Abstract
Skin cancer is the most prevalent genre of all cancers. Melanoma, being the deadliest of all skin cancers, calls for the requirement of an automated Artificial Intelligence-based skin diagnosis system to assist physicians with early diagnosis. We propose a fusion of conventional therapeutic approaches and deep learning frameworks to identify skin lesions. The work explores the scope of employing image data, handcrafted lesion features, and patient-centric metadata together to diagnose skin cancers effectively. We combined the image features transfer-learned from EfficientNets, colour and texture information extracted from the images, and patients’ preprocessed metadata to produce the final hybrid model. They were fed to a multi-input single-output (MISO) model to fine-tune an artificial neural network classifier. Multiple MISO models were trained with their backbones substituted with EfficientNets B4 through B7. The predicted labels from these, along with a separate set of models trained with only image data and metadata were ensembled using majority soft voting. We experimented with weighing the models based on their contribution to ensemble accuracy and ensemble sensitivity. Each model was trained and evaluated using the well-known ISIC2018 and ISIC2019 datasets. The extreme imbalance in the datasets necessitates the use of appropriate evaluation metrics. ISIC2018 tested 90.49% sensitive and 97.76% specific, whereas the larger and more divergent dataset ISIC2019 rated 85.58% sensitive and 98.29% specific. The network is by far the finest compared to most other research in the field.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Skin lesions are abnormal skin cells that occur due to increased exposure to the sun’s harmful ultraviolet rays. WHO reports that, on a global scale, one in every three cases of cancer diagnosed currently is skin cancer. Melanoma infections are the most malignant of all forms of skin cancers. However, skin lesions have an interesting 5-year melanoma survival rate of 95% on early detection that falls to 20% if left untreated. The importance of early detection of Melanoma has been highlighted due to the rising number of cases and the ever-increasing mortality rate. Skin lesions are detectable only through an expert visual inspection. Using Machine learning and Deep learning concepts, skin lesion diagnosis could be automated efficiently with the availability of high-resolution dermoscopic images. The International Skin Imaging Collaboration (ISIC) initiated the ISIC grand challenge happening every year since 2016, aiming the researchers’ community all around the world to contribute towards the cause of efficient skin cancer detection and analysis [16].
There are over 2,000 kinds of skin cancers identified. The general hierarchy of lesions is shown in the supplementary material (Figure S1). Broadly categorized as benign and malignant, the former lesions are non-cancerous skin growths that are mere birthmarks or rashes uniformly formed on human skin. In contrast, malignant lesions are the fast-growing and irregular-shaped cancerous category. Physicians perform clinical tests based on certain standard procedural rules such as the ABCDE rule (asymmetry, border, shape, colour, diameter, and evolution of lesion), the CASH rule (colour, architecture, symmetry, and homogenous nature of the lesion), or the Glasgow’s 7-point checklist which includes seven yes/ no questions related to changes in size, shape irregularities, and infection scales. The past decade saw the importance of automating skin cancer detection for early and accurate diagnosis with the availability of high resolution dermoscopic images [6]. Table 1 briefs the various state-of-the-art models published in reputed journals over the years.
1.1 Machine learning tools for diagnosis
The conventional therapeutic approaches based on dermoscopy and lesion verification rules have influenced diagnosis automation using digital dermoscopic images. Zghal et al. [28], and Monika et al. [20] extracted features similar to ABCDE and CASH from affected regions of skin lesion images. These were then used to train different machine learning classifiers for effective automated diagnosis. A limited supply of look-alike data explains the appreciable performance of these models. In Ghalejoogh et al., patterns were captured using texture descriptors from grey converted dermoscopic images that were fed into an ensemble classifier [9]. Hameed et al. concluded that as the complexity of classifiers increased, so does the capability of a model in recognizing classes [13]. Most models were evaluated using the accuracy metric that should never be chosen as the evaluation metric for imbalanced datasets. Furthermore, the abundance of skin lesion images with high inter-class similarity makes it difficult to identify unique and distinguishable custom characteristics.
1.2 Deep learning tools for automation
Deep neural architectures are known to project high efficiency with the increase in the number of layers to capture the latent dynamics of input data [3]. Moreover, The inconveniences of tweaking the hyperparameters of a freshly created network are reduced by transferring the knowledge of pre-trained models. Kassem et al. [18] explore the impact of GoogLeNet in the ISIC2019 dataset by fine-tuning all architecture layers of the network [18]. Nahata et al. performed a comparative analysis of skin lesion classification using several pre-trained networks [21]. The lack of sufficient data was observed to underfit these over-constrained pre-trained models. Researchers also use metadata included in the dataset to enhance detection rates. They are often processed in DenseNets.
The current research trend curve appears to be biased toward aggregating predictions from numerous pre-trained models to improve performance and decrease outcome uncertainties. This strategy named ensemble technique has also been explored widely in the latest ISIC skin lesion detection challenges. Ha et al. present their winning approach to the ISIC Melanoma detection challenge 2020. The image datasets were augmented and trained using an ensemble of networks in a 5-fold validation strategy [12]. However, the task here is to categorize lesions as benign or malignant, while real-life situations demand the diagnosis of more specific lesion types. Gessert et al. describe the winning solution to the ISIC 2019 skin lesion classification challenge. The authors explored an ensemble of multiresolution networks trained over extensive data augmentation, and loss balancing of data [7]. Gessert et al. also project their runner-up solution to the ISIC 2018 challenge that combines multiple networks in a 5-fold cross-validation scheme [8]. The usage of unscaled images during training ensured detailed feature extraction at the cost of computation. The method adopted by Harangi et al. had the disadvantage of the limited supply of data [15]. It is common practice to combine several publicly available datasets to train complex pre-trained networks [7, 12, 24]. Gong et al. addressed the data imbalance by generating fake sample images of classes with fewer samples using General Adversarial Networks (GAN)s to produce a highly accurate but insensitive model [10]. The model’s poor true positive rate (sensitivity) explains how the model is biased towards the greater class.
Melanoma diagnosis poses several obstacles and opportunities. High interclass characteristic resemblances only make it a more challenging task. We noted that pre-trained models outperformed machine learning techniques showcasing promising results. Rather than using a conventional single neural network, combining the individual performances of several networks seeks to value the goodness of each network outcome and generates remarkable predictions. Furthermore, choosing an optimal evaluation metric suitable for imbalanced data seems to influence research in the area. They are often misleading if chosen wrong.
We merged machine learning and deep learning concepts to create the best representations of skin lesions to perform their categorization. The classification task was accomplished by combining several multi-input models using the weighted ensemble strategy. The architecture was trained and tested on public standard datasets to authenticate the model’s novelty. It has also been compared with the state-of-the-art models from the literature. This model overcame the challenges and delivered a strong performance.
The main contributions of the work could be summarized as:
-
The fusion of neural network features, extracted features, and patient metadata to classify skin lesion dermoscopic images.
-
A weighted majority voting strategy based on ensembled accuracy and ensembled sensitivity of the participating models is explored.
-
The method is proven by performance comparison with benchmarked datasets and state-of-the-art models.
-
Overall, we have developed an automated skin lesion analysis approach that is reliable, and time-efficient capable of identifying even the rarest cases of skin cancers.
2 Datasets
This research aims to combine image data, lesion-specific handcrafted features, and patient-specific metadata in an ensemble of networks to diagnose skin lesions. We used the well-known ISIC2018, and ISIC2019 datasets of the International Skin Imaging Collaboration (ISIC) challenge [17]. Both datasets have patient-specific metadata associated with each of the skin lesion images contained in the dataset. They were validated separately to compare the model’s performance on entirely different datasets.
2.1 Data statistics
Specifications of both repositories are given in supplementary material (Table S1). The ISIC2018 dataset has 7 skin lesion classes of which 5 are benign, and 2 belong to the cancerous category, namely Melanoma and Basal Cell Carcinoma [26]. Besides the class divisions from ISIC2018, ISIC2019 has an additional cancerous category, the Squamous Cell Carcinoma [4, 5, 26]. We split the two datasets into the ratio 8:1:1, 8 parts assigned to training, and the rest split among validation and test sets. Table 2 sets down the number of lesion images belonging to each class under the train, validation, and test sets. A huge imbalance in the two datasets was observed, with more than half of the data belonging to the Melanocytic Nevus category. Approximately 66% of data in ISIC2018 belong to the nevi benign class, whereas it is around 50% in ISIC2019. This means that even if the entire dataset is categorized as the most frequent class, the model would be as accurate as the ratio of the largest occurring class.
The distribution of skin cancer patients under different categories of metadata values from ISIC2018 and ISIC2019 are illustrated in the supplementary material (Figure S2). Networks reduce entire images into their most abstract representations, and handcrafted features assume the human way of looking at problems, whereas metadata corresponds to an entirely different dimension, ’patient’. Metadata also prevents over-fitting caused due to the intense training of image data alone. Moreover, a physician’s diagnosis would always include patient data, and it is only intuitive that metadata adds to the performance of an artificially intelligent model.
3 Methodology
We propose a hybrid approach involving deep learning and machine learning techniques. Handcrafted features from skin lesion images and the clinical metadata included in the dataset are trained alongside their corresponding images.
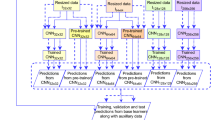
Figure 1 schematically depicts the colour-coded outline of our skin lesion diagnosis model. The blocks in blue represent the image training network. The images were passed through EfficientNets and compressed into abstract representations named feature maps. The pooling layer followed by batch normalization and dropout layers avoids the overfitting of the network to the training data. The feature maps were then flattened and passed onto the classification layers. The orange blocks illustrate the extraction of handcrafted features. The raw images were preprocessed using the dull razor method to remove human hair strands and noise particles. Lesions were then segmented using a UNet segmentation architecture from which colour and texture features were handcrafted by computing the colour variegation and GLCM statistics. A total of 8 distinct features were extracted from each image. Blocks in red elaborate on the metadata preprocessing procedure. We identified and preprocessed useful columns in the metadata file, which were further combined with the handcrafted features into a single vector of feature values. They were then passed through a pair of dense layers. Features from this layer and the flattened features from EfficientNets were concatenated and passed through a multi-input fully connected network with two layers. The final layer with the softmax activation function produces a probability distribution vector for all classes. The predictions from multiple models backed by different variants of EfficientNets were ensembled using soft majority voting and weighted majority voting techniques.
3.1 Image training
We fine-tuned EfficientNets to skin cancer-specific datasets ISIC2018 and ISIC2019. EfficientNets were published by Google in the year 2019 [25]. It is a convolutional neural network that employs a novel compound scaling technique to increase the efficiency and accuracy of the network. Compound scaling is an aggregative strategy where the depth, width as well as resolution of the network are scaled uniformly using a single compound coefficient 𝜃 (1). EfficientNet variants are generated by incrementing the value of 𝜃.
EfficientNet B4, B5, B6, and B7 variants were exercised in this work. As only the skeletal architecture of EfficientNets was required, we intentionally removed the final layers at the head of the networks. It was observed that the space and time complexities increased in the higher variants of EfficientNet.
3.2 Handcrafted Feature Extraction
Here, we draw out image-specific features from the skin lesion dataset to assist the classification process. The following subsections describe each step in detail.
3.2.1 Image Pre-processing
All images were preprocessed to enhance the image quality such that unwanted distortions are reduced, and aspects important to the application are improved. Since human skin hair is an artifact of distortion that may hinder the proper extraction of features, we employed the dull razor hair removal algorithm. Figure 2 illustrates the flow of the dull razor hair removal procedure.
Initially, a black hat transformation is applied that uncovers minute components in an image. The grey-converted image I is morphologically closed with a structuring element k of size 5 × 5. It is further subtracted from the original image (2). A threshold t is obtained in the process that best describes the separation of highlighted objects from the background.
Next, a binary threshold is applied to the black hat transformed output BT(I) based on the threshold t as in (3). Conceptually, binary 1 represents noise or hair segments, whereas binary 0 represents skin regions, including the lesion areas. Finally, the binary thresholded mask TIp is inpainted into the original image with a masking radius value of 3. It fills in the masked pixels with the surrounding pixels from the original image.
3.2.2 Lesion segmentation
We custom-trained the UNet architecture using the dataset available. UNet is an asymmetric and fully convolutional network comprising an encoder and a decoder. While the encoder reduces spatial dimension and records the image context using a series of convolution and pooling operations, the decoder regenerates binary masks from feature maps using transposed convolution operations. The knowledge from previous encoder layers is incorporated into the decoder layers through skip connections.
Our encoder and decoder had five blocks each. Each block included a pair of convolution, batch normalization, and pooling layers followed by ReLU activations. The network was optimized with the Adam optimizer at a learning rate of 0.001. The binary cross-entropy loss function calculated the prediction error among the intensity values of the predicted mask and the ground truth intensities. We used the ISIC2018 dataset split in the ratio 8:2 to train and validate the network. The dataset has ground-truth masks associated with all its images, while ISIC2019 has none. For this reason, the trained UNet model was simply used to generate masks for the ISIC2019 dataset.
Mean pixel Accuracy (Acc_pix) and Intersection over Union (IoU) metrics were used to evaluate the UNet (4). While mean accuracy is the average number of correctly predicted pixels in the generated binary mask, IoU calculates the percentage of area covered by the predicted and ground truth masks.
3.2.3 Colour variegation
We extracted the colour variegation to represent the occurrence of various hues and colour tones in each lesion. Colour variegation in skin lesions increases as they turn more cancerous and could potentially be an influential discriminator of skin lesions.
Assuming the image is in the RGB colour space, the standard deviation of the intensity distribution across each channel was calculated and normalized separately (5). The value quantifies the dispersion of image intensities concerning their mean.
3.2.4 Grey level co-occurrence matrix
GLCM [14] estimates the textural characteristics of an image using second-order statistical characteristics. It is a histogram of co-occurring greyscale values at a predetermined offset. Each element (i,j) in the matrix is the frequency with which a grey level i co-occurs with grey level j at a distance d in the direction 𝜃. Since the non-cancerous classes of skin lesion have close textural patterns, we considered four angles 𝜃 = \(0,\ \frac {\pi }{2},\ \pi ,\ \frac {3\pi }{2}\) with a pixel spacing of 1 to extract GLCM features. Further, the contrast, energy, homogeneity, dissimilarity and correlation second-order statistics (denoted as S1, S2, S3, S4, and S5) were measured from the normalized GLCM.
We calculated the five statistics separately for each of the co-occurrence matrices (ie, about all angles of co-occurrence enquiry) denoted as \(S_{k}\rvert _{0}\), \(S_{k}\rvert _{\frac {\pi }{2}}\), \(S_{k}\rvert _{\pi }\), \(S_{k}\rvert _{\frac {3\pi }{2}}\), where k = (1 to 5). A single representation of each statistic \(\mu _{S_{k}}\) was obtained by averaging similar statistics across all co-occurrence matrices (6).
3.3 Metadata encoding
The lack of standard procedures in collecting metadata leaves a lot of missing values in the data. We undertook various data cleaning and pre-processing measures to identify and handle such dispensable data for the smooth functioning of the model.
-
1.
Feature selection- the insignificant attributes that could misguide the classification such as lesion id and diagnosis type were removed from both datasets.
-
2.
Handling missing values- The mean substitution and the maximum frequency imputation techniques were employed to handle missing fields in the numerical and categorical attributes. While the mean substitution method filled all the zeroes and non-numerical values with the average value across the column, maximum frequency imputation filled missing values with the most frequently occurring category in the entire attribute column.
-
3.
Metadata encoding- Unlike numerical values, categorical values need to be converted to numerals to process them. One-hot encoding was implemented where additional columns were generated based on the unique categorical values. Binary 1 in a one-hot encoded row indicates the existence of a category while the rest of the categories are set to 0.
3.4 Combined feature normalization
The preprocessed metadata and the extracted features were combined to produce a total of 23 features for ISIC2018 and 28 features for ISIC 2019. The value difference is due to the extra number of anatomical site categories in the ISIC2019 dataset. Attributes with a wide range of values affect a model to bias towards them. To equalize the contribution of all features, we used Min-Max normalization, ensuring that the entire feature set values are transformed to the [0,1] scale similar to the one-hot encoded features (7). The scaling aids the training of networks to be more stable and quicker.
3.5 Architecture design
To evaluate a scenario, the human brain tries to connect information acquired from the different senses. Similarly, a network architecture capable of processing multiple input data from the same source is anticipated to outperform its single input counterparts. A Multi-Input Single-Output (MISO) model was built to take in image data and their corresponding numerical metadata and give out categorical lesion classes.
As depicted in Fig. 3, the EfficientNet model was trained using the lesion images. The compiled metadata was transformed into their latent representations using a multilayer perceptron with two dense layers comprising 256 neurons in the second branch. Each layer was followed by Batch normalization and ReLU activation layers. A dropout of 25% after the first dense layer tried to generalize metadata into the network, thereby avoiding possible overfitting of data. We designed a custom generator that generates mini-batches of skin lesion images and their corresponding metadata for network training. A similar custom test data generator was also defined for the sequential generation of singular data.
A total of 1280 features from the CNN branch and an additional 256 features from the MLP branch were combined to train the classification layers. The initial fully connected dense layer followed by the Batch Normalization layer, ReLU activation, and a Dropout rate of 40% transformed the (1280 + 256) input features into their 1024 latent representations. A second dense layer with the softmax activation deduced confidence scores of each class. The predictions are obtained in probability values that correspond to the confidence with which the input data belongs to a specific skin lesion class.
The function σ generates a vector of n confident scores, one for each skin lesion class l (8). The label corresponding to the maximum probability score is the final predicted skin lesion class.
3.6 Majority voting ensemble technique
Neural networks have very high variance as the hyperparameters of a model keep tuning each time the network is trained. This can affect the performance of a model. We combined the predictions from several trained models based on majority voting to reduce variance and improve predictions.
Majority voting
This soft voting method asserts that the final predicted skin lesion class would be the label associated with the maximum of the summed up probability values for each class label i = 1 to n across all models j = 1 to m (9).
Weighted Majority Voting:
This technique demands that weights be assigned to each model based on their contribution to the most error-free predictions. Unlike simple majority voting, confidence scores generated by model j are weighted wj times prior to voting (Equation 10).
Estimation of model weights
An optimal set of model weights needs to be computed before weighted majority voting. A grid search is performed in the vector of all possible weight combination the models could be assigned to. The process searches for the unique weight combination that produces the finest prediction set.
We evaluated the predictions for each combination of weights using (10). Simultaneously, we computed the accuracy and sensitivity for the new predictions and stored them alongside the combinations worked. Once all combinations were operated with, the weight vectors Wa and Ws associated with the maximum accuracy and the maximum test positive rate were determined to be the optimal model weights (11).
3.7 Evaluation metrics
Our model was assessed using Mean Sensitivity, Specificity, and the Balanced Accuracy metrics to focus on individual classes. These are determined from a confusion matrix plotted against the actual lesion classes for the predicted lesions. True positives and negatives are the numbers of lesion classes rightly predicted, while false positives and negatives are those that are incorrectly predicted. Sensitivity measures the true positive rate, evaluating the model’s capability to rightly categorize persons suffering from a disease class. Conversely, specificity or the true negative rate measures the model capacity to categorize persons without the condition.
Using (12) and (13), we calculated TPR and TNR for each class i, which was further averaged to obtain the mean sensitivity and mean specificity. Balanced accuracy (BA) was computed by averaging the true positive and true negative rates (14). BA gives a sense of how sensitive and specific the model concerns disease diagnosis.
4 Results and discussion
We trained and tested the proposed approach on the train and test splits of the ISIC2018 and the ISIC2019 datasets separately. There are three separate modules (the image training, the handcrafted feature extraction, and the metadata preprocessing modules) conjoined by the classification entity.
4.1 Network settings
The hyperparameters of the model set for training are given in supplementary material (Table S2). Each model was trained for 40 epochs with a batch size of 32 per step. The model was transfer-learned initially for 10 epochs at a learning rate of 1e − 3. Here, all the layers except the final classification layers were frozen. Later, they were fine-tuned for another 30 epochs at a learning rate of 1e − 4 to capture data-specific information within the network parameters. This is done by unfreezing a preset number of layers as given in the supplementary material (Table S3). We reduced the learning rate by half for effective learning during the process, whenever the validation loss did not improve for three continuous epochs. The Adam optimizer was configured with the following settings: alpha rate (initial step size of descent)= 0.001, beta_1, and beta_2 (exponential decay rates of the first and second-moment computations)= 0.9 and 0.999. The classification error was computed as the Categorical Cross Entropy loss function. It compares the two probability distributions (true and predicted) and determines the difference between them during the training process (15).
We performed data augmentation of images to regularize the model and avoid over-fitting problems. Each image was transformed by randomly rotating, translating, and flipping them by a factor of 0.1 each. Another strategy used for data balancing was introducing class weights during the training of the two datasets. Table 3 displays the weights assigned to each class. The model considers the minority classes with greater weightage, thereby maintaining a balance in the data.
4.2 Data preparation
All the images were initially resized to 224 × 224 to maintain integrity in comparing the different variants of EfficientNets. Before extracting features, the images were preprocessed using the dull razor hair removal algorithm. We customized the UNet architecture with a validation accuracy of 70.23% and IoU of 75.21%. The segmented masks were superimposed on the original image to obtain the region of interest (i.e. lesion areas). The colour and texture feature corresponding to the images were extracted and stored alongside the preprocessed metadata. Figure 4 shows the initial records of the normalized and combined metadata.
4.3 Training and validation
The following experiments were conducted by feeding data from a single dataset at a time.
-
1.
We trained four MISO models (pillared by EfficientNets B4 through B7) using image data, handcrafted feature (denoted as h_feat), and metadata (denoted as meta).
-
2.
Similarly, a separate set of multi-input models was trained that accepted only images and the processed metadata to compare the performance exempting handcrafted features.
-
3.
Further, the predictions from the model sets were ensembled using the majority voting technique.
-
4.
An optimal set of model weight vectors [Wa] and [Ws] were obtained by performing a grid search on every possible combination of weights where each model could be assigned values in the range [0, 1.0].
-
5.
We then weighted and ensembled the predictions to produce systems that are most accurate (max acc) and most sensitive (max tpr).
The training and validation loss curves of the first set of experiments on ISIC2018 and ISIC2019 are presented in Fig. 5. Similarly, the training and validation accuracy plots of the same experiments are provided in the supplementary material (Figure S3). The training curve depicts how well the model fits the training data, while the validation curve describes the behaviour of the trained model on unseen data [2]. It could be inferred that the validation was consistent with training, thereby eliminating any possibility that the model is overfitted to the train data. Moreover, the models improved after the initial 10 epochs (i.e., transfer learning), explaining how the latent features of the lesion images were captured in the fine-tuning phase. It was also observed that the models had converged at around 30 epochs as the validation curve remained static later. Any further training would be in vain as it simply would increase the computational complexity.
4.4 Model evaluation
ISIC2018- The most sensitive system was the ensemble model weighted based on maximum sensitivity with a remarkable mean sensitivity of 90.50% and balanced accuracy of 94.13% (Table 4). It was observed that certain models exhibit similar performance. However, the significance of each model relies hugely on the optimal weight vectors Wa and Ws. It was also noticed that the simple voting strategy could only perform almost equivalent to some of the individual models. The weighted ensemble based on maximum accuracy had performed comparatively better than the individual models. The normalized confusion matrices of the different ensemble methods exhibit the percentage of each category classified (Fig. 6). The Receiver Operating Characteristic (ROC) curves at different classification thresholds for the predictions using ensemble models as a one-class versus rest-of-the-classes case were also plotted. We perceived that the classes with the least representation (DF and VASC) were accurately captured in the TPR-based weighted voting technique. However, the significance of a skin cancer detection model lies in detecting cancerous classes (BCC and MEL). They were categorized remarkably by recognizing exactly 47 out of 50 BCC cases and 94 out of the 112 Melanoma cases.
ISIC2019
- Here, the ensemble strategy based on maximum sensitivity outperformed the other models by a slight margin (Table 5). However, we could infer that the ensemble strategy, in general, outmatches the individual networks. In contrast, their independent performances were limited to 89.00%, and the ensemble performed with \(\sim \)91.00% accuracy. The normalized confusion matrices and ROC curves of the three ensembles are plotted in Fig. 7. They perform similarly to their ISIC2018 counterparts, except for classification performance on the new class SCC. Squamous cell carcinoma is a cancerous category that contributes to the significance of the model. The model’s reduced sensitivity towards SCC could have occurred due to its high visual similarity with the BCC and BKL categories. Overall, the model exhibits immense potential in identifying skin lesions with an accuracy of 91.93%.
The performance of the model with the two datasets is comparable. From the ROC curves, the Area under the Curve (AUC) of each model was computed to be over 98.00%. The disparity in performance evaluation might be since ISIC2019 is a composite of different standard skin cancer datasets, whereas ISIC2018 is relatively homogeneous.
4.5 Discussion
The proposed model results conclude the hybrid ensemble approach to be an effective skin lesion classification method. To illustrate the quality of research, we compared the model with relevant works in the area performed using the same data set. Entries in the ISIC 2018 challenge were evaluated with the balanced accuracy metric. Gessert et al. secured the second position with a performance of 85.1% by employing an ensemble model integrated with metadata [8]. Milton et al. [19] and Shahin et al. [22] also experimented with ensembles on the open-sourced ISIC2018 dataset. Almaraz-Damian et al. showcase the significance of the fusion of handcrafted features with an image training network [1]. Our model outperforms all methods by a margin of at least 6% (Table 6).
We also compared the performance of the proposed hybrid multi-input single-output model on the ISIC2019 dataset with a few of the top submissions in the 2019 skin lesion analysis challenge (Table 7). Mean sensitivity and specificity being the evaluation metrics of the competition, Gessert et al. won the challenge with an exceptional tpr-fpr rate by employing ensembles of multiresolution EfficientNets [7]. While Valiuddin et al. [27] and Guissous et al. [11] failed to implement any data balancing schemes, data augmentation as the only data proportioning strategy could not keep up the network performance in Steppan et al. [24].
Our proposed model outperformed the state-of-the-art models that were compared with. Including the patient and lesion-centric data in the model brought forth the much-needed edge during classification. It was fascinating how metadata and custom features influenced skin cancer detection. However, it was also discovered that the suggested framework works better on the smaller dataset ISIC2018, which is likely due to the diverse nature of the bigger dataset. Moreover, the prediction of Melanoma in both datasets is a whopping 84% and 85%, respectively. This is way higher given that the clinical melanoma recognition rate is only 70% [23]. A downside to the model could be that many of the benign Nevi class is being categorized as Melanoma. However, this does not pose any potential risk as the automated system is proposed as a physicians’ aid to initial diagnosis.
5 Conclusion
We present a novel Artificial Intelligence-based classification model for skin lesion detection and classification based on ensembles of networks. Deep learning has made it possible to build and deploy intelligent medical diagnosis and classification systems using all kinds of imaging modalities available at present. Moreover, they have proven beneficial in improving diagnostic accuracy. The hybrid model combines lesion images, their custom-made features, and relevant patient metadata for the effective diagnosis of the various skin cancer classes. It is trained and tested on the well-known, highly imbalanced International Skin Imaging Collaboration (ISIC) challenge datasets of 2018 and 2019. We extracted highly representational handcrafted features from lesion images by implementing segmentation and feature extraction algorithms. Various data balancing and regularization techniques were performed to enhance the model sensitivity towards all classes. Transfer learning and fine-tuning fit well for the compiled architecture training. The suggested weighted majority voting strategy wrings out the goodness of each network and escalates the model performance way more than anticipated. Furthermore, we could infer that the ensemble model is consistent as it predicts well on the two datasets trained and evaluated separately.
No other works incorporate handcrafted features from the lesion images into the training process, along with image data and the patient metadata. The suggested study outperforms the state-of-the-art models against which it was assessed. It, however, appears to function better on the smaller dataset by a slight margin, probably due to the diverse nature of the larger dataset.
The technique could be extended by exploring other ensemble techniques such as the k-fold cross-validation and integrated stacking. Besides the colour and texture features extracted from skin lesions, other representational features such as the boundary symmetry and circularity of lesions might contribute to the model capability. Additionally, it would be interesting to examine the performance of the proposed model with other imaging modalities requiring a similar task.
References
Almaraz-Damian J. -A., Ponomaryov V., Sadovnychiy S., Castillejos-Fernandez H. (2020) Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual information measures. Entropy 22(4):484
Anand H. S., Vinod Chandra S. S. (2016) Association rule mining using treap. International Journal of Machine Learning and Cybernetics 9(4):589–597
Aswathy A. L., Anand H. S., Vinod Chandra S. S. (2021) Covid-19 diagnosis and severity detection from ct-images using transfer learning and back propagation neural network. Journal of Infection and Public Health 14(10):1435–1445
Codella N.C., Gutman D., Celebi M.E., Helba B., Marchetti M.A., Dusza S.W., Kalloo A., Liopyris K., Mishra N., Kittler H., et al. (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 168–172. IEEE
Combalia M., Codella N. C., Rotemberg V., Helba B., Vilaplana V., Reiter O., Carrera C., Barreiro A., Halpern A. C., Puig S., et al. (2019) Bcn20000: Dermoscopic lesions in the wild. arXiv:1908.02288
Dugonik B., Dugonik A., Marovt M., Golob M. (2020) Image quality assessment of digital image capturing devices for melanoma detection. Appl. Sci. 10(8):2876
Gessert N., Nielsen M., Shaikh M., Werner R., Schlaefer A. (2020) Skin lesion classification using ensembles of multi-resolution efficientnets with meta data. MethodsX 7:100864
Gessert N., Sentker T., Madesta F., Schmitz R., Kniep H., Baltruschat I., Werner R., Schlaefer A. (2018) Skin lesion diagnosis using ensembles, unscaled multi-crop evaluation and loss weighting. arXiv:1808.01694
Ghalejoogh G. S., Kordy H. M., Ebrahimi F. (2020) A hierarchical structure based on stacking approach for skin lesion classification. Expert Syst. Appl. 145:113127
Gong A., Yao X., Lin W. (2020) Classification for dermoscopy images using convolutional neural networks based on the ensemble of individual advantage and group decision. IEEE Access 8:155337–155351
Guissous A. E. (2019) Skin lesion classification using deep neural network. arXiv:1911.07817
Ha Q., Liu B., Liu F. (2020) Identifying melanoma images using efficientnet ensemble: Winning solution to the siim-isic melanoma classification challenge. arXiv:2010.05351
Hameed N., Shabut A. M., Ghosh M. K., Hossain M. A. (2020) Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert Syst. Appl. 141:112961
Haralick R. M., Shanmugam K., Dinstein I. H. (1973) Textural features for image classification. IEEE Transactions on systems, man, and cybernetics (6), pp. 610–621
Harangi B. (2018) Skin lesion classification with ensembles of deep convolutional neural networks. Journal of biomedical informatics 86:25–32
ISIC Challenge. https://challenge.isic-archive.com/
ISIC Challenge Datasets. https://challenge.isic-archive.com/data/
Kassem M. A., Hosny K. M., Fouad M. M. (2020) Skin lesions classification into eight classes for isic 2019 using deep convolutional neural network and transfer learning. IEEE Access 8:114822–114832
Milton M. A. A. (2019) Automated skin lesion classification using ensemble of deep neural networks in isic 2018: Skin lesion analysis towards melanoma detection challenge. arXiv:1901.10802
Monika M. K., Vignesh N. A., Kumari C. U., Kumar M., Lydia E. L. (2020) Skin cancer detection and classification using machine learning. Materials Today: Proceedings 33:4266–4270
Nahata H., Singh S. P. (2020) Deep learning solutions for skin cancer detection and diagnosis. Machine Learning with Health Care Perspective, pp. 159–182
Shahin A.H., Kamal A., Elattar M.A. (2018) Deep ensemble learning for skin lesion classification from dermoscopic images. In: 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), pp. 150–153 . IEEE
Sondermann W., Zimmer L., Schadendorf D., Roesch A., Klode J., Dissemond J. (2016) Initial misdiagnosis of melanoma located on the foot is associated with poorer prognosis. Medicine 95(29)
Steppan J., Hanke S. (2021) Analysis of skin lesion images with deep learning. arXiv:2101.03814
Tan M., Le Q. (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114 . PMLR
Tschandl P., Rosendahl C., Kittler H. (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data 5(1):1–9
Valiuddin M. (2019) Using the efficientnet convolutional neural network architecture for skin lesion analysis and melanoma detection a submission for the ISIC2019 challenge
Zghal N. S., Derbel N. (2020) Melanoma skin cancer detection based on image processing. Current Medical Imaging 16(1):50–58
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by the author.
Conflict of interests
The authors declare that they have no conflict of interest.
Consent for publication
Yes
Additional information
Availability of data and materials
The datasets are downloaded from the ISIC repository https://challenge.isic-archive.com/data/
Code availability
http://mirworks.in/downloads.php
Consent to participate
Not applicable
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharafudeen, M., S., V.C.S. Detecting skin lesions fusing handcrafted features in image network ensembles. Multimed Tools Appl 82, 3155–3175 (2023). https://doi.org/10.1007/s11042-022-13046-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13046-0