Abstract
Convolutional Neural Networks (CNNs) are widely employed in the medical imaging field. In dermoscopic image analysis, the large amount of data provided by the International Skin Imaging Collaboration (ISIC) encouraged the development of several machine learning solutions to the skin lesion images classification problem. This paper introduces an ensemble of image-only based and image-and-metadata based CNN architectures to classify skin lesions as melanoma or non-melanoma. In order to achieve this goal, we analyzed how models performance are affected by the amount of available data, image resolution, data augmentation pipeline, metadata importance and target choice. The proposed solution achieved an AUC score of 0.9477 on the official ISIC2020 test set. All the experiments were performed employing the ECVL and EDDL libraries, developed within the european DeepHealth project.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Melanoma is a malignant tumor of the skin which shows a constantly growing incidence all over the world. Numerous studies suggest it doubled in the last 10 years. According to epidemiological studies presented by the GLOBOCAN (Global Cancer Observatory), melanoma has reached more than 320 000 new cases per year worldwide [32], with a slightly higher incidence among men (170 000 cases versus 150 000 cases among women). In particular, in Italy the estimate of melanomas is nearly 7 300 new cases per year among men and 6 700 among women, mainly affecting people aged between 30 and 60 years [21].
Although melanoma represents only a small percentage of tumors affecting the skin, it is responsible for most of skin cancer deaths. According to the statistics of 2020 [32], in Europe the mortality rate for melanoma cancer is 46.2%. Early diagnosis plays an essential role in treatment. Unfortunately, the recent COVID-19 pandemic and the consequent lockdown had an overall negative impact regarding timely diagnosis, leading to the increase of waiting time in the hospitals booking agendas as well as to medical examinations canceled by patients themselves due to their fear of COVID-19 infections.
The need for early and accurate diagnosis combined with the new possibilities offered by modern digital dermatoscopes, justifies the significant increase of publications regarding the use of machine learning algorithms in the field of skin imaging, a narrow branch of medical imaging focused on analysis and classification of images representing skin lesions. Although each lesion may differ from others in minimal details, deep convolutional neural networks have recently shown their great potential in melanoma recognition.
The aim of this article is to present an ensemble of Convolutional Neural Networks realized by means of the ECVL (European Computer Vision Library) and EDDL (European Distributed Deep Learning Library), developed within the H2020 DeepHealth project [9], that integrates five different models that vary in architecture, input data (i.e., image and metadata or only image), and data preprocessing to address the skin lesion classification problem. A brief analysis of the state-of-the-art solutions in the medical field is also included.
2 Related Work
Dermoscopic Images. Skin cancer is the most prevalent type of cancer and melanoma is its most deadly form [25, 26]. When recognized and treated in its earliest stages, melanoma is readily curable with minor surgery. Skin cancer is mainly diagnosed visually with an initial clinical screening followed by dermoscopy, biopsy and histopathological examination.
As detailed in the next subsection, the extensive use of dermoscopic images combined with the recent advancements in deep learning based computer vision have resulted in a great effort to develop different approaches to solve the problem of automated skin lesion classification. In the last years, multiple organizations such as the International Skin Imaging Collaboration (ISIC) have released dermoscopic datasets labeled with their skin lesion categories and associated with image-level and clinical-level metadata. The main goal is to achieve standards for the use of digitized images of skin lesions to assist the recognition of skin cancer, with a particular focus on melanoma, through clinical decision support and automated diagnosis. Specifically, ISIC Archive contains over 150 000 total images, of which approximately 70 000 are public [3]: the 2019 and 2020 challenge training sets count respectively 25 331 and 33 126 images; 10 982 more images are from the 2020 test set.
The dataset used to perform the experiments described in this paper is explored in Sect. 3.
CNNs for Classification. Machine learning and deep learning are widely used in the medical imaging field. In particular, Convolutional Neural Networks are the most common deep learning approach for medical image analysis and retrieval [5], detection of cancer forms (such as those affecting breasts, lungs, prostate, colon, liver, and brain) [20], renal biopsy [29], maxillofacial imagery [10, 11, 22], and many other application fields [8, 12, 13, 27, 28, 30, 31]. CNNs implemented in the work described in this paper are ResNet [18], ResNeXt [35], ResNeSt [37] and EfficientNet [33].
Proposed in 2014 by He et al., ResNet is successful in introducing the Residual Block and its Skip Connection to solve the vanishing gradient problem [7], resulting in the improvement of scaling up techniques to achieve better accuracy.
In 2016, Xie et al. proposed ResNeXt, a ResNet variant in which the building blocks are split in parallel (introducing the cardinality dimension), and summed together for the output.
The following year, SE-Net (Squeeze-and-Excitation Net) [19] improved the representative power of the network thanks to its SE block, a mechanism that allows the network to perform the recalibration of the features, in order to be able to selectively emphasize the informative features and suppress the less useful ones.
ResNeSt inherits the SE-block from Se-Net and the cardinality from ResNeXt to introduce a model that integrates the channel-level attention strategy with the multi-path network structure.
Finally, in 2019 EfficientNet achieved state-of-the-art accuracy on ImageNet providing a new scaling up method that uniformly increases the dimensions of depth, width, and resolution. EfficientNet is still one of the best performing networks, challenged in the last period only by the boost of Vision Transformers [14] even in the image classification task.
An in-depth description of how some of these models have been implemented in an ensemble method is provided in Sect. 4.
Deep Learning in Skin Imaging. With the support of massive parallel architecture (Graphic Processing Unites, GPUs), deep learning techniques increased their popularity and recently have been proposed also for dermoscopic image analysis [6, 36].
Esteva et al. studied pre-trained CNNs for skin cancer classification over a dataset of 129 450 clinical images and compared the performance of those neural networks to the accuracy of expert dermatologists. The results of this study demonstrated an artificial intelligence capable of classifying skin cancer with a level of competence comparable to dermatologists [15].
Mahbod et al. used a pre-trained AlexNet and VGG-16 architecture to extract deep features from dermoscopic images for skin lesion classification. The proposed architecture showed to achieve very good classification performance, yielding an area under the receiver operating characteristic curve (AUC) of 83.83% for melanoma classification and of 97.55% for seborrheic keratosis classification [24].
The winners of the ISIC 2019 Skin Lesion Classification Challenge [4] proposed an ensemble of multi-resolution models, in which EfficientNets prevail, in order to perform the tasks of skin lesion classification using only-image and image-and-metadata as input data, combined with extensive data augmentation and loss balancing [16]. On the ISIC official test set their method is ranked first for both tasks with an AUC score of 0.636 and 0.634 respectively.
Wei et al. suggested a lightweight skin cancer recognition model with feature discrimination based on fine-grained classification principle. Compared to the existing multi-CNN fusion method, the framework can achieve an approximate (or even higher) model performance with a lower number of model parameters end-to-end [34].
3 Dataset
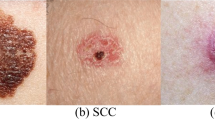
Since 2016, ISIC hosts challenges to engage the computer science community to improve dermatologic diagnostic accuracy through machine learning solutions. In addition, ISIC periodically expands its ISIC Archive, an open source platform with publicly available images of skin lesions. The dermoscopic images contained in the archive are associated with clinical metadata, such as patient’s age and sex, lesion’s anatomic site, diagnosis and the relative benign/malignant target, and more. In the 2019 challenge the diagnosis feature corresponds to 9 ground-truth category of the skin lesion (Fig. 1): Melanoma (MEL), Melanocytic Nevus (NV), Benign Keratosis Lesion (BKL), Basal Cell Carcinoma (BCC), Actinic Keratosis (AK), Squamous Cell Carcinoma (SCC), Vascular Lesion (VASC), Dermatofibroma (DF) plus the unknown (UNK) class (without samples in the training set), to which the images that cannot be classified in the other categories belong.
The current most numerous datasets provided by ISIC are the ones associated to the 2019 and 2020 challenges. As detailed in Table 1, both datasets are heavily unbalanced with respect to the melanoma class, to which belong only the 8.78% of samples. For the sake of completeness, the distribution of each skin lesion category is reported in Table 2.
Since deep learning models perform better with large amount of data, the experiments described in Sect. 5 use the concatenation of both 2019 and 2020 training images as a single dataset. Three new datasets are created from the original, each containing the images resized to \(512\times 512\), \(768\times 768\) and \(1024\times 1024\) respectively. In this way it is possible to compare the performance of models in classifying images starting from different resolutions. Moreover, during the training step, images are pre-processed with a pipeline of Data Augmentation techniques (it could be the Full pipeline or the Reduced pipeline), implemented to reduce overfitting. The transformations in the pipelines are applied to the image with a specific probability and they are chosen considering that dermoscopic images present very high resolution, but low colour variability. For this reason, the transforms mostly emphasize contrast and contours. All of these transformations have been performed through the augmentation functionalities of ECVL. They have to be wrapped into a container, that can be a SequentialContainer (i.e., all the augmentations in the container are performed sequentially) or a OneOfContainer (i.e., only one of the augmentations in the container is performed with a certain probability). Containers can be nested in order to apply the needed set of augmentations.
Full Pipeline. It consists of a sequence of: Transpose, Vertical Flip, Mirroring, Scale, Rotate and Resize, which are operators that do not alter the semantic content; changing the Brightness and Contrast of the image randomly; Coarse Dropout, which removes rectangular regions in the image; Normalize and finally a blurring and a distorting transformations (Fig. 2).
Reduced Pipeline. It is a reduced version of the Full pipeline, consisting only of Transpose, Vertical Flip, Mirroring and Normalize transformations.
3.1 Metadata
ISIC provides the metadata associated to each image. From those information, we obtained our own set of metadata, some of which are related to the image itself, such as image_name (i.e., the name of the image), filepath (i.e., path where the image is stored), fold (i.e., fold index to which the image is assigned by the Cross Validation Stratified 5-Fold strategy) and is_ext (which is 1 if the image belongs to the 2019 dataset, 0 otherwise). However, others are related to the patient and its skin lesion, such as patient_id, sex, age_approx, anatom_site (i.e., the location of the lesion on the patient’s body), diagnosis (i.e., skin lesion category or unknown), benign_malignant (where benign stands for non-melanoma, while malignant stands for melanoma), target (i.e., the corresponding binary numeric value of the bening_malignant feature).
Since the images originally belong to two different datasets, it is necessary to map the values of the diagnosis feature from the 2020 to the 2019 diagnosis, to maintain consistency between the labels and the metadata.
Feature Importance. Feature importance is a class of techniques that score the features provided as input to a predictive model, based on how relevant they are in predicting the target. In particular, the Gradient Boosting algorithm shows that the age_approx feature is the most significant in target prediction [38]. Afterwards, the features anatom_site and sex.
For this reason, even models that classify images by integrating information extrapolated from metadata are included in the ensemble described in Sect. 5.1. For example, Table 3 shows that an image-and-metadata EfficientNet-b3 model performs better than the same model receiving only dermoscopic images as input data.
4 Classification Models
During the experimental phase, different Convolutional Neural Network models proved to be effective solutions for the skin lesion images classification task: ResNet-152, ResNest-101, ResNeXt-50, EfficientNet-b3, EfficientNet-b4, EfficientNet-b5, EfficientNet-b6, EfficientNet-b7.
As in a typical image classification problem, each tested CNN model has been loaded in EDDL pretrained on ImageNet and its last layer is replaced so that the output dimension equals the target dimension. Experiments showed better results when using diagnosis as target instead of benign_malignant. Table 4 illustrates that ResNet-152 trained to predict 9 diagnoses achieved more effective results than the same model trained on binary target. In fact, a CNN trained on a binary target must become particularly efficient at distinguish melanoma from all the others lesions. On the other hand, a CNN with a 9-dimensional output layer is trained to learn all the skin lesion categories and their details, so that the model can gather more information. In this case, to validate the performance of the network trained with a 9-dimensional target we decided to measure its accuracy on the validation set (described in Sect. 5) and to compare it with the accuracy achieved with a binary target. In fact, the official ISIC2020 Challenge metric (AUC) provided to measure only the model’s capability of classifying skin lesions as melanoma or non-melanoma, i.e. with binary target.
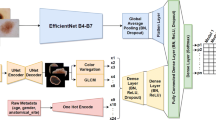
The architecture of the model that processes images and metadata is composed by a CNN that performs feature extraction from the dermoscopic images, a Fully Connected Neural Network (FCNN) for clinical metadata processing and a Fully Connected (Dense) layer followed by a Softmax layer, which takes both the outputs of the CNN and of the FCNN in input to return the class distribution (Fig. 3). Specifically, the metadata-FCNN is composed of two blocks, each containing a Dense layer followed by batch normalization and ReLU activation, interspersed with a Dropout. Its 128-dimensional output is concatenated with the CNN’s feature vector (which size depends on the number of output features of the CNN’s last layer, excluding the prediction head), followed by the final classification layer.
5 Experimental Results
The dataset used for training and evaluating the proposed models includes 58 457 dermoscopic images, as the concatenation of the ISIC2019 and ISIC2020 datasets. Images are resized to different resolutions, ranging from \(256\times 256\) to \(768\times 768\).
The Cross Validation Stratified 5-Folds technique splits the training set into 5 smaller sets (i.e., 5 folds) according to three criteria: folds are created such that they had the same class distribution as the entire dataset, images belonging to the same patient are put into the same fold and the patient’s images count distribution is balanced within the folds, i.e. some patients have as many as 115 images and some patients have as few as 2 images, so each fold has an equal number of patients with 115 images, with 100, with 70, with 50, with 5, with 2, etc. This makes validation more reliable. Then, each model is trained using \(k-1\) folds as training data, with the aim of minimizing the Cross-Entropy loss, and it is validated on the remaining part of the data computing the model’s accuracy. The ADAM algorithm [23] is used to optimize the loss, and the learning rate is set to \(3\times 10^{-5}\).
As shown in Table 4, a model trained to predict a 9-dimensional target performs better than the same model trained to predict a binary target. For this reason, the diagnosis feature is used as label. All the models have been trained for 18 epochs, which was sufficient to achieve satisfactory results.
Table 5 reports the comparison between the AUC score achieved by an EfficientNet-b3 model which receives input data processed with the Full pipeline and then with the Reduced Pipeline. As expected, performance improves if images are transformed with the Full pipeline. Moreover, experiments have shown that good diversity can be achieved by inserting models that process metadata into the final ensemble.
5.1 Compact Models Ensemble
Ensemble methods use multiple learning algorithms to obtain better predictive performance than the one that could be obtained from any of the constituent learning algorithms alone. In this paper, the final ensemble is a simple average of the five more reliable models’ probability ranks. Specifically, for each model predictions on the official ISIC2020 test set, the rank function is used to compute numerical data ranks (1 through n, where n is equal to the test set size) based on target probability predicted for each image. Equal probabilities are assigned a rank that is the average of the ranks of those values and then the rank values are expressed as a percentage. Finally, ensemble method consists in computing the new target value for each test sample as the average of the models ranks associated to the sample.
Each of the five multi-class classifiers employed in the ensemble is evaluated considering the melanoma class vs. the rest. Since the ground truth of the ISIC2020 test set has not yet been released, the only metric available is the AUC, provided by the challenge once the predictions have been submitted. Table 6 shows the configuration of the models ensemble that achieved our best AUC score. On the ISIC 2020 Skin Lesion Classification Challenge Leaderboard [4], our ensemble ranked fourth with a more compact models ensemble compared to the top three and with a difference of only 0.0013 from the AUC score of the winning proposal (Table 7).
The winning strategy [17] differs in the choice of augmentations techniques and hyperparameters and it is important to underscore that it consists of an ensemble of 18 models unlike ours which has only 5.
Instead, the second-place-winner [1] built an ensemble of 15 image-only-models, in which EfficientNets prevail, trained with 3-dimensional target: melanoma, nevus and other classes.
Finally, the third-place-winner [2] built an ensemble of 8 image-and-metadata models, in which EfficientNets prevail.
6 Conclusions
This paper introduces an effective method to solve the dermoscopic images classification task. The proposed solution takes advantages of the larger number of training images obtained from the union of the ISIC2019 and the ISIC2020 archives. In addition, replacing the last layer of the models with a 9-dimensional layer to use the diagnosis attribute as target has proven to be a successful approach. Finally, the ensemble of different Convolutional Neural Network models, with and without metadata, further increases the AUC of the classification. These experiments led to a final compact ensemble that achieved a very satisfactory AUC score on the test set with a relatively low computational effort, compared to the competitors.
Future research directions will hence include an investigation to find a reliable solution that can assist dermatologists in their diagnosis.
References
ISIC 2020–2nd place solution. https://github.com/i-pan/kaggle-melanoma/blob/master/documentation.pdf
ISIC 2020–3rd place solution. https://github.com/Masdevallia/3rd-place-kaggle-siim-isic-melanoma-classification
ISIC Archive. https://www.isic-archive.com
ISIC Leaderboards. https://challenge.isic-archive.com/leaderboards/live/
Allegretti, S., Bolelli, F., Pollastri, F., Longhitano, S., Pellacani, G., Grana, C.: Supporting skin lesion diagnosis with content-based image retrieval. In: 2020 25th International Conference on Pattern Recognition (ICPR), pp. 8053–8060 (2021)
Barata, C., Celebi, M.E., Marques, J.S.: Explainable skin lesion diagnosis using taxonomies. Pattern Recogn. 110, 107413 (2021)
Bengio, Y., Simard, P., Frasconi, P., et al.: Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Networks 5(2), 157–166 (1994)
Bigazzi, R., Landi, F., Cascianelli, S., Baraldi, L., Cornia, M., Cucchiara, R.: Focus on impact: indoor exploration with intrinsic motivation. IEEE Robot. Autom. Lett. 7, 2985–2992 (2022)
Cancilla, M., et al.: The deephealth toolkit: a unified framework to boost biomedical applications. In: 2020 25th International Conference on Pattern Recognition (ICPR), pp. 9881–9888. IEEE (2021)
Cipriano, M., et al.: Deep Segmentation of the Mandibular Canal: a New 3D Annotated Dataset of CBCT Volumes, pp. 1–11. IEEE Access (2022)
Cipriano, M., Allegretti, S., Bolelli, F., Pollastri, F., Grana, C.: Improving segmentation of the inferior alveolar nerve through deep label propagation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1–10. IEEE (2022)
Cornia, M., Baraldi, L., Cucchiara, R.: Smart: training shallow memory-aware transformers for robotic explainability. In: Proceedings of the International Conference on Robotics and Automation (2020)
Cornia, M., Baraldi, L., Serra, G., Cucchiara, R.: Sam: pushing the limits of saliency prediction models. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (2018)
Dosovitskiy, A., et al.: An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
Esteva, A., et al.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639), 115–118 (2017)
Gessert, N., Nielsen, M., Shaikh, M., Werner, R., Schlaefer, A.: Skin lesion classification using loss balancing and ensembles of multi-resolution EfficientNets. línea], ISIC Chellange (2019)
Ha, Q., Liu, B., Liu, F.: Identifying melanoma images using EfficientNet ensemble: winning solution to the SIIM-ISIC melanoma classification challenge. CoRR abs/2010.05351 (2020). https://arxiv.org/abs/2010.05351
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141 (2018)
Hu, Z., Tang, J., Wang, Z., Zhang, K., Zhang, L., Sun, Q.: Deep learning for image-based cancer detection and diagnosis - a survey. Pattern Recogn. 83, 134–149 (2018)
Istituto Superiore di Sanità: L’epidemiologia per la sanità pubblica - melanoma. https://www.epicentro.iss.it/melanoma/
Jaskari, J., et al.: Deep learning method for mandibular canal segmentation in dental cone beam computed tomography volumes. Sci. Rep. 10(1), 1–8 (2020)
Kingma, D.P., Ba, J.: Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Mahbod, A., Schaefer, G., Wang, C., Ecker, R., Ellinge, I.: Skin lesion classification using hybrid deep neural networks. In: ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1229–1233. IEEE (2019)
Melanoma Research Alliance: Melanoma Statistics. https://www.curemelanoma.org/about-melanoma/melanoma-101/melanoma-statistics-2/
Pellacani, G., Grana, C., Seidenari, S.: Algorithmic reproduction of asymmetry and border cut-off parameters according to the ABCD rule for dermoscopy. J. Eur. Acad. Dermatol. Venereol. 20(10), 1214–1219 (2006)
Pollastri, F., Bolelli, F., Paredes, R., Grana, C.: Augmenting data with GANs to segment melanoma skin lesions. Multimedia Tools Appl. 79, 15575–15592 (2019)
Pollastri, F., Cipriano, M., Bolelli, F., Grana, C.: Long-range 3D self-attention for MRI prostate segmentation. In: 2022 IEEE 18th International Symposium on Biomedical Imaging (ISBI), pp. 1–5. IEEE, March 2021
Pollastri, F., et al.: Confidence calibration for deep renal biopsy immunofluorescence image classification. In: 2020 25th International Conference on Pattern Recognition (ICPR), pp. 1–8. IEEE (2021)
Pollastri, F., et al.: A Deep Analysis on High ResolutionDermoscopic Images Classification. IET Comput. Vision 15(7), 514–526 (2021)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sung, H., et al.: Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Can. J. Clin. 71(3), 209–249 (2021)
Tan, M., Le, Q.: EfficientNet: rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114 (2019)
Wei, L., Ding, K., Hu, H.: Automatic skin cancer detection in dermoscopy images based on ensemble lightweight deep learning network. IEEE Access 8, 99633–99647 (2020)
Xie, S., Girshick, R., Dollár, P., Tu, Z., He, K.: Aggregated residual transformations for deep neural networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1492–1500 (2017)
Yuan, Y., Chao, M., Lo, Y.: Automatic skin lesion segmentation using deep fully convolutional networks with Jaccard distance. IEEE Trans. Med. Imaging 36(9), 1876–1886 (2017)
Zhang, H., et al.: Resnest: Split-attention networks. arXiv preprint arXiv:2004.08955 (2020)
Zhanshan, L., Zhaogeng, L.: Feature selection algorithm based on XGBoost. J. Commun. 40(10), 101 (2019)
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 825111, DeepHealth Project.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Giovanetti, A., Canalini, L., Perliti Scorzoni, P. (2022). A Compact Deep Ensemble for High Quality Skin Lesion Classification. In: Mazzeo, P.L., Frontoni, E., Sclaroff, S., Distante, C. (eds) Image Analysis and Processing. ICIAP 2022 Workshops. ICIAP 2022. Lecture Notes in Computer Science, vol 13373. Springer, Cham. https://doi.org/10.1007/978-3-031-13321-3_45
Download citation
DOI: https://doi.org/10.1007/978-3-031-13321-3_45
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13320-6
Online ISBN: 978-3-031-13321-3
eBook Packages: Computer ScienceComputer Science (R0)