima M, Fukuda K, Fukuda N (2001) A manual of standardized terminology, techniques and scoring system for sleep stages of human subjects, the rechtschaffen & kales (1968) standard. Psychiatr Clin Neurosci 55:305–310. https://doi.org/10.1046/j.1440-1819.2001.00810.x
Hsu YL, Yang YT, Wang JS, Hsu CY (2013) Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 104:105–114. https://doi.org/10.1016/j.neucom.2012.11.003
Huang CS, Lin CL, Yang WY, Ko LW, Liu SY, Lin CT (2013) Applying the fuzzy cmeans based dimension reduction to improve the sleep classification system, 2013 IEEE Int. Conf. Fuzzy Syst, pp. 1–5,https://doi.org/10.1109/FUZZ-IEEE.2013.6622495
Hussein R, Palangi H, Ward RK, Wang ZJ (2019) Optimized deep neural network architecture for robust detection of epileptic seizures using EEG signals. Clin Neurophysiol 130(1):25–37. https://doi.org/10.1016/j.clinph.2018.10.010
Ieracitano C, Mammone N, Bramanti A, Hussain A, Morabito FC (2018) A convolutional neural network approach for classification of dementia stages based on 2D-spectral representation of EEG recordings. Neurocomputing. https://doi.org/10.1016/j.neucom.2018.09.071
Ieracitano C, Mammone N, Bramanti A, Hussain A, Morabito FC (2018) A convolutional neural network approach for classification of dementia stages based on 2D-spectral representation of EEG recordings. Neurocomputing 323:96–107
Imtiaz SA, Rodriguez-Villegas E (2015) Automatic sleep staging using state machine-controlled decision trees. Conf Proc IEEE Eng Med Biol Soc 2015:378–381. https://doi.org/10.1109/EMBC.2015.7318378
Isaac F-V, Elena H-P, Moret-Bonillo V (2018) A convolutional network for the classification of sleep stages. Proceedings. 2:1174. https://doi.org/10.3390/proceedings2181174
Jadhav P, Rajguru G, Datta D, Mukhopadhyay S (2020) Automatic sleep stage classification using time–frequency images of CWT and transfer learning using convolution neural network. Biocybernetics and Biomedical Engineering. 10.1016/j.bbe.2020.01.010
Jiao Z, Gao X, Wang Y, Li J, Xu H (2018) Deep convolutional neural networks for mental load classification based on EEG data. Pattern Recogn 76:582–595. https://doi.org/10.1016/j.patcog.2017.12.002
Jolliffe I (2011) Principal component analysis. International encyclopedia of statistical science, 1094–1096. https://doi.org/10.1007/978-3-642-04898-2_455.
Kalbkhani H, Ghasemzadeh PG, Shayesteh M (2018) Sleep Stages Classification from EEG Signal based on Stockwell Transform IET Signal Processing 2018
Khalighi S, Sousa T, Oliveira D, Pires G, Nunes U (2011) Efficient feature selection for sleep staging based on maximal overlap discrete wavelet transform and SVM. Annu Int Conf IEEE Eng Med Biol Soc 2011:3306–3309. https://doi.org/10.1109/IEMBS.2011.6090897
Khalighi S, Sousa T, Santos JM, Nunes U (2015) ISRUC-sleep: a comprehensive public dataset for sleep researchers. Comput Methods Prog Biomed 2016(124):180–192. https://doi.org/10.1016/j.cmpb.2015.10.013
Kononenko I (1994) Estimating attributes: analysis and extensions of RELIEF. Lect Notes Comput Sci 171–182. https://doi.org/10.1007/3-540-57868-4_57
Korkalainen H (2020) Accurate deep learning-based sleep staging in a clinical population with suspected obstructive sleep Apnea.IEEE Journal of Biomedical and Health Informatics 24(7), 2073–2081. https://doi.org/10.1109/JBHI.2019.2951346.
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst
Lajnef T, Chaibi S, Ruby P, Aguera PE, Eichenlaub JB, Samet M, Kachouri A (2015) Jerbi K (2015) learning machines and sleeping brains: automatic sleep stage classification using decision-tree multi-class support vector machines. J Neurosci Methods 250:94–105. https://doi.org/10.1016/j.jneumeth.2015.01.022
Lajnef T, Chaibi S, Ruby P (2015) Learning machines and sleeping brains: automatic sleep stage classification using decision-tree multi-class support vector machines. J Neurosci Methods 250:94–105. https://doi.org/10.1016/j.jneumeth.2015.01.022
Li Y, Luo ML, Li K (2016) A multiwavelet-based time-varying model identification approach for time–frequency analysis of EEG signals. Neurocomputing 193:106–114. https://doi.org/10.1016/j.neucom.2016.01.062
Li X, La R, Wang Y, Niu J, Zeng S, Sun S, Zhu J (2019) EEG-based mild depression recognition using convolutional neural network. Med Biol Eng Comput 57(6):1341–1352. https://doi.org/10.1007/s11517-019-01959-2
Li X, La R, Wang Y, Niu J, Zeng S, Sun S, Zhu J (2019) EEG-based mild depression recognition using convolutional neural network. Med biol Eng Comput. https://doi.org/10.1007/s11517-019-01959-2
Lovrek I, Howlett RJ, Jain LC. (2008) Knowledge-based intelligent information and engineering systems. Lect Notes Comput Sci https://doi.org/10.1007/978-3-540-85565-1
Memar P, Faradji F (2018) A novel multi-class EEG-based sleep stage classification system. IEEE Trans Neural Syst Rehabil Eng. 26(1):84–95. https://doi.org/10.1109/TNSRE.2017.2776149
Michielli N, Acharya UR, Molinari F (2019) Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput Biol Med 106:71–81. https://doi.org/10.1016/j.compbiomed.2019.01.013
Mienye ID, Sun Y, Wang Z (2020). Improved sparse autoencoder based artificial neural network approach for prediction of heart disease Informatics in Medicine Unlocked, 100307. https://doi.org/10.1016/j.imu.2020.100307
Mousavi Z, Yousefi Rezaii T, Sheykhivand S, Farzamnia A, Razavi SN (2019). Deep convolutional neural network for classification of sleep stages from single-channel EEG signals. J Neurosci Methods, 108312. https://doi.org/10.1016/j.jneumeth.2019
Nagabushanam P, Thomas George S, Radha S (2019) EEG signal classification using LSTM and improved neural network algorithms. Soft Computing https://doi.org/10.1007/s00500-019-04515-0
Najdi S, Gharbali AA, Fonseca JM (2017) Feature transformation based on stacked sparse auto encoders for sleep stage classification. Technological Innovation for Smart Systems 2017:191–200. https://doi.org/10.1007/978-3-319-56077-9_18
Nakamura T, Adjei T, Alqurashi Y, Looney D, Morrell MJ, Mandic DP (2017) Complexity science for sleep stage classification from EEG. In proceedings of the international joint conference on neural networks, Anchorage, AK, USA, 14–19 may 2017.
Nannapas B, Pichayoot O, Pitshaporn L, Payongkit L, Busarakum C, Jaimchariyatam Nattapong, Chuangsuwanich Ekapol, Chen Wei, Phan Huy, Dilokthanakul Nat, Wilaiprasitporn Theerawit. (2020). MetaSleepLearner: a pilot study on fast adaptation of bio-signals-based sleep stage classifier to new individual subject using Meta-learning.
Nejedly P, Cimbalnik J, Klimes P, Plesinger F, Halamek J, Kremen V, Viscor I, Brinkmann BH, Pail M, Brazdil M, Worrell G, Jurak P (2019) Intracerebral EEG Artifact Identification Using Convolutional Neural Networks. Neuroinformatics. 17(2):225–234. https://doi.org/10.1007/s12021-018-9397-6
Oh SL, Ng EYK, Tan RS, Acharya UR (2018) Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput Biol Med 102:278–287. https://doi.org/10.1016/j.compbiomed.2018.06.002
Tsinalis Orestis, Matthews Paul, Guo Yike, Zafeiriou Stefanos (2016) Automatic Sleep Stage Scoring with Single-Channel EEG Using Convolutional Neural Networks
Parrino L, Grassi A, Milioli G (2014) Cyclic alternating pattern in polysomnography: what is it and what does it mean? Curr Opin Pulm Med 20(6):533–541. https://doi.org/10.1097/MCP.0000000000000100
Peng H, Long F, Ding C (2005) Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27:1226–1238. https://doi.org/10.1109/TPAMI.2005.159
Penzel T, Conradt R (2000) Computer based sleep recording and analysis. Sleep Med Rev 4(2):131–148. https://doi.org/10.1053/smrv.1999.0087
Powers DM (2011) Evaluation: from precision, recall and f-measure to roc, informedness, markedness and correlation
Pudil P, Novovičová J, Kittler J (1994) Floating search methods in feature selection. Pattern Recogn Lett 15:1119–1125. https://doi.org/10.1016/0167-8655(94)90127-9
Rahman MM, Bhuiyan MIH, Hassan AR (2018) Sleep stage classification using single-channel EOG. Comput Biol Med 102:211–220. https://doi.org/10.1016/j.compbiomed.2018.08.022
Rechtschaffen A (1968) A manual for standardized terminology, techniques and scoring system for sleep stages in human subjects. Brain inform service 1968
Reynolds CF, O’Hara R (2013) DSM-5 sleep-wake disorders classification: overview for use in clinical practice. Am J Psychiatr 170(10):1099–1101. https://doi.org/10.1176/appi.ajp.2013.13010058
Rosenberg RS, Van Hout S (2013) The American Academy of sleep medicine inter-scorer reliability program: sleep stage scoring. J Clin Sleep Med 09:81–87. https://doi.org/10.5664/jcsm.2350
Sanders TH, McCurry M, Clements MA (2014) Sleep stage classification with cross frequency coupling. Annu Int Conf IEEE Eng Med Biol Soc 4579-82. https://doi.org/10.1109/EMBC.2014.6944643.
Seifpour S, Niknazar H, Mikaeili M, Nasrabadi AM (2018) A new automatic sleep staging system based on statistical behavior of local Extrema using single channel EEG signal. Expert Syst Appl 104:277–293. https://doi.org/10.1016/j.eswa.2018.03.020
Şen B, Peker M, Çavuşoğlu A (2014) A comparative study on classification of sleep stage based on EEG signals using feature selection and classification algorithms. J Med Syst 38:18. https://doi.org/10.1007/s10916-014-0018-0
Sharma M, Goyal D, Achuth PV, Acharya UR (2018) An accurate sleep stages classification system using a new class of optimally time-frequency localized three-band wavelet filter bank. Computers in Biology and Medicine 98:58–75. https://doi.org/10.1016/j.compbiomed.2018.04.025
Shen H, Ran F, Xu M, Guez A, Li A, Guo A (2020) An automatic sleep stage classification algorithm using improved model based essence features. Sensors 20(17):4677. https://doi.org/10.3390/s20174677
Shuyuan X, Bei W, Jian Z, Qunfeng Z, Junzhong Z, Nakamura M (2015) An improved K-means clustering algorithm for sleep stages classification, 2015 54th Annu. Conf.Soc. Instrum. Control Eng. Japan, pp. 1222–1227m https://doi.org/10.1109/SICE.2015.7285326Ijim
Silveiral T, Kozakevicius J, Rodrigues R (2016) Single-channel EEG sleep stage classification based on a streamlined set of statistical features in wavelet domain. International Federation For. Medical and Biological Engineering.
Simões H, Pires G, Nunes U, Silva V (n.d.) Feature Extraction and Selection for Automatic Sleep Staging using EEG. In Proceedings of the 7th International Conference on Informatics in Control, Automation and Robotics, 3, 128–133. https://doi.org/10.5220/0002950601280133
Sors A, Bonnet S, Mirek S, Vercueil L, Payen JF (2018) A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed Signal Process Control 42:107–114. https://doi.org/10.1016/j.bspc.2017.12.001
Sousa T, Cruz A, Khalighi S, Pires G, Nunes U (2015) A two-step automatic sleep stage classification method with dubious range detection. Comput Biol Med 59:42–53
Sturm I, Lapuschkin S, Samek W, Müller KR (2016) Interpretable deep neural networks for single-trial EEG classification. J Neurosci Methods 274:141–145. https://doi.org/10.1016/j.jneumeth.2016.10.008
Sun C, Chen C, Fan J, Li W, Zhang Y, Chen W (2019) A hierarchical sequential neural network with feature fusion for sleep staging based on EOG and RR signals. J Neural Eng 16:066020. https://doi.org/10.1088/1741-2552/ab39ca
Sun C, Fan J, Chen C, Li W, Chen W (2019) A two-stage neural network for sleep stage classification based on feature learning, sequence learning, and data augmentation. IEEE Access 7:109386–109397. https://doi.org/10.1109/ACCESS.2019.2933814
Supratak A, Dong H, Wu C, Guo Y (2017) DeepSleepNet: a model for automatic sleep stage scoring based on raw Single-Channel EEG. IEEE Trans Neural Syst Rehabil Eng 25(11):1998–2008. https://doi.org/10.1109/TNSRE.2017.2721116
Tagluk ME, Sezgin N, Akin M (2010) Estimation of sleep stages by an artificial neural network employing EEG, EMG and EOG. J Med Syst 34:717–725. https://doi.org/10.1007/s10916-009-9286-5
Tang Z, Li C, Sun S (2017) Single-trial EEG classification of motor imagery using deep convolutional neural networks. International Journal for Light and Electron Optics, 130, 11–18. https://doi.org/10.1016/j.ijleo.2016.10.117
Tripathy RK, Rajendra Acharya U (2018) Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybernetics and Biomedical Engineering 38:890–902. https://doi.org/10.1016/j.bbe.2018.05.005
Tzimourta KD, Tsimbaris A, Tzioukalia K, Tzallas AT, Tsipouras MG, Asktrakas LG, Giannakeas N (2018) EEG based automatic sleep stage classification. Biomed J Sci tech res. https://doi.org/10.26717/BJSTR.2018.07.001535.
Wan H, Wang HB, and Liu J (2019) A novel Gaussian mixture model for classification," 2019 IEEE international conference on systems, man and cybernetics (SMC), Bari, Italy, pp. 3298–3303. https://doi.org/10.1109/SMC.2019.8914215.
Wang Q, Zhao D,Wang Y,Hou X (2019) Ensemble learning algorithm based on multi-parameters for sleep staging. Medical & Biological Engineering & Computing. 10. 1007/s11517-019-01978-z
Wei L, Lin Y, Wang J, Ma Y (2017) Time-frequency convolutional neural network for automatic sleep stage classification based on single-channel EEG, 2017 IEEE 29th Int. Conf. Tools with Artif. Intell, 88–95. https://doi.org/10.1109/ICTAI.2017.00025
Yan R, Zhang C, Spruyt K, Wei L, Wang Z, Tian L, Cong F (2019) Multi-modality of polysomnography signals’ fusion for automatic sleep scoring. Biomedical Signal Processing and Control 49:14–23. https://doi.org/10.1016/j.bspc.2018.10.001
Yildirim Ö (2018) A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput Biol Med 96:189–202. https://doi.org/10.1016/j.compbiomed.2018.03.016
Yildirim O, Baloglu U, Acharya U (2019) A deep learning model for automated sleep stages classification using PSG signals. Int J Environ Res Public Health 16(4):599. https://doi.org/10.3390/ijerph16040599
Zhang T (2021) Sleep staging using plausibility score: a novel feature selection method based on metric learning. IEEE Journal of Biomedical and Health Informatics 25(2):577–590. https://doi.org/10.1109/JBHI.2020.2993644
Zhang S, Li X, Zong M (2017) Learning k, for KNN classification, ACM trans. Intell.Syst Technol 8(3) 43(1–19)
Zhang X, Xu M, Li Y, Su M, Xu Z, Wang C, Han D (2020) Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep and Breathing 24:581–590. https://doi.org/10.1007/s11325-019-02008-w
Zhu G, Li Y, Wen P (2014) Analysis and classification of sleep stages based on difference visibility graphs from a single-channel EEG. IEEE Journal of Biomedical and Health Informatics 18(6), 1813–1821, https://doi.org/10.1109/JBHI.2014.2303991.
Zhu T, Luo W, Yu F (2020) Convolution-and attention-based neural network for automated sleep stage classification. Int J Environ Res Public Health 17(11):4152. https://doi.org/10.3390/ijerph17114152
Funding
This study is not funded by any organization.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
All procedures performed in studies involving human partici-pants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflict of interest
All the authors in the paper have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Satapathy, S.K., Loganathan, D. Automated classification of multi-class sleep stages classification using polysomnography signals: a nine- layer 1D-convolution neural network approach. Multimed Tools Appl 82, 8049–8091 (2023). https://doi.org/10.1007/s11042-022-13195-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13195-2